Peach Buds’ Microbiome Profiling Reveals Cultivar-Specific Signatures Associated with TCSB Susceptibility
Abstract
1. Introduction
2. Results
2.1. Sequencing Output and ASVs Recovery
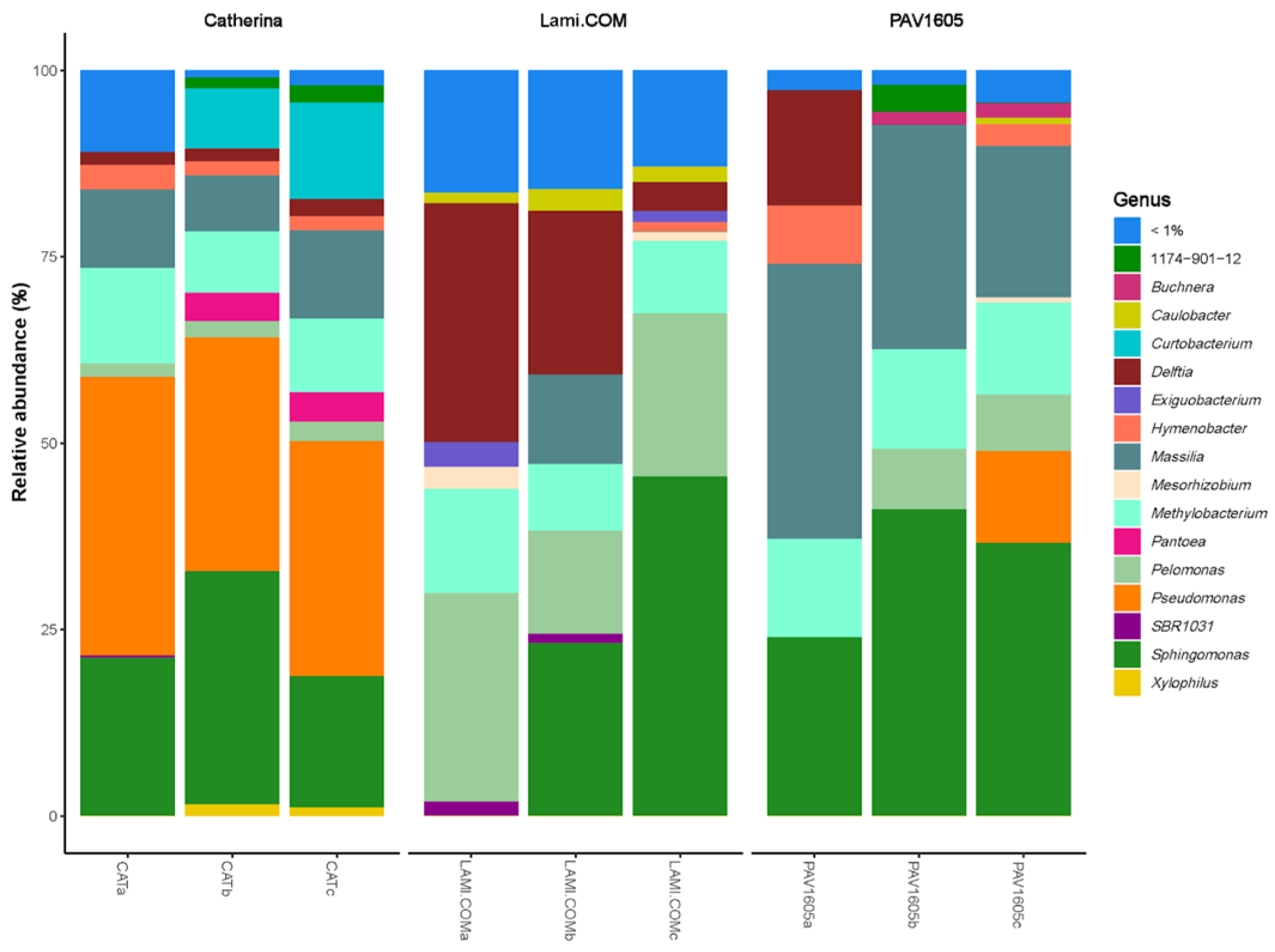
2.2. Bacterial Community Composition
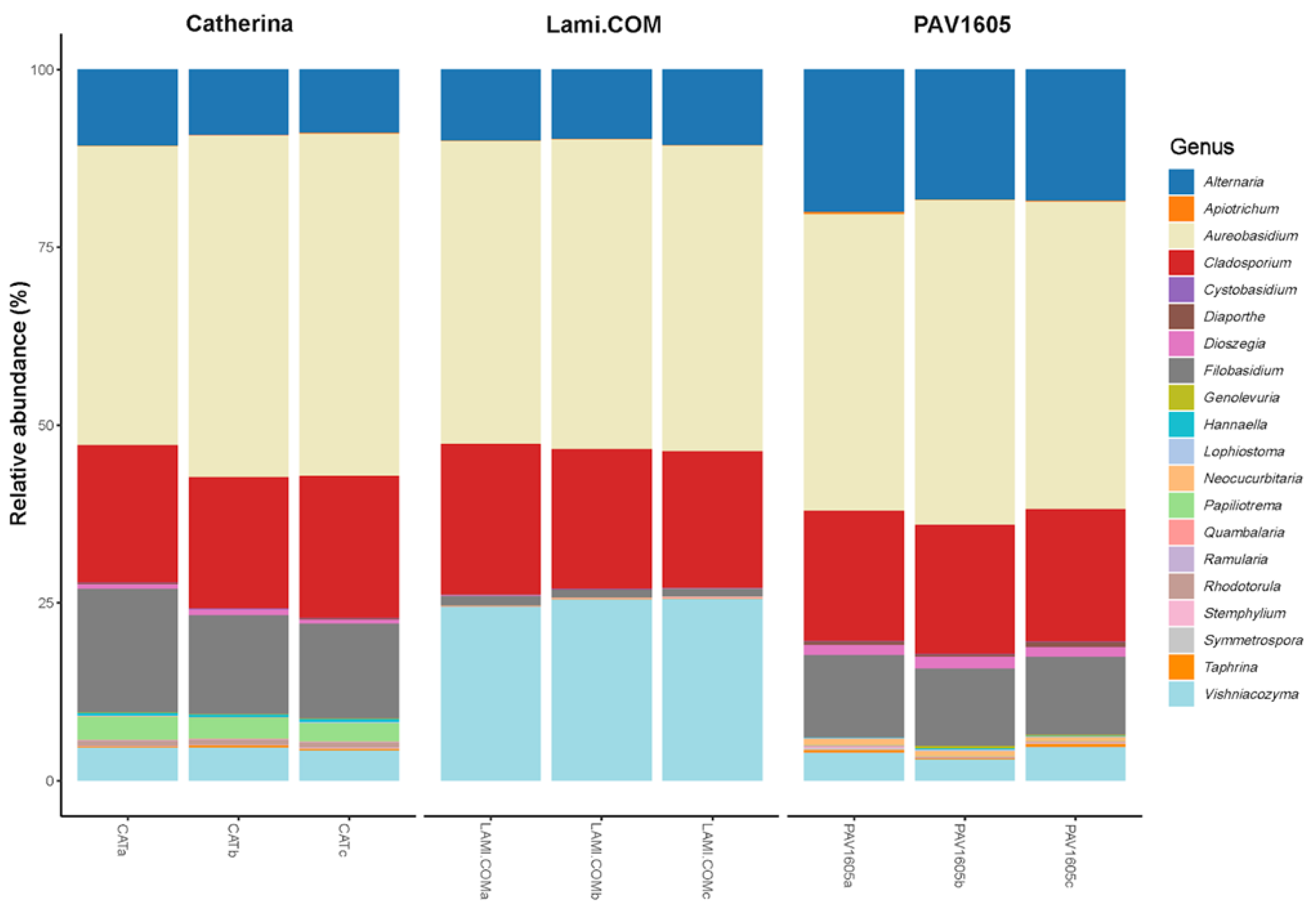
2.3. Fungal Community Composition
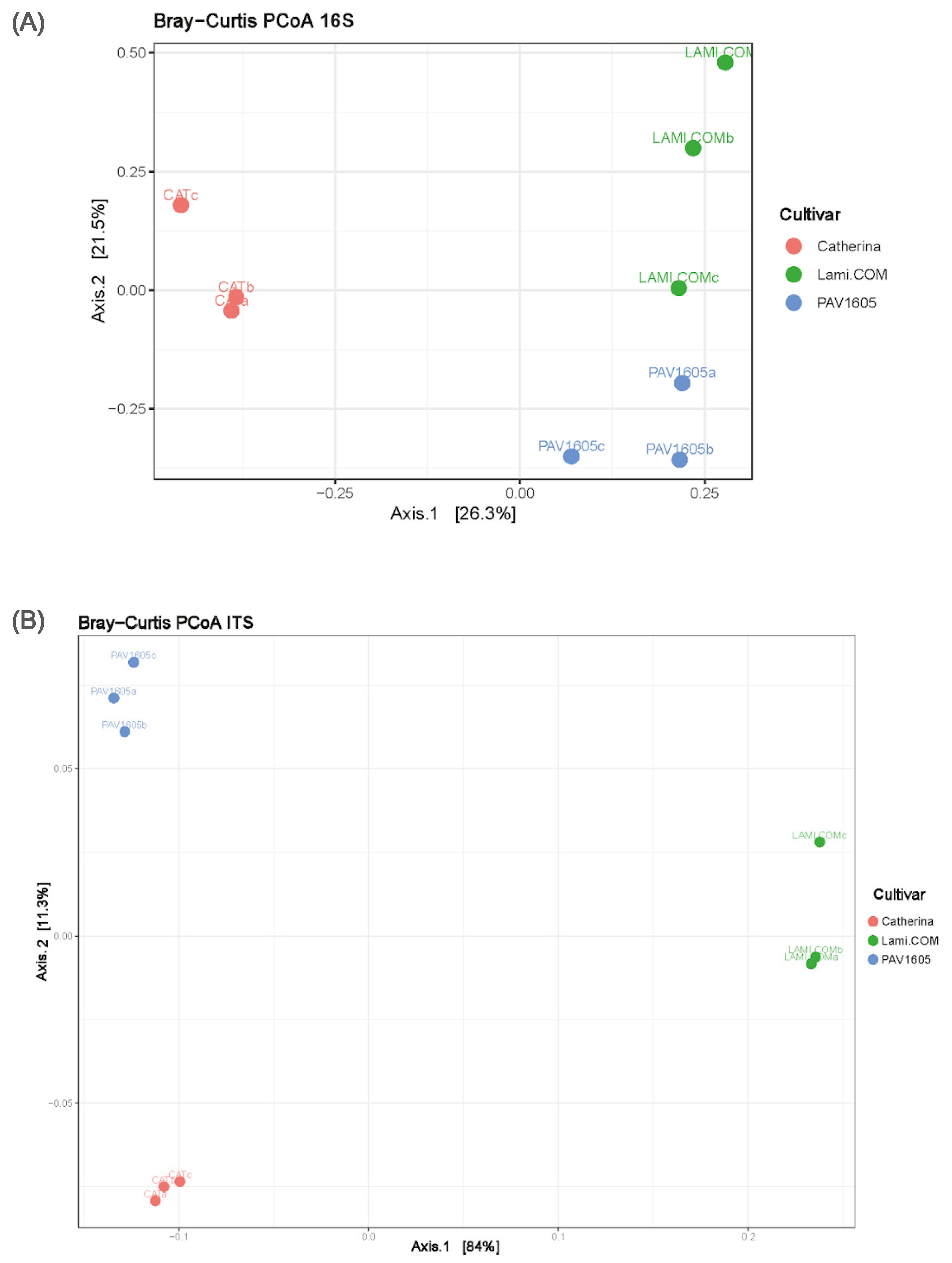
2.4. Communities’ Diversities and Dissimilarities
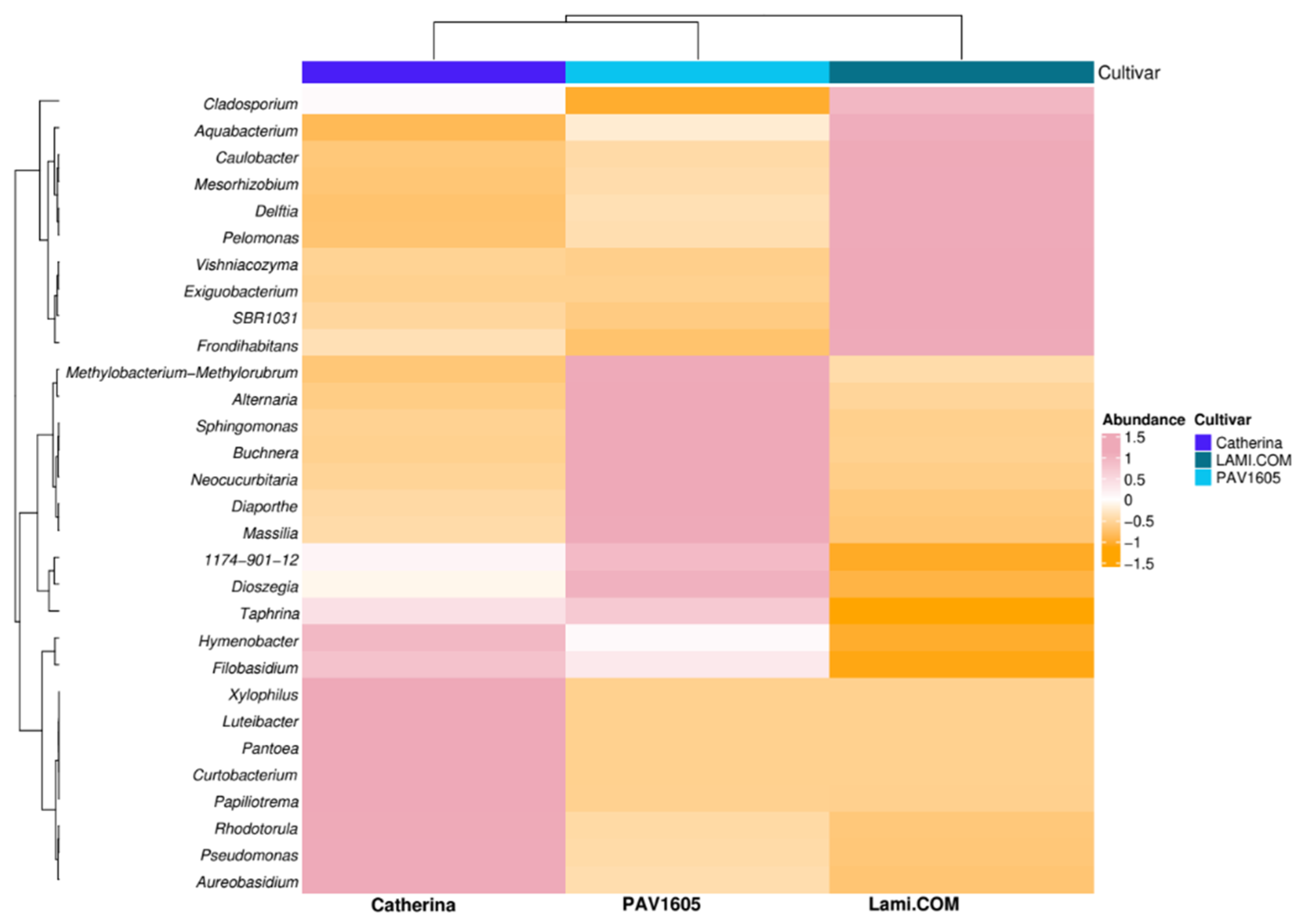
2.5. Differential Abundance of Key Microbial Taxa
3. Discussion
4. Materials and Methods
4.1. Peach Buds Collection
4.2. DNA Extraction and Sequencing
4.3. PCR Amplification and Library Preparation
4.4. Data Summary and Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ristaino, J.B.; Anderson, P.K.; Bebber, D.P.; Brauman, K.A.; Cunniffe, N.J.; Fedoroff, N.V.; Finegold, C.; Garrett, K.A.; Gilligan, C.A.; Jones, C.M.; et al. The Persistent Threat of Emerging Plant Disease Pandemics to Global Food Security. Proc. Natl. Acad. Sci. USA 2021, 118, e2022239118. [Google Scholar] [CrossRef]
- Mwangi, R.W.; Mustafa, M.; Charles, K.; Wagara, I.W.; Kappel, N. Selected Emerging and Reemerging Plant Pathogens Affecting the Food Basket: A Threat to Food Security. J. Agric. Food Res. 2023, 14, 100827. [Google Scholar] [CrossRef]
- Corredor-Moreno, P.; Saunders, D.G.O. Expecting the Unexpected: Factors Influencing the Emergence of Fungal and Oomycete Plant Pathogens. New Phytol. 2020, 225, 118–125. [Google Scholar] [CrossRef]
