Regulatory Effects of GPR158 Overexpression in Trabecular Meshwork Cells of the Eye’s Aqueous Outflow Pathways
Abstract
1. Introduction
2. Results
2.1. Profiling of GPR158-Regulated Gene Expression
2.2. IPA Upstream Stress Regulator Analysis
2.2.1. Upstream Stress Regulators: Dexamethasone and TGFB1 (Fibrosis)
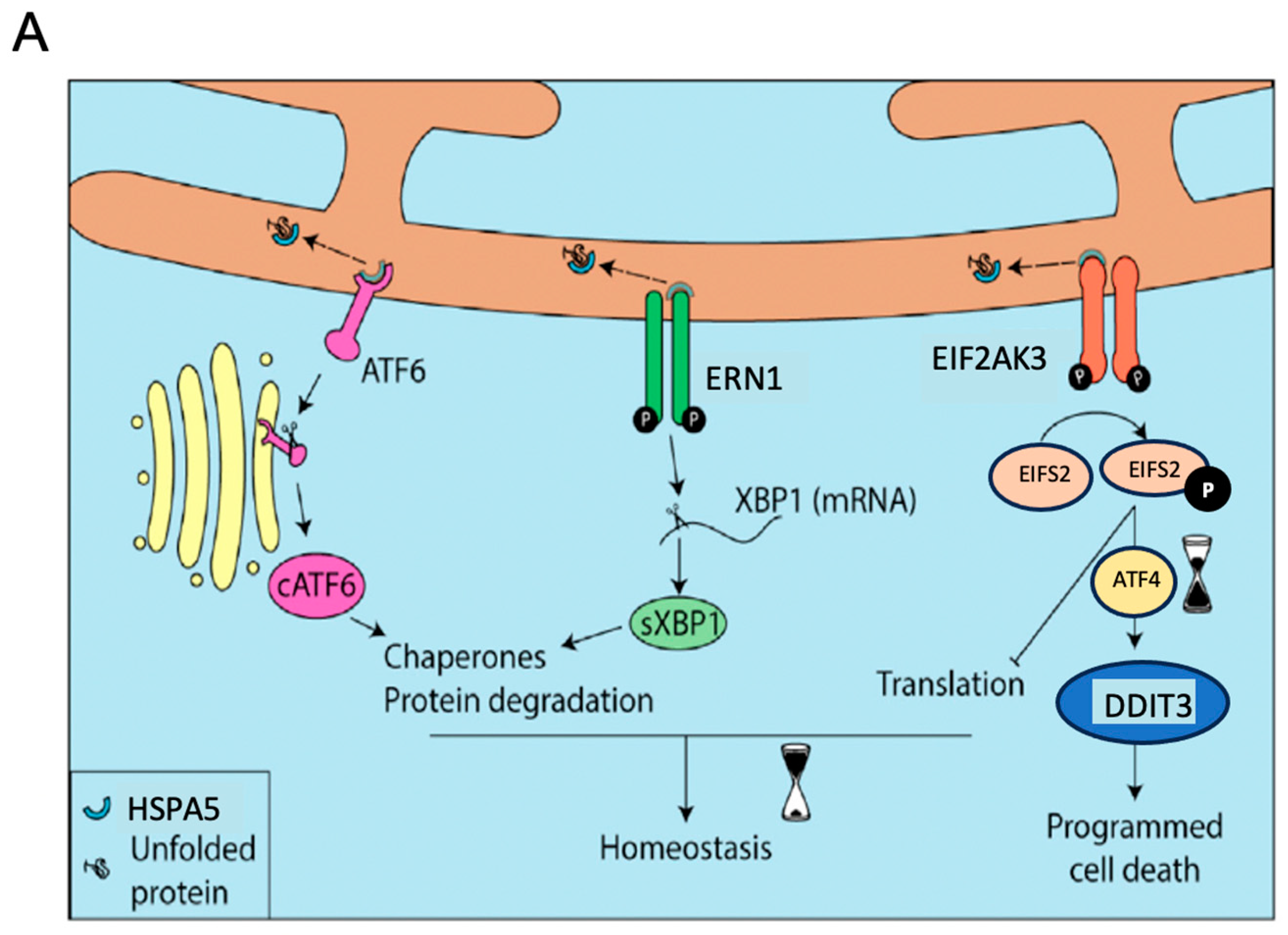
2.2.2. Upstream Stress Regulators: XBP1 and ATF4 (UPR)
2.2.3. Upstream Stress Regulator: TP53
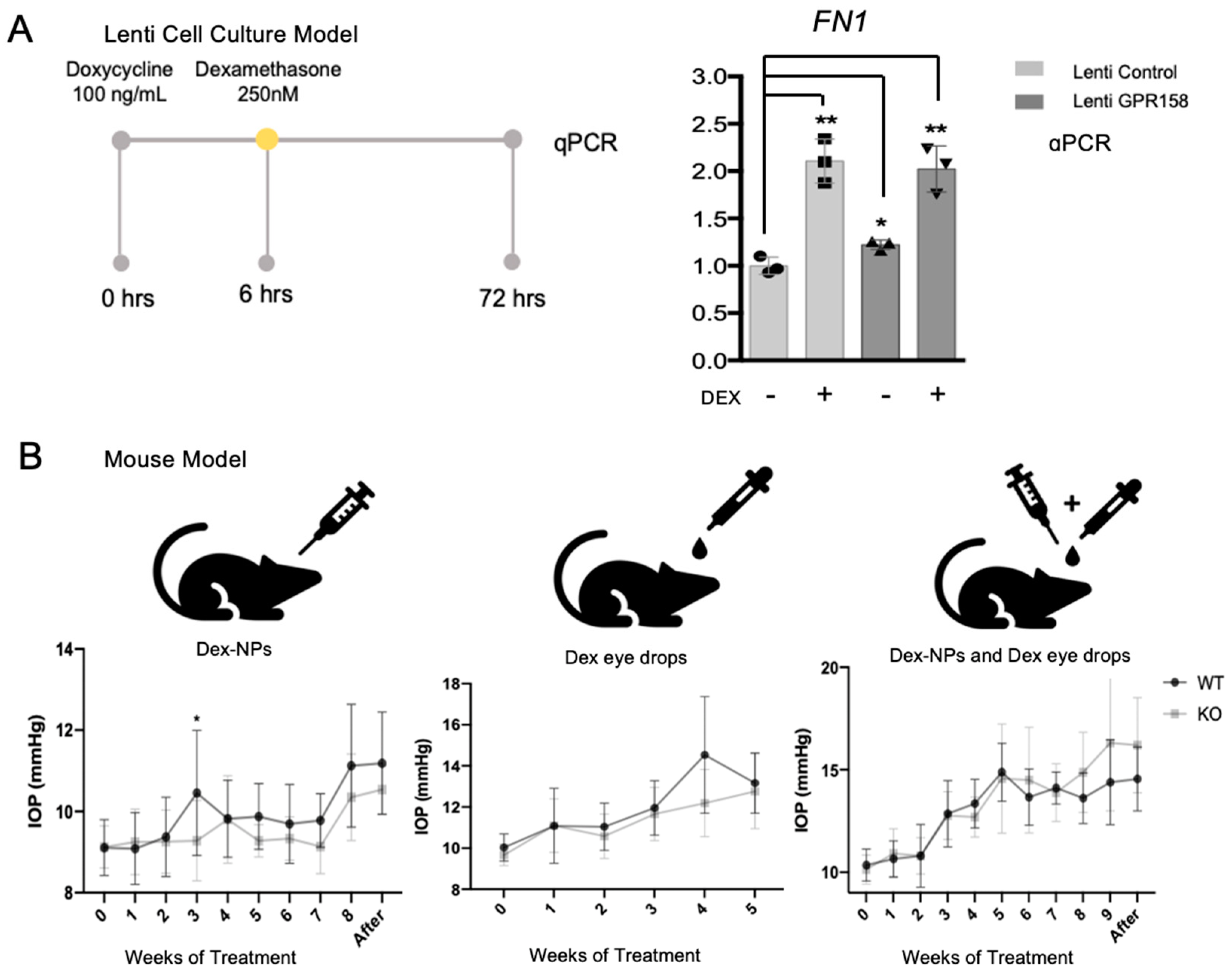
2.3. Functional Studies
2.4. Upstream GPCR Regulator ADRB1 and Other Genes Linked to GPCR Signaling
3. Discussion
4. Materials and Methods
4.1. Antibodies
4.2. Immortalized Trabecular Meshwork Cell Line and Lentivirus-Transformed GPR158 Overexpression Model
4.3. Human Trabecular Meshwork Primary Cell Strains and Transient Transfection
4.4. Microarray-Based Gene Expression Profiling
4.5. Ingenuity Pathway Analysis
4.6. Quantitative Polymerase Chain Reaction (qPCR)
4.7. Immunoblotting
4.8. Calcification Assay
4.9. Mouse Model of Steroid-Induced Ocular Hypertension
4.10. Statistical Analysis and Reproducibility
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharif, N.A. Therapeutic Drugs and Devices for Tackling Ocular Hypertension and Glaucoma, and Need for Neuroprotection and Cytoprotective Therapies. Front. Pharmacol. 2021, 12, 729249. [Google Scholar] [CrossRef] [PubMed]
- Epstein, D.L.; Allingham, R.R.; Schuman, J.S. Chandler and Grant’s Glaucoma, 4th ed.; Williams and Wilkins: Baltimore, Maryland, 1996. [Google Scholar]
- Grant, W.M. Experimental aqueous perfusion in enucleated human eyes. Arch. Ophthalmol. 1963, 69, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, J.; Murphy, C.; Polansky, J.; Juster, R. Age-related changes in trabecular meshwork cellularity. Investig. Ophthalmol. Vis. Sci. 1981, 21, 714–727. [Google Scholar]
- Alvarado, J.A.; Wood, I.; Polansky, J.R. Human trabecular cells. II. Growth pattern and ultrastructural characteristics. Investig. Ophthalmol. Vis. Sci. 1982, 23, 464–478. [Google Scholar]
- Alvarado, J.; Murphy, C.; Juster, R. Trabecular meshwork cellularity in primary open-angle glaucoma and nonglaucomatous normals. Ophthalmology 1984, 91, 564–579. [Google Scholar] [CrossRef]
- Alvarado, J.A.; Alvarado, R.G.; Yeh, R.F.; Franse-Carman, L.; Marcellino, G.R.; Brownstein, M.J. A new insight into the cellular regulation of aqueous outflow: How trabecular meshwork endothelial cells drive a mechanism that regulates the permeability of Schlemm’s canal endothelial cells. Br. J. Ophthalmol. 2005, 89, 1500–1505. [Google Scholar] [CrossRef]
- Adams, C.M.; Stacy, R.; Rangaswamy, N.; Bigelow, C.; Grosskreutz, C.L.; Prasanna, G. Glaucoma—Next Generation Therapeutics: Impossible to Possible. Pharm. Res. 2018, 36, 25. [Google Scholar] [CrossRef] [PubMed]
- Acott, T.S.; Kelley, M.J. Extracellular matrix in the trabecular meshwork. Exp. Eye Res. 2008, 86, 543–561. [Google Scholar] [CrossRef]
- Ethier, C.R. The inner wall of Schlemm’s canal. Exp. Eye Res. 2002, 74, 161–172. [Google Scholar] [CrossRef]
- Johnson, M. What controls aqueous humour outflow resistance? Exp. Eye Res. 2006, 82, 545–557. [Google Scholar] [CrossRef]
- Maepea, O.; Bill, A. Pressures in the juxtacanalicular tissue and Schlemm’s canal in monkeys. Exp. Eye Res. 1992, 54, 879–883. [Google Scholar] [CrossRef]
- Lutjen-Drecoll, E.; Shimizu, T.; Rohrbach, M.; Rohen, J.W. Quantitative analysis of ‘plaque material’ in the inner- and outer wall of Schlemm’s canal in normal- and glaucomatous eyes. Exp. Eye Res. 1986, 42, 443–455. [Google Scholar] [CrossRef]
- Schuman, J.S.; Wang, N.; Eisenberg, D.L. Leukemic glaucoma: The effects on outflow facility of chronic lymphocytic leukemia lymphocytes. Exp. Eye Res. 1995, 61, 609–617. [Google Scholar] [CrossRef]
- Underwood, J.L.; Murphy, C.G.; Chen, J.; Franse-Carman, L.; Wood, I.; Epstein, D.L.; Alvarado, J.A. Glucocorticoids regulate transendothelial fluid flow resistance and formation of intercellular junctions. Am. J. Physiol. 1999, 277, C330–C342. [Google Scholar] [CrossRef]
