Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach
Abstract
:1. Introduction
2. Results and Discussion
2.1. Study of Pharmacokinetic Properties of the Ligands by a Predictive Model
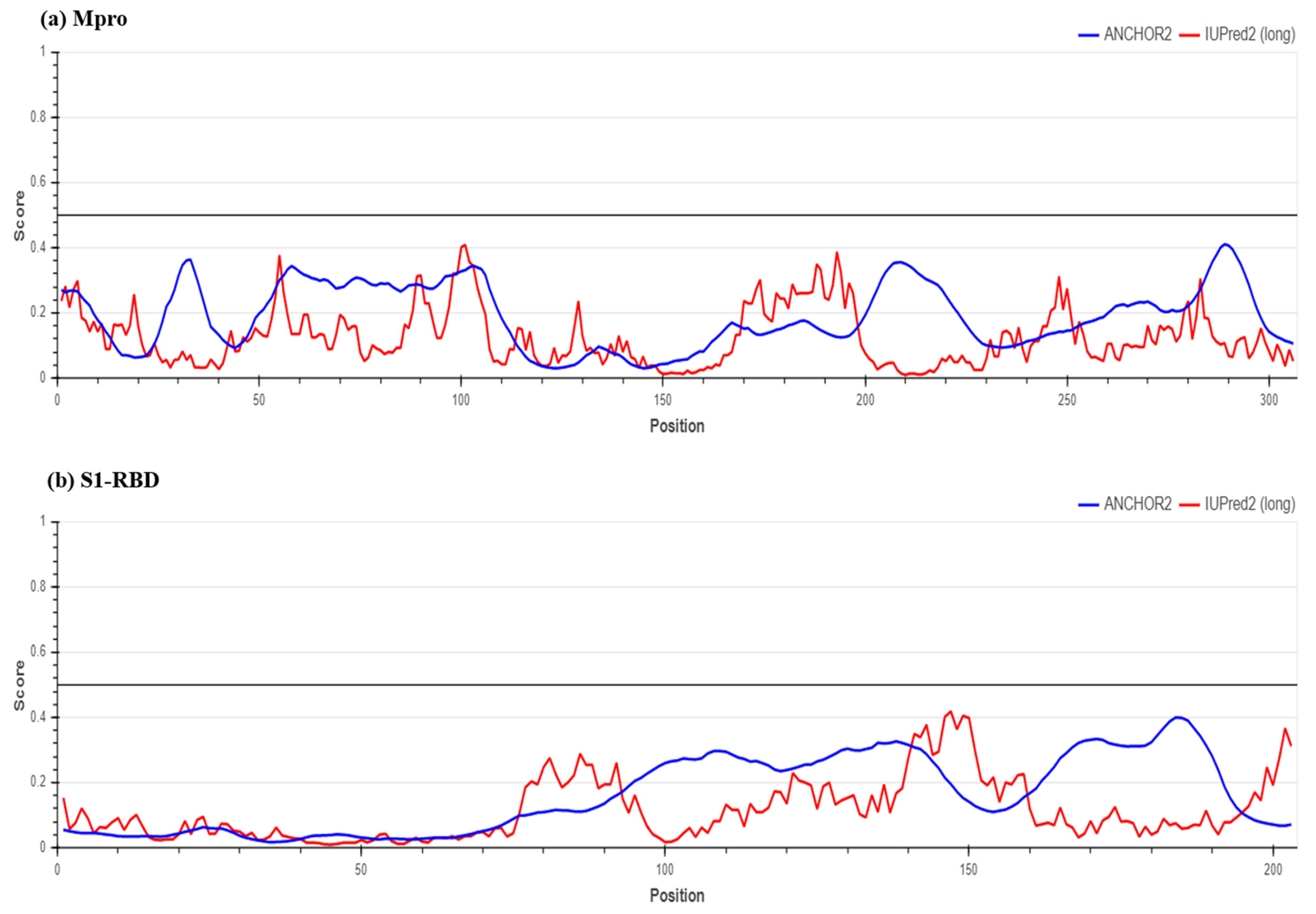
2.2. Determination of Protein Stability
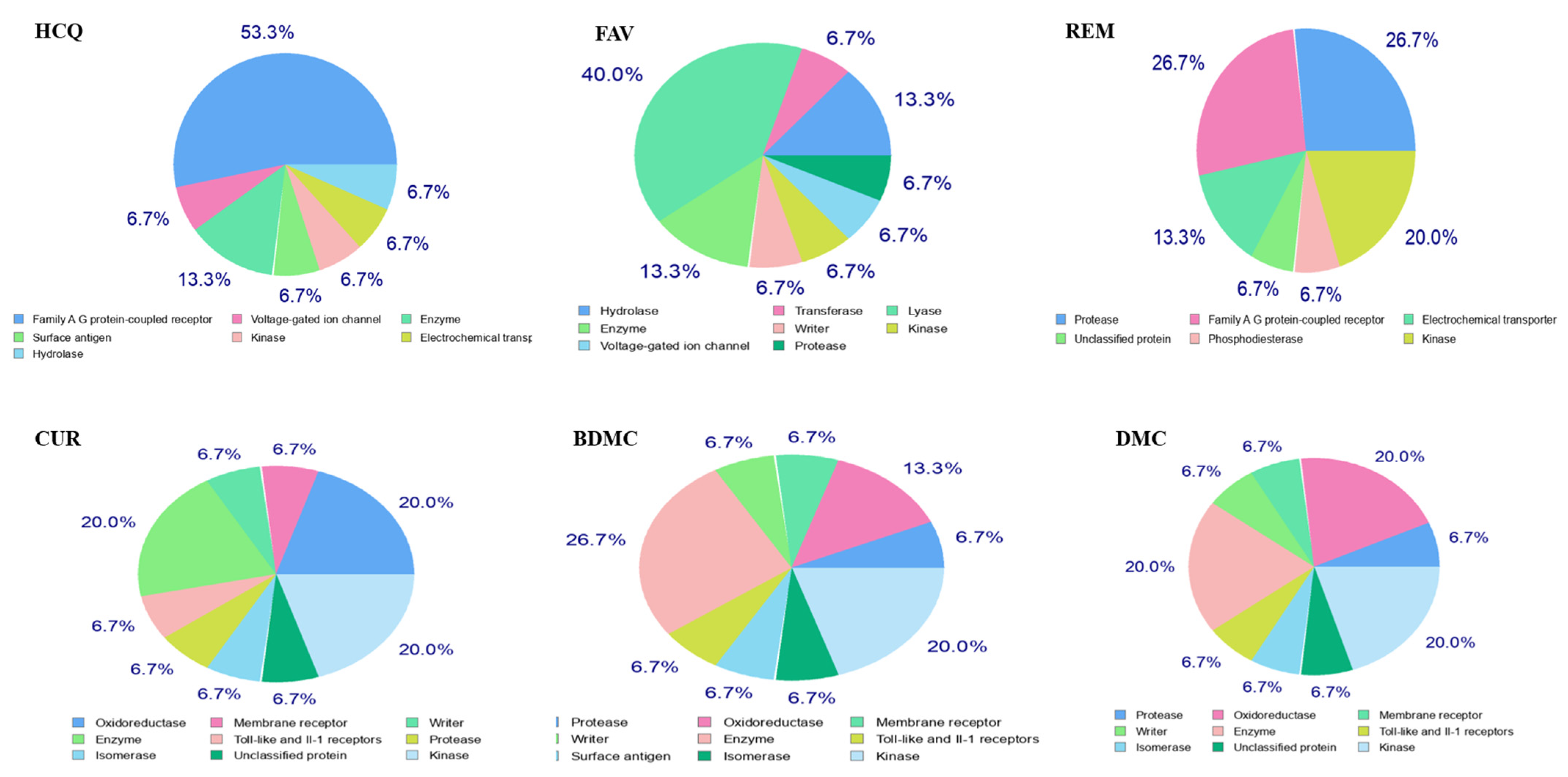
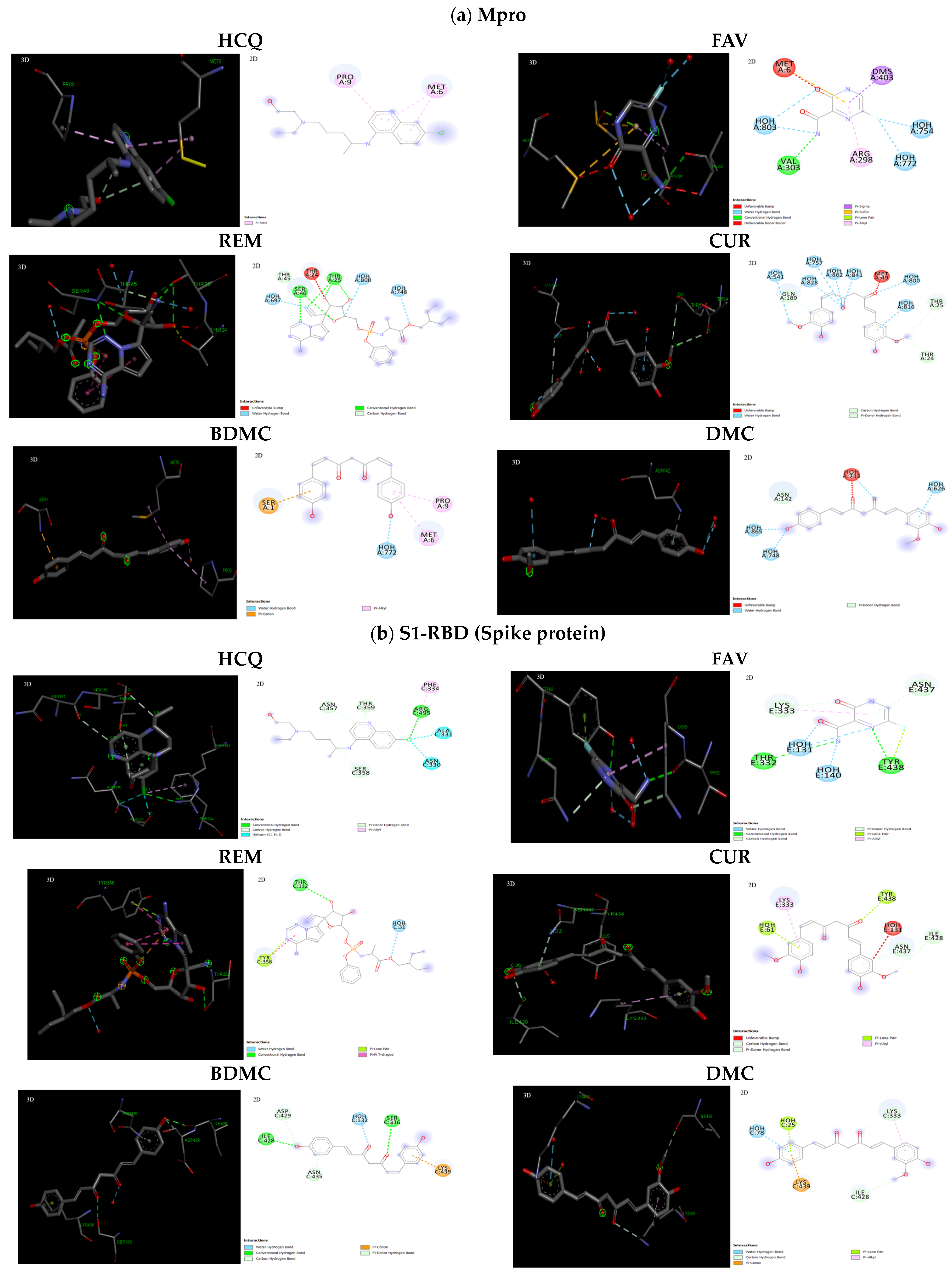
2.3. Interaction of Ligands with Receptors of SARS-CoV-2
3. Materials and Methods
3.1. Selection of Ligands and Proteins
3.2. Analysis of Pharmacokinetic Properties of the Selected Molecules
3.3. Prediction of Protein Stability
3.4. Molecular Docking
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.C.; Shih, T.P.; Ko, W.C.; Tang, H.J.; Hsueh, P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Gautret, P.; Lagier, J.C.; Parola, P.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; Dupont, H.T.; et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 56, 105949. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; McGoogan, J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020, 323, 1239–1242. [Google Scholar] [CrossRef]