- Martino, I.; Tabone, G.; Giordano, R.; Gullino, M.L.; Guarnaccia, V. First Report of Diaporthe eres Causing Stem Blight and Dieback on Highbush Blueberry (Vaccinium corymbosum) in Italy. Plant Dis. 2023, 107, 1236. [Google Scholar] [CrossRef]
- Brugneti, F.; Rossini, L.; Cirilli, M.; Nardecchia, A.; Onofri, M.; Mazzaglia, A.; Turco, S. Elucidation of “Twig Canker and Shoot Blight” (TCSB) in Peach Caused by Diaporthe amygdali in the North of Italy in Emilia-Romagna. Physiol. Plant. 2025, 177, e70428. [Google Scholar] [CrossRef]
- Occhibove, F.; Chapman, D.S.; Mastin, A.J.; Parnell, S.S.R.; Agstner, B.; Mato-Amboage, R.; Jones, G.; Dunn, M.; Pollard, C.R.J.; Robinson, J.S.; et al. Eco-Epidemiological Uncertainties of Emerging Plant Diseases: The Challenge of Predicting Xylella Fastidiosa Dynamics in Novel Environments. Phytopathology 2020, 110, 1740–1750. [Google Scholar] [CrossRef] [PubMed]
- Giraud, T.; Gladieux, P.; Gavrilets, S. Linking the Emergence of Fungal Plant Diseases with Ecological Speciation. Trends Ecol. Evol. 2010, 25, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Romero, F.; Cazzato, S.; Walder, F.; Vogelgsang, S.; Bender, S.F.; van der Heijden, M.G.A. Humidity and High Temperature Are Important for Predicting Fungal Disease Outbreaks Worldwide. New Phytol. 2022, 234, 1553–1556. [Google Scholar] [CrossRef] [PubMed]
- Seidel, D.; Wurster, S.; Jenks, J.D.; Sati, H.; Gangneux, J.-P.; Egger, M.; Alastruey-Izquierdo, A.; Ford, N.P.; Chowdhary, A.; Sprute, R.; et al. Impact of Climate Change and Natural Disasters on Fungal Infections. Lancet Microbe 2024, 5, e594–e605. [Google Scholar] [CrossRef] [PubMed]