- Tripathi, R.C. Mechanism of the aqueous outflow across the trabecular wall of Schlemm’s canal. Exp. Eye Res. 1971, 11, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Vranka, J.A.; Kelley, M.J.; Acott, T.S.; Keller, K.E. Extracellular matrix in the trabecular meshwork: Intraocular pressure regulation and dysregulation in glaucoma. Exp. Eye Res. 2015, 133, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Stamer, W.D.; Braakman, S.T.; Zhou, E.H.; Ethier, C.R.; Fredberg, J.J.; Overby, D.R.; Johnson, M. Biomechanics of Schlemm’s canal endothelium and intraocular pressure reduction. Prog. Retin. Eye Res. 2015, 44, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Braunger, B.M.; Fuchshofer, R.; Tamm, E.R. The aqueous humor outflow pathways in glaucoma: A unifying concept of disease mechanisms and causative treatment. Eur. J. Pharm. Biopharm. 2015, 95, 173–181. [Google Scholar] [CrossRef]
- Filla, M.S.; Faralli, J.A.; Peotter, J.L.; Peters, D.M. The role of integrins in glaucoma. Exp. Eye Res. 2016. [Google Scholar] [CrossRef]
- Gagen, D.; Faralli, J.A.; Filla, M.S.; Peters, D.M. The role of integrins in the trabecular meshwork. J. Ocul. Pharmacol. Ther. 2014, 30, 110–120. [Google Scholar] [CrossRef]
- Lutjen-Drecoll, E. Morphological changes in glaucomatous eyes and the role of TGFbeta2 for the pathogenesis of the disease. Exp. Eye Res. 2005, 81, 1–4. [Google Scholar] [CrossRef]
- Shepard, A.R.; Millar, J.C.; Pang, I.H.; Jacobson, N.; Wang, W.H.; Clark, A.F. Adenoviral gene transfer of active human transforming growth factor-{beta}2 elevates intraocular pressure and reduces outflow facility in rodent eyes. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2067–2076. [Google Scholar] [CrossRef]
- Wordinger, R.J.; Fleenor, D.L.; Hellberg, P.E.; Pang, I.H.; Tovar, T.O.; Zode, G.S.; Fuller, J.A.; Clark, A.F. Effects of TGF-beta2, BMP-4, and gremlin in the trabecular meshwork: Implications for glaucoma. Investig. Ophthalmol. Vis. Sci. 2007, 48, 1191–1200. [Google Scholar] [CrossRef]
- Zode, G.S.; Clark, A.F.; Wordinger, R.J. Bone morphogenetic protein 4 inhibits TGF-beta2 stimulation of extracellular matrix proteins in optic nerve head cells: Role of gremlin in ECM modulation. Glia 2009, 57, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; McNatt, L.G.; Pang, I.H.; Millar, J.C.; Hellberg, P.E.; Hellberg, M.H.; Steely, H.T.; Rubin, J.S.; Fingert, J.H.; Sheffield, V.C.; et al. Increased expression of the WNT antagonist sFRP-1 in glaucoma elevates intraocular pressure. J. Clin. Investig. 2008, 118, 1056–1064. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Millar, J.C.; Wang, W.H.; Silverman, S.M.; Liu, Y.; Wordinger, R.J.; Rubin, J.S.; Pang, I.H.; Clark, A.F. Existence of the canonical Wnt signaling pathway in the human trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 2012, 53, 7043–7051. [Google Scholar] [CrossRef] [PubMed]
- Borras, T.; Comes, N. Evidence for a calcification process in the trabecular meshwork. Exp. Eye Res. 2009, 88, 738–746. [Google Scholar] [CrossRef]
- Peters, J.C.; Bhattacharya, S.; Clark, A.F.; Zode, G.S. Increased Endoplasmic Reticulum Stress in Human Glaucomatous Trabecular Meshwork Cells and Tissues. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3860–3868. [Google Scholar] [CrossRef]
- Zode, G.S.; Kuehn, M.H.; Nishimura, D.Y.; Searby, C.C.; Mohan, K.; Grozdanic, S.D.; Bugge, K.; Anderson, M.G.; Clark, A.F.; Stone, E.M.; et al. Reduction of ER stress via a chemical chaperone prevents disease phenotypes in a mouse model of primary open angle glaucoma. J. Clin. Investig. 2015, 125, 3303. [Google Scholar] [CrossRef] [PubMed]
- Kasetti, R.B.; Maddineni, P.; Millar, J.C.; Clark, A.F.; Zode, G.S. Increased synthesis and deposition of extracellular matrix proteins leads to endoplasmic reticulum stress in the trabecular meshwork. Sci. Rep. 2017, 7, 14951. [Google Scholar] [CrossRef]