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit vaccines against emerging pathogenic human coronaviruses. Front. Microbiol. 2020, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jiang, S.; Du, L. Prospects for a MERS-CoV spike vaccine. Expert Rev. Vaccines 2018, 17, 677–686. [Google Scholar] [CrossRef]
- Du, L.; Yang, Y.; Zhou, Y.; Lu, L.; Li, F.; Jiang, S. MERS-CoV spike protein: A key target for antivirals. Expert Opin. Ther. Targets 2017, 21, 131–143. [Google Scholar] [CrossRef]
- Wang, Q.; Wong, G.; Lu, G.; Yan, J.; Gao, G.F. MERS-CoV spike protein: Targets for vaccines and therapeutics. Antiviral Res. 2016, 133, 165–177. [Google Scholar] [CrossRef]
- Liu, S.; Xiao, G.; Chen, Y.; He, Y.; Niu, J.; Escalante, C.R.; Xiong, H.; Farmar, J.; Debnath, A.K.; Tien, P.; et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: Implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 2004, 363, 938–947. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; Pu, J.; Voronin, D.; Jiang, S.; Zhou, Y.; Du, L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020, 17, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Anand, K.; Ziebuhr, J.; Wadhwani, P.; Mesters, J.R.; Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767. [Google Scholar] [CrossRef] [PubMed]
- Hilgenfeld, R. From SARS to MERS: Crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014, 281, 4085–4096. [Google Scholar] [CrossRef]
- Jeon, S.; Ko, M.; Lee, J.; Choi, I.; Byun, S.Y.; Park, S.; Shum, D.; Kim, S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020, 64, e00819–e00820. [Google Scholar] [CrossRef]
- Biot, C.; Daher, W.; Chavain, N.; Fandeur, T.; Khalife, J.; Dive, D.; De Clercq, E. Design and synthesis of hydroxyferroquine derivatives with antimalarial and antiviral activities. J. Med. Chem. 2006, 49, 2845–2849. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef]