- Waheed, A.; Haxim, Y.; Islam, W.; Ahmad, M.; Muhammad, M.; Alqahtani, F.M.; Hashem, M.; Salih, H.; Zhang, D. Climate Change Reshaping Plant-Fungal Interaction. Environ. Res. 2023, 238, 117282. [Google Scholar] [CrossRef]
- Cremonini, L.; Randi, P.; Fazzini, M.; Nardino, M.; Rossi, F.; Georgiadis, T. Causes and Impacts of Flood Events in Emilia-Romagna (Italy) in May 2023. Land 2024, 13, 1800. [Google Scholar] [CrossRef]
- Pereira, D.S.; Phillips, A.J.L. Exploring the Diversity and Ecological Dynamics of Palm Leaf Spotting Fungi—A Case Study on Ornamental Palms in Portugal. J. Fungi 2025, 11, 43. [Google Scholar] [CrossRef] [PubMed]
- Montes, N.; Vijayan, V.; Pagán, I. Host Population Structure for Tolerance Determines the Evolution of Plant–Virus Interactions. New Phytol. 2021, 231, 1570–1585. [Google Scholar] [CrossRef]
- Cirilli, M.; Rossini, L.; Geuna, F.; Palmisano, F.; Minafra, A.; Castrignanò, T.; Gattolin, S.; Ciacciulli, A.; Babini, A.R.; Liverani, A.; et al. Genetic Dissection of Sharka Disease Tolerance in Peach (P. persica L. Batsch). BMC Plant Biol. 2017, 17, 192. [Google Scholar] [CrossRef]
- van Mölken, T.; Stuefer, J.F. The Potential of Plant Viruses to Promote Genotypic Diversity via Genotype × Environment Interactions. Ann. Bot. 2011, 107, 1391–1397. [Google Scholar] [CrossRef]
- Busby, P.E.; Newcombe, G.; Dirzo, R.; Whitham, T.G. Differentiating Genetic and Environmental Drivers of Plant–Pathogen Community Interactions. J. Ecol. 2014, 102, 1300–1309. [Google Scholar] [CrossRef]