- Kasetti, R.B.; Patel, P.D.; Maddineni, P.; Patil, S.; Kiehlbauch, C.; Millar, J.C.; Searby, C.C.; Raghunathan, V.; Sheffield, V.C.; Zode, G.S. ATF4 leads to glaucoma by promoting protein synthesis and ER client protein load. Nat. Commun. 2020, 11, 5594. [Google Scholar] [CrossRef] [PubMed]
- Zode, G.S.; Sharma, A.B.; Lin, X.; Searby, C.C.; Bugge, K.; Kim, G.H.; Clark, A.F.; Sheffield, V.C. Ocular-specific ER stress reduction rescues glaucoma in murine glucocorticoid-induced glaucoma. J. Clin. Investig. 2014, 124, 1956–1965. [Google Scholar] [CrossRef] [PubMed]
- Kasetti, R.B.; Maddineni, P.; Patel, P.D.; Searby, C.; Sheffield, V.C.; Zode, G.S. Transforming growth factor beta2 (TGFbeta2) signaling plays a key role in glucocorticoid-induced ocular hypertension. J. Biol. Chem. 2018, 293, 9854–9868. [Google Scholar] [CrossRef]
- Iglesias, A.I.; Springelkamp, H.; Ramdas, W.D.; Klaver, C.C.; Willemsen, R.; van Duijn, C.M. Genes, pathways, and animal models in primary open-angle glaucoma. Eye 2015, 29, 1285–1298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Y.; Allingham, R.R. Major review: Molecular genetics of primary open-angle glaucoma. Exp. Eye Res. 2017, 160, 62–84. [Google Scholar] [CrossRef] [PubMed]
- Gharahkhani, P.; Jorgenson, E.; Hysi, P.; Khawaja, A.P.; Pendergrass, S.; Han, X.; Ong, J.S.; Hewitt, A.W.; Segre, A.V.; Rouhana, J.M.; et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 2021, 12, 1258. [Google Scholar] [CrossRef]
- Fini, M.E.; Schwartz, S.G.; Gao, X.; Jeong, S.; Patel, N.; Itakura, T.; Price, M.O.; Price, F.W., Jr.; Varma, R.; Stamer, W.D. Steroid-induced ocular hypertension/glaucoma: Focus on pharmacogenomics and implications for precision medicine. Prog. Retin. Eye Res. 2017, 56, 58–83. [Google Scholar] [CrossRef]
- Patel, N.; Itakura, T.; Gonzalez, J.M., Jr.; Schwartz, S.G.; Fini, M.E. GPR158, an orphan member of G protein-coupled receptor Family C: Glucocorticoid-stimulated expression and novel nuclear role. PLoS ONE 2013, 8, e57843. [Google Scholar] [CrossRef] [PubMed]
- Itakura, T.; Webster, A.; Chintala, S.K.; Wang, Y.; Gonzalez, J.M., Jr.; Tan, J.C.; Vranka, J.A.; Acott, T.; Craft, C.M.; Sibug Saber, M.E.; et al. GPR158 in the Visual System: Homeostatic Role in Regulation of Intraocular Pressure. J. Ocul. Pharmacol. Ther. 2019, 35, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Itakura, T.; Jeong, S.; Liao, C.P.; Roy-Burman, P.; Zandi, E.; Groshen, S.; Pinski, J.; Coetzee, G.A.; Gross, M.E.; et al. Expression and Functional Role of Orphan Receptor GPR158 in Prostate Cancer Growth and Progression. PLoS ONE 2015, 10, e0117758. [Google Scholar] [CrossRef] [PubMed]
- Levoye, A.; Dam, J.; Ayoub, M.A.; Guillaume, J.L.; Jockers, R. Do orphan G-protein-coupled receptors have ligand-independent functions? New insights from receptor heterodimers. EMBO Rep. 2006, 7, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Milligan, G. G protein-coupled receptor hetero-dimerization: Contribution to pharmacology and function. Br. J. Pharmacol. 2009, 158, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, C.; Xie, K.; Masuho, I.; Fajardo-Serrano, A.; Lujan, R.; Martemyanov, K.A. Orphan Receptor GPR158 Is an Allosteric Modulator of RGS7 Catalytic Activity with an Essential Role in Dictating Its Expression and Localization in the Brain. J. Biol. Chem. 2015, 290, 13622–13639. [Google Scholar] [CrossRef] [PubMed]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Reviews. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef]