- Reinoso, H.; Luna, V.; Pharis, R.P.; Bottini, R. Dormancy in Peach (Prunus persica) Flower Buds. V. Anatomy of Bud Development in Relation to Phenological Stage. Can. J. Bot. 2002, 80, 656–663. [Google Scholar] [CrossRef]
- Bhattarai, N.; Saud, S.; Atreya, P.N.; Dhakal, S.; Bohara, A.K. Assessment of Phenological and Physicochemical Characteristics of Peach (Prunus persica L.) Varieties in Mustang, Nepal. Arch. Agric. Environ. Sci. 2024, 9, 793–799. [Google Scholar] [CrossRef]
- Thomidis, T.; Michailides, T.J.; Exadaktylou, E. Phoma Glomerata (Corda) Wollenw. & Hochapfel a New Threat Causing Cankers on Shoots of Peach Trees in Greece. Eur. J. Plant Pathol. 2011, 131, 171–178. [Google Scholar] [CrossRef]
- Froelich, M.H.; Schnabel, G. Investigation of Fungi Causing Twig Blight Diseases on Peach Trees in South Carolina. Plant Dis. 2019, 103, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.-X.; Schnabel, G.; Hu, M.; De Cal, A. Global Distribution and Management of Peach Diseases. Phytopathol. Res. 2022, 4, 30. [Google Scholar] [CrossRef]
- He, Z.; Abeywickrama, P.D.; Wu, L.; Zhou, Y.; Zhang, W.; Yan, J.; Shang, Q.; Zhou, Y.; Li, S. Diversity of Cytospora Species Associated with Trunk Diseases of Prunus Persica (Peach) in Northern China. J. Fungi 2024, 10, 843. [Google Scholar] [CrossRef]
- Uddin, W.; Stevenson, K.L.; Pardo-Schultheiss, R.A. Pathogenicity of a Species of Phomopsis Causing a Shoot Blight on Peach in Georgia and Evaluation of Possible Infection Courts. Plant Dis. 1997, 81, 983–989. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, W.; Abeywickrama, P.D.; He, Z.; Zhang, Z.; Li, Y.; Li, S.; Fan, Z.; Yan, J. Diversity and Virulence of Diaporthe Species Associated with Peach Trunk Diseases in China. Plants 2024, 13, 3238. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, H.; Cheng, Y.; Li, J.; Liu, J.; Li, G. First Report of Neofusicoccum Parvum Causing Gummosis of Peach Trees in Hubei Province, Central China. Plant Dis. 2019, 103, 2673. [Google Scholar] [CrossRef]
- Miller, S.A.; Testen, A.L.; Jacobs, J.M.; Ivey, M.L.L. Mitigating Emerging and Reemerging Diseases of Fruit and Vegetable Crops in a Changing Climate. Phytopathology 2024, 114, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Bashir, I.; War, A.F.; Rafiq, I.; Reshi, Z.A.; Rashid, I.; Shouche, Y.S. Phyllosphere Microbiome: Diversity and Functions. Microbiol. Res. 2022, 254, 126888. [Google Scholar] [CrossRef]
- Xue, R.; Liu, S.; Stirling, E.; Wang, Y.; Zhao, K.; Matsumoto, H.; Wang, M.; Xu, J.; Ma, B. Core Community Drives Phyllosphere Bacterial Diversity and Function in Multiple Ecosystems. Sci. Total Environ. 2023, 896, 165187. [Google Scholar] [CrossRef]
- Dong, C.-J.; Wang, L.-L.; Li, Q.; Shang, Q.-M. Bacterial Communities in the Rhizosphere, Phyllosphere and Endosphere of Tomato Plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef]
- Mehmood, N.; Saeed, M.; Zafarullah, S.; Hyder, S.; Rizvi, Z.F.; Gondal, A.S.; Jamil, N.; Iqbal, R.; Ali, B.; Ercisli, S.; et al. Multifaceted Impacts of Plant-Beneficial Pseudomonas spp. in Managing Various Plant Diseases and Crop Yield Improvement. ACS Omega 2023, 8, 22296–22315. [Google Scholar] [CrossRef]