- Byrns, M.C.; Jin, Y.; Penning, T.M. Inhibitors of type 5 17beta-hydroxysteroid dehydrogenase (AKR1C3): Overview and structural insights. J. Steroid Biochem. Mol. Biol. 2011, 125, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Liton, P.B.; Liu, X.; Stamer, W.D.; Challa, P.; Epstein, D.L.; Gonzalez, P. Specific targeting of gene expression to a subset of human trabecular meshwork cells using the chitinase 3-like 1 promoter. Investig. Ophthalmol. Vis. Sci. 2005, 46, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.Y.; Ivanov, S.; Ivanova, A.; Ghosh, S.; Cote, M.A.; Keefe, K.; Coca-Prados, M.; Stanbridge, E.J.; Lerman, M.I. Expression of cell surface transmembrane carbonic anhydrase genes CA9 and CA12 in the human eye: Overexpression of CA12 (CAXII) in glaucoma. J. Med. Genet 2003, 40, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.R.; Rowlette, L.L.; Caballero, M.; Yang, P.; Hernandez, M.R.; Borras, T. Tissue differential microarray analysis of dexamethasone induction reveals potential mechanisms of steroid glaucoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 473–485. [Google Scholar] [CrossRef]
- Rozsa, F.W.; Reed, D.M.; Scott, K.M.; Pawar, H.; Moroi, S.E.; Kijek, T.G.; Krafchak, C.M.; Othman, M.I.; Vollrath, D.; Elner, V.M.; et al. Gene expression profile of human trabecular meshwork cells in response to long-term dexamethasone exposure. Mol. Vis. 2006, 12, 125–141. [Google Scholar] [PubMed]
- Ishibashi, T.; Takagi, Y.; Mori, K.; Naruse, S.; Nishino, H.; Yue, B.Y.; Kinoshita, S. cDNA microarray analysis of gene expression changes induced by dexamethasone in cultured human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3691–3697. [Google Scholar]
- Nehme, A.; Lobenhofer, E.K.; Stamer, W.D.; Edelman, J.L. Glucocorticoids with different chemical structures but similar glucocorticoid receptor potency regulate subsets of common and unique genes in human trabecular meshwork cells. BMC Med. Genom. 2009, 2, 58. [Google Scholar] [CrossRef]
- Danias, J.; Gerometta, R.; Ge, Y.; Ren, L.; Panagis, L.; Mittag, T.W.; Candia, O.A.; Podos, S.M. Gene expression changes in steroid-induced IOP elevation in bovine trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8636–8645. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Keller, K.E.; Bradley, J.M.; Vranka, J.A.; Acott, T.S. Segmental versican expression in the trabecular meshwork and involvement in outflow facility. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5049–5057. [Google Scholar] [CrossRef]
- Picht, G.; Welge-Luessen, U.; Grehn, F.; Lutjen-Drecoll, E. Transforming growth factor beta 2 levels in the aqueous humor in different types of glaucoma and the relation to filtering bleb development. Graefes. Arch Clin. Exp. Ophthalmol. 2001, 239, 199–207. [Google Scholar] [CrossRef]
- Tripathi, R.C.; Li, J.; Chan, W.F.; Tripathi, B.J. Aqueous humor in glaucomatous eyes contains an increased level of TGF-beta 2. Exp. Eye Res. 1994, 59, 723–727. [Google Scholar] [CrossRef]
- Flugel-Koch, C.; Ohlmann, A.; Fuchshofer, R.; Welge-Lussen, U.; Tamm, E.R. Thrombospondin-1 in the trabecular meshwork: Localization in normal and glaucomatous eyes, and induction by TGF-beta1 and dexamethasone in vitro. Exp. Eye Res. 2004, 79, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Call, M.K.; Yuan, Y.; Zhang, Y.; Fischesser, K.; Liu, C.Y.; Kao, W.W. Dexamethasone induces cross-linked actin networks in trabecular meshwork cells through noncanonical wnt signaling. Investig. Ophthalmol. Vis. Sci. 2013, 54, 6502–6509. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.; Comes, N.; Borras, T. Presence of an established calcification marker in trabecular meshwork tissue of glaucoma donors. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3184–3194. [Google Scholar] [CrossRef][Green Version]
- Xue, W.; Wallin, R.; Olmsted-Davis, E.A.; Borras, T. Matrix GLA protein function in human trabecular meshwork cells: Inhibition of BMP2-induced calcification process. Investig. Ophthalmol. Vis. Sci. 2006, 47, 997–1007. [Google Scholar] [CrossRef]
- Hu, H.; Tian, M.; Ding, C.; Yu, S. The C/EBP Homologous Protein (CHOP) Transcription Factor Functions in Endoplasmic Reticulum Stress-Induced Apoptosis and Microbial Infection. Front. Immunol. 2018, 9, 3083. [Google Scholar] [CrossRef]