- Raio, A.; Puopolo, G. Pseudomonas chlororaphis Metabolites as Biocontrol Promoters of Plant Health and Improved Crop Yield. World J. Microbiol. Biotechnol. 2021, 37, 99. [Google Scholar] [CrossRef]
- Labancová, E.; Šípošová, K.; Kučerová, D.; Horváthová, Á.; Schusterová, H.; Vivodová, Z.; Vadkertiová, R.; Kollárová, K. The Tremellaceous Yeast: Papiliotrema Terrestris—As the Growth Stimulant of Maize Plants. J. Plant Growth Regul. 2023, 42, 3835–3850. [Google Scholar] [CrossRef]
- Ianiri, G.; Barone, G.; Palmieri, D.; Quiquero, M.; Gaeta, I.; De Curtis, F.; Castoria, R. Transcriptomic Investigation of the Interaction between a Biocontrol Yeast, Papiliotrema Terrestris Strain PT22AV, and the Postharvest Fungal Pathogen Penicillium Expansum on Apple. Commun. Biol. 2024, 7, 359. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Bakker, P.A.H.M. Interactions between Plants and Beneficial Pseudomonas spp.: Exploiting Bacterial Traits for Crop Protection. Antonie Leeuwenhoek 2007, 92, 367–389. [Google Scholar] [CrossRef]
- Chi, Z.; Kong, C.-C.; Wang, Z.-Z.; Wang, Z.; Liu, G.-L.; Hu, Z.; Chi, Z.-M. The Signaling Pathways Involved in Metabolic Regulation and Stress Responses of the Yeast-like Fungi Aureobasidium spp. Biotechnol. Adv. 2022, 55, 107898. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Zajc, J.; Stenberg, J.A. Aureobasidium spp.: Diversity, Versatility, and Agricultural Utility. Horticulturae 2023, 9, 59. [Google Scholar] [CrossRef]