- Kim, I.; Xu, W.; Reed, J.C. Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 2008, 7, 1013–1030. [Google Scholar] [CrossRef]
- Satyanarayana, A.; Kaldis, P. Mammalian cell-cycle regulation: Several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene 2009, 28, 2925–2939. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.S.; Ma, W.; Mao, D.Y.; Benchimol, S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol. Cell. Biol. 2005, 25, 7423–7431. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Kalyanaraman, V.; Gautam, N. Differential ability to form the G protein betagamma complex among members of the beta and gamma subunit families. J. Biol. Chem. 1996, 271, 7141–7146. [Google Scholar] [CrossRef]
- Ponten, F.; Jirstrom, K.; Uhlen, M. The Human Protein Atlas--a tool for pathology. J. Pathol. 2008, 216, 387–393. [Google Scholar] [CrossRef]
- Kasukawa, T.; Masumoto, K.H.; Nikaido, I.; Nagano, M.; Uno, K.D.; Tsujino, K.; Hanashima, C.; Shigeyoshi, Y.; Ueda, H.R. Quantitative expression profile of distinct functional regions in the adult mouse brain. PLoS ONE 2011, 6, e23228. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, D.M.; Murphy, A.J.; Frendewey, D.; Gale, N.W.; Economides, A.N.; Auerbach, W.; Poueymirou, W.T.; Adams, N.C.; Rojas, J.; Yasenchak, J.; et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat. Biotechnol. 2003, 21, 652–659. [Google Scholar] [CrossRef]
- Khrimian, L.; Obri, A.; Ramos-Brossier, M.; Rousseaud, A.; Moriceau, S.; Nicot, A.S.; Mera, P.; Kosmidis, S.; Karnavas, T.; Saudou, F.; et al. Gpr158 mediates osteocalcin’s regulation of cognition. J. Exp. Med. 2017, 214, 2859–2873. [Google Scholar] [CrossRef]
- Sutton, L.P.; Orlandi, C.; Song, C.; Oh, W.C.; Muntean, B.S.; Xie, K.; Filippini, A.; Xie, X.; Satterfield, R.; Yaeger, J.D.W.; et al. Orphan receptor GPR158 controls stress-induced depression. eLife 2018, 7, e33273. [Google Scholar] [CrossRef]
- Tao, Y.X.; Conn, P.M. Chaperoning G protein-coupled receptors: From cell biology to therapeutics. Endocr. Rev. 2014, 35, 602–647. [Google Scholar] [CrossRef]
- Lakin, N.D.; Jackson, S.P. Regulation of p53 in response to DNA damage. Oncogene 1999, 18, 7644–7655. [Google Scholar] [CrossRef]
- Abuetabh, Y.; Wu, H.H.; Chai, C.; Al Yousef, H.; Persad, S.; Sergi, C.M.; Leng, R. DNA damage response revisited: The p53 family and its regulators provide endless cancer therapy opportunities. Exp. Mol. Med. 2022, 54, 1658–1669. [Google Scholar] [CrossRef]
- Crawley, L.; Zamir, S.M.; Cordeiro, M.F.; Guo, L. Clinical options for the reduction of elevated intraocular pressure. Ophthalmol. Eye Dis. 2012, 4, 43–64. [Google Scholar] [CrossRef]
- Erickson-Lamy, K.A.; Nathanson, J.A. Epinephrine increases facility of outflow and cyclic AMP content in the human eye in vitro. Investig. Ophthalmol. Vis. Sci. 1992, 33, 2672–2678. [Google Scholar]
- Busch, M.J.; Kobayashi, K.; Hoyng, P.F.; Mittag, T.W. Adenylyl cyclase in human and bovine trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 1993, 34, 3028–3034. [Google Scholar]
- Crider, J.Y.; Sharif, N.A. Adenylyl cyclase activity mediated by beta-adrenoceptors in immortalized human trabecular meshwork and non-pigmented ciliary epithelial cells. J. Ocul. Pharmacol. Ther. 2002, 18, 221–230. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe-Curtin, D.; Buckley, C. Review of alpha adrenoceptor function in the eye. Eye 1989, 3 Pt 4, 472–476. [Google Scholar] [CrossRef]
- Huang, Y.; Gil, D.W.; Vanscheeuwijck, P.; Stamer, W.D.; Regan, J.W. Localization of alpha 2-adrenergic receptor subtypes in the anterior segment of the human eye with selective antibodies. Investig. Ophthalmol. Vis. Sci. 1995, 36, 2729–2739. [Google Scholar]
- Wax, M.B.; Molinoff, P.B.; Alvarado, J.; Polansky, J. Characterization of beta-adrenergic receptors in cultured human trabecular cells and in human trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 1989, 30, 51–57. [Google Scholar]
- Stamer, W.D.; Huang, Y.; Seftor, R.E.; Svensson, S.S.; Snyder, R.W.; Regan, J.W. Cultured human trabecular meshwork cells express functional alpha 2A adrenergic receptors. Investig. Ophthalmol. Vis. Sci. 1996, 37, 2426–2433. [Google Scholar]
- Nakahara, M.; Shimozawa, M.; Nakamura, Y.; Irino, Y.; Morita, M.; Kudo, Y.; Fukami, K. A novel phospholipase C, PLC(eta)2, is a neuron-specific isozyme. J. Biol. Chem. 2005, 280, 29128–29134. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wing, M.R.; Sondek, J.; Harden, T.K. Molecular cloning and characterization of PLC-eta2. Biochem. J. 2005, 391, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.J.; Morgan, K.; Farquharson, C.; Millar, R.P. Phospholipase C-eta enzymes as putative protein kinase C and Ca2+ signalling components in neuronal and neuroendocrine tissues. Neuroendocrinology 2007, 86, 243–248. [Google Scholar] [CrossRef]
- Kanemaru, K.; Nakahara, M.; Nakamura, Y.; Hashiguchi, Y.; Kouchi, Z.; Yamaguchi, H.; Oshima, N.; Kiyonari, H.; Fukami, K. Phospholipase C-eta2 is highly expressed in the habenula and retina. Gene. Expr. Patterns 2010, 10, 119–126. [Google Scholar] [CrossRef]
- Laboute, T.; Zucca, S.; Holcomb, M.; Patil, D.N.; Garza, C.; Wheatley, B.A.; Roy, R.N.; Forli, S.; Martemyanov, K.A. Orphan receptor GPR158 serves as a metabotropic glycine receptor: mGlyR. Science 2023, 379, 1352–1358. [Google Scholar] [CrossRef]
- Orlandi, C.; Posokhova, E.; Masuho, I.; Ray, T.A.; Hasan, N.; Gregg, R.G.; Martemyanov, K.A. GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J. Cell Biol. 2012, 197, 711–719. [Google Scholar] [CrossRef]
- Filla, M.S.; Liu, X.; Nguyen, T.D.; Polansky, J.R.; Brandt, C.R.; Kaufman, P.L.; Peters, D.M. In vitro localization of TIGR/MYOC in trabecular meshwork extracellular matrix and binding to fibronectin. Investig. Ophthalmol. Vis. Sci. 2002, 43, 151–161. [Google Scholar]
- Polansky, J.R.; Weinreb, R.N.; Baxter, J.D.; Alvarado, J. Human trabecular cells. I. Establishment in tissue culture and growth characteristics. Investig. Ophthalmol. Vis. Sci. 1979, 18, 1043–1049. [Google Scholar]
- Polansky, J.R.; Wood, I.S.; Maglio, M.T.; Alvarado, J.A. Trabecular meshwork cell culture in glaucoma research: Evaluation of biological activity and structural properties of human trabecular cells in vitro. Ophthalmology 1984, 91, 580–595. [Google Scholar] [CrossRef]
- Nguyen, T.D.; Chen, P.; Huang, W.D.; Chen, H.; Johnson, D.; Polansky, J.R. Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J. Biol. Chem. 1998, 273, 6341–6350. [Google Scholar] [CrossRef]
- Stamer, W.D.; Seftor, R.E.; Williams, S.K.; Samaha, H.A.; Snyder, R.W. Isolation and culture of human trabecular meshwork cells by extracellular matrix digestion. Curr. Eye Res. 1995, 14, 611–617. [Google Scholar] [CrossRef]
- Stamer, D.W.; Roberts, B.C.; Epstein, D.L.; Allingham, R.R. Isolation of primary open-angle glaucomatous trabecular meshwork cells from whole eye tissue. Curr. Eye Res. 2000, 20, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Keller, K.E.; Bhattacharya, S.K.; Borras, T.; Brunner, T.M.; Chansangpetch, S.; Clark, A.F.; Dismuke, W.M.; Du, Y.; Elliott, M.H.; Ethier, C.R.; et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp. Eye Res. 2018, 171, 164–173. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Buie, L.K.; Karim, M.Z.; Smith, M.H.; Borras, T. Development of a model of elevated intraocular pressure in rats by gene transfer of bone morphogenetic protein 2. Investig. Ophthalmol. Vis. Sci. 2013, 54, 5441–5455. [Google Scholar] [CrossRef] [PubMed]
- Duma, D.; Collins, J.B.; Chou, J.W.; Cidlowski, J.A. Sexually dimorphic actions of glucocorticoids provide a link to inflammatory diseases with gender differences in prevalence. Sci. Signal 2010, 3, ra74. [Google Scholar] [CrossRef]