- Arrigoni, E.; Antonielli, L.; Pindo, M.; Pertot, I.; Perazzolli, M. Tissue Age and Plant Genotype Affect the Microbiota of Apple and Pear Bark. Microbiol. Res. 2018, 211, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Makino, A.; Nakai, R.; Yoneda, Y.; Toyama, T.; Tanaka, Y.; Meng, X.-Y.; Mori, K.; Ike, M.; Morikawa, M.; Kamagata, Y.; et al. Isolation of Aquatic Plant Growth-Promoting Bacteria for the Floating Plant Duckweed (Lemna minor). Microorganisms 2022, 10, 1564. [Google Scholar] [CrossRef]
- Luo, B.; Sun, H.; Zhang, Y.; Gu, Y.; Yan, W.; Zhang, R.; Ni, Y. Habitat-Specificity and Diversity of Culturable Cold-Adapted Yeasts of a Cold-Based Glacier in the Tianshan Mountains, Northwestern China. Appl. Microbiol. Biotechnol. 2019, 103, 2311–2327. [Google Scholar] [CrossRef]
- Villarreal, P.; Carrasco, M.; Barahona, S.; Alcaíno, J.; Cifuentes, V.; Baeza, M. Antarctic Yeasts: Analysis of Their Freeze-Thaw Tolerance and Production of Antifreeze Proteins, Fatty Acids and Ergosterol. BMC Microbiol. 2018, 18, 66. [Google Scholar] [CrossRef]
- Tsuji, M.; Tanabe, Y.; Vincent, W.F.; Uchida, M. Vishniacozyma ellesmerensis sp. nov., a Psychrophilic Yeast Isolated from a Retreating Glacier in the Canadian High Arctic. Int. J. Syst. Evol. Microbiol. 2019, 69, 696–700. [Google Scholar] [CrossRef]