- Li, G.; Lee, C.; Agrahari, V.; Wang, K.; Navarro, I.; Sherwood, J.M.; Crews, K.; Farsiu, S.; Gonzalez, P.; Lin, C.W.; et al. In vivo measurement of trabecular meshwork stiffness in a corticosteroid-induced ocular hypertensive mouse model. Proc. Natl. Acad. Sci. USA 2019, 116, 1714–1722. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Millar, J.C.; Pang, I.H.; Wax, M.B.; Clark, A.F. Noninvasive measurement of rodent intraocular pressure with a rebound tonometer. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4617–4621. [Google Scholar] [CrossRef]



| GPR158 Dose | Exp 1 | Exp 2A | Exp 2B |
|---|---|---|---|
| Doxycycline | Low | High | High |
| Duration | 72 h | 24 h | 96 h |
| IPA Top Upstream Regulators | |||
| Dexamethasone | p = 2.71 × 10−9 | p = 4.55 × 10−17 | p = 1.62 × 10−6 |
| activated | |||
| TGFB1 | p = 1.19 × 10−9 | p = 1.02 × 10−23 | p = 2.54 × 10−16 |
| inhibited | |||
| XBP1 | p = 1.09 × 10−5 | ||
| inhibited | |||
| ATF4 | p = 5.46 × 10−6 | ||
| TP53 | p = 1.13 × 10−5 | p = 4.84 × 10−18 | p = 3.41 × 10−15 |
| inhibited | activated | ||
| Gene | Fold Change in Gene Expression | Change Direction Predicted | Change Direction Observed | ||
|---|---|---|---|---|---|
| Exp 1 Low, 72 h | Exp 2A High, 24 h | Exp 2B High, 96 h | |||
| Dexamethasone regulated | |||||
| Acute | |||||
| AKR1C1/C2 | −1.302 | −1.991 | −1.503 | up | down |
| AKR1C3 | −1.443 | −1.625 | up | down | |
| AKR1C4 * | −1.313 | −2.206 | −1.576 | up * | down |
| CA12 ** | −1.317 | −1.727 | −1.362 | up | down |
| CHI3L1 | −1.370 | −1.571 | up | down | |
| Extracellular matrix | |||||
| ADAMTS1 | −1.229 | up | down | ||
| COL1A1 | −1.250 | up | down | ||
| COL4A1 | −1.276 | −1.256 | up | down | |
| COL4A5 | +1.256 | up | up | ||
| COL10A1 | +1.686 | +3.183 | down | up | |
| LAMA4 | −1.218 | −1.218 | −1.212 | up | down |
| LAMA5 | −1.212 | −1.394 | +1.220 | up | regulates |
| VCAN | −1.235 | −1.570 | −1.786 | down | down |
| Growth factors and matricellular proteins | |||||
| CTGF | −1.144 | −1.320 | up | down | |
| CYR61 | −1.211 | −1.292 | +1.315 | regulates | regulates |
| GDF15 | +1.354 | +1.374 | down | up | |
| TGFB2 | −1.443 | up | down | ||
| THBS1 | −1.242 | −1.363 | +1.249 | up | regulates |
| WNT5A | −1.264 | up | down | ||
| WNT5B *** | −1.212 | −1.376 | --- | down | |
| TGFB1 regulated | |||||
| Extracellular matrix | |||||
| COL1A1 | −1.250 | up | down | ||
| COL3A1 | −1.594 | up | down | ||
| COL4A1 | −1.276 | −1.256 | up | down | |
| COL5A1 | −1.254 | up | down | ||
| COL6A3 | −1.539 | up | down | ||
| VCAN | −1.570 | −1.786 | down | down | |
| Growth factors and matricellular proteins | |||||
| BMP4 | −1.212 | −1.382 | down | down | |
| DACT3 **** | +1.457 | +1.228 | --- | up | |
| DKK3 | +1.262 | up | up | ||
| GDF15 | +1.354 | +1.374 | up | up | |
| THBS1 | −1.242 | −1.363 | +1.249 | up | regulates |
| Gene | Fold Change in Gene Expression | Change Direction Predicted | Change Direction Observed | ||
|---|---|---|---|---|---|
| Exp 1 Low, 72 h | Exp 2A High, 24 h | Exp 2B High, 96 h | |||
| Calcification related | |||||
| ACTG2 | −1.204 | −1.414 | up | down | |
| ALPL | +1.340 | up | up | ||
| ALPP | +1.578 | up | up | ||
| BGLAP | --- | --- | |||
| BMP2 | up | --- | |||
| BMP4 | −1.212 | −1.382 | up | down | |
| COL1A1 | −1.250 | up | down | ||
| COL10A1 | +1.686 | +3.183 | up | up | |
| ECM1 | +1.319 | up | up | ||
| MGP | −1.221 | down | down | ||
| OSTF1 | +1.203 | down | up | ||
| SLC8A2 | +2.174 | +1.777 | down | up | |
| TNFRSF11B | −1.215 | down | down | ||
| WNT5A | −1.264 | up | down | ||
| WNT5B | −1.212 | −1.376 | up | down | |
| Gene | Fold Change in Gene Expression | Change Direction Predicted | Change Direction Observed | ||
|---|---|---|---|---|---|
| Exp 1 Low, 72 h | Exp 2A High, 24 h | Exp 2B High, 96 h | |||
| TP53-regulated | |||||
| Cell cycle progression | |||||
| CCNA1 ***** | −1.956 | --- | down | ||
| CCNA2 | −1.368 | down | down | ||
| CCNB2 | −1.269 | regulates | down | ||
| CCND1 | −1.292 | regulates | down | ||
| CCNE1 | −1.258 | regulates | down | ||
| CDC25C | −1.371 | down | down | ||
| MYC | −1.707 | −1.268 | down | down | |
| Cell cycle arrest | |||||
| BRSK1 | +1.475 | +1.335 | up | up | |
| CDC42EP5 | +1.708 | +1.547 | up | up | |
| CDKN1A | −1.186 | +1.264 | up | up | |
| GAS1 | −1.516 | −1.513 | regulates | down | |
| KLF6 | −1.818 | up | down | ||
| MAPRE3 | −1.357 | +1.219 | up | regulates | |
| SIRT4 | +1.433 | +1.306 | up | up | |
| TP53 | up | --- | |||
| TXNIP | −1.427 | +9.300 | up | regulates | |
| Mitosis | |||||
| CDC20 | −1.255 | down | down | ||
| CENPF | −2.203 | down | down | ||
| CEP55 | −1.236 | down | down | ||
| AURKA | −1.259 | down | down | ||
| PEG10 | −1.235 | −1.410 | down | down | |
| Gene | Fold Change in Gene Expression | Function | ||
|---|---|---|---|---|
| Exp 1 Low, 72 h | Exp 2A High, 24 h | Exp 2B High, 96 h | ||
| GPCR signaling | ||||
| GNB5 | +1.281 | G protein | ||
| GNG7 | +1.360 | +1.26 | G protein | |
| RGS4 | −2.778 | −2.21 | GTPase activating | |
| PLCG1 | −1.387 | Phospholipase C | ||
| PLCH2 | +1.265 | Phospholipase C | ||
| Antigen | Where Purchased | Catalogue Number | Species |
|---|---|---|---|
| GPR158 | Sigma-Aldrich Corporation, St Louis, MO, USA | Cat# HPA013185 | Rabbit |
| MYOC | Santa Cruz Biotechnology, Santa Cruz, CA, USA | Cat# sc-137233 | Mouse |
| TP53 | Cell Signaling Technology, Danvers, MA, USA | Cat# IC12 | Mouse |
| COL10A1 | Invitrogen, Waltham, MA, USA | Cat# PA5-49198 | Rabbit |
| SLCA8 | Origene Technologies, Rockville, MD, USA | Cat# TA328916 | Rabbit |
| HSPA5 (GRP78/BiP) | Abcam, Cambridge, UK | Cat# ab212054 | Mouse |
| ATF4 | Cell Signaling Technology, Danvers, MA, USA | Cat# 11815 | Rabbit |
| DDIT3 (CHOP) | Cell Signaling Technology, Danvers, MA, USA | Cat# 5554 | Rabbit |
| ACTB | Abcam, Cambridge, UK | Cat# ab6276 | Mouse |
| GAPDH | Cell Signaling Technology, Danvers, MA, USA | Cat# 97166 | Mouse |
| Gene | Primer Sequence |
|---|---|
| TGFB2 | Forward: GCGACGAAGAGTACTACGCC Reverse: TGGCATCAAGGTACCCACAG |
| COL10A1 | Forward: CAAGGCACCATCTCCAGG AA Reverse: AAAGGGTAT TTGTGGCAGCATATT |
| CA12 | Forward: ACTGCGGCAGGACTGAGTCT Reverse: CACAATACAGATGCCAAGAATGC |
| TP53 | Forward: TTTTCCCCTCCCATGTGCTC Reverse: TGGACGGTGGCTCTAGACTT |
| FN1 | Forward: AGCGGACCTACCTAGGCAAT Reverse: GGTTTGCGATGGTACAGCTT |
| HSPA5 | Forward: CCAAGAGAGGGTTCTTGAATCTCG Reverse: ATGGGCCAGCCTGGATATACAACA |
| sXBP1 | Forward: CTGAGTCCGAATCAGGTGCAG Reverse: ATCCATGGGGAGATGTTCTGG |
| ATF4 | Forward: CAGCACAGCCCCTCTACCA Reverse: GCCCGCCTTAGCCTTGTC |
| DDIT3 | Forward: AGAACCAGGAAACGGAAACAGA Reverse: TCTCCTTCATGCGCTGCTTT |
| ACTB | Forward: GTCATTCCAAATATGAGATGCGT Reverse: GCTATCACCTCCCCTGTGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suarez, M.F.; Itakura, T.; Pany, S.; Jeong, S.; Chintala, S.K.; Raizman, M.B.; Riesinger, S.; Lazarova, T.; Echenique, J.; Serra, H.M.; et al. Regulatory Effects of GPR158 Overexpression in Trabecular Meshwork Cells of the Eye’s Aqueous Outflow Pathways. Stresses 2023, 3, 629-652. https://doi.org/10.3390/stresses3030044
Suarez MF, Itakura T, Pany S, Jeong S, Chintala SK, Raizman MB, Riesinger S, Lazarova T, Echenique J, Serra HM, et al. Regulatory Effects of GPR158 Overexpression in Trabecular Meshwork Cells of the Eye’s Aqueous Outflow Pathways. Stresses. 2023; 3(3):629-652. https://doi.org/10.3390/stresses3030044
Chicago/Turabian StyleSuarez, Maria Fernanda, Tatsuo Itakura, Satyabrata Pany, Shinwu Jeong, Shravan K. Chintala, Michael B. Raizman, Steven Riesinger, Tsvetelina Lazarova, José Echenique, Horacio M. Serra, and et al. 2023. "Regulatory Effects of GPR158 Overexpression in Trabecular Meshwork Cells of the Eye’s Aqueous Outflow Pathways" Stresses 3, no. 3: 629-652. https://doi.org/10.3390/stresses3030044
APA StyleSuarez, M. F., Itakura, T., Pany, S., Jeong, S., Chintala, S. K., Raizman, M. B., Riesinger, S., Lazarova, T., Echenique, J., Serra, H. M., Stamer, W. D., & Fini, M. E. (2023). Regulatory Effects of GPR158 Overexpression in Trabecular Meshwork Cells of the Eye’s Aqueous Outflow Pathways. Stresses, 3(3), 629-652. https://doi.org/10.3390/stresses3030044