- Cheng, F.S.; Brown, S.K.; Weeden, N.F. A DNA Extraction Protocol from Various Tissues in Woody Species. HortScience 1997, 32, 921–922. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Turco, S.; Brugneti, F.; Giubilei, I.; Silvestri, C.; Petrović, M.; Drais, M.I.; Cristofori, V.; Speranza, S.; Mazzaglia, A.; Contarini, M.; et al. A Bud’s Life: Metabarcoding Analysis to Characterise Hazelnut Big Buds Microbiome Biodiversity. Microbiol. Res. 2024, 287, 127851. [Google Scholar] [CrossRef]
- Cardacino, A.; Turco, S.; Balestra, G.M. Seasonal Dynamics of Kiwifruit Microbiome: A Case Study in a KVDS-Affected Orchard. Microbiol. Res. 2025, 292, 128044. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardacino, A.; Tastekin, T.; Brugneti, F.; Cirilli, M.; Mazzaglia, A.; Turco, S. Peach Buds’ Microbiome Profiling Reveals Cultivar-Specific Signatures Associated with TCSB Susceptibility. Stresses 2025, 5, 60. https://doi.org/10.3390/stresses5030060
Cardacino A, Tastekin T, Brugneti F, Cirilli M, Mazzaglia A, Turco S. Peach Buds’ Microbiome Profiling Reveals Cultivar-Specific Signatures Associated with TCSB Susceptibility. Stresses. 2025; 5(3):60. https://doi.org/10.3390/stresses5030060
Chicago/Turabian StyleCardacino, Antonella, Taner Tastekin, Federico Brugneti, Marco Cirilli, Angelo Mazzaglia, and Silvia Turco. 2025. "Peach Buds’ Microbiome Profiling Reveals Cultivar-Specific Signatures Associated with TCSB Susceptibility" Stresses 5, no. 3: 60. https://doi.org/10.3390/stresses5030060
APA StyleCardacino, A., Tastekin, T., Brugneti, F., Cirilli, M., Mazzaglia, A., & Turco, S. (2025). Peach Buds’ Microbiome Profiling Reveals Cultivar-Specific Signatures Associated with TCSB Susceptibility. Stresses, 5(3), 60. https://doi.org/10.3390/stresses5030060