- Schrezenmeier, E.; Dörner, T. Mechanisms of action of hydroxychloroquine and chloroquine: Implications for rheumatology. Nat. Rev. Rheumatol. 2020, 16, 155–166. [Google Scholar] [CrossRef]
- Zhou, D.; Dai, S.M.; Tong, Q. COVID-19: A recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020, 75, 1667–1670. [Google Scholar] [CrossRef]
- Vincent, M.J.; Bergeron, E.; Benjannet, S.; Erickson, B.R.; Rollin, P.E.; Ksiazek, T.G.; Seidah, N.G.; Nichol, S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005, 2, 69. [Google Scholar] [CrossRef]
- Furuta, Y.; Gowen, B.B.; Takahashi, K.; Shiraki, K.; Smee, D.F.; Barnard, D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013, 100, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Yang, M.; Liu, D.; Chen, J.; Shu, D.; Xia, J.; Liao, X.; Gu, Y.; Cai, Q.; Yang, Y.; et al. Experimental treatment with favipiravir for COVID-19: An open-label control study. Engineering 2020, 6, 1192–1198. [Google Scholar] [CrossRef] [PubMed]
- Yavuz, S.; Ünal, S. Antiviral treatment of COVID-19. Turk. J. Med. Sci. 2020, 50, 611–619. [Google Scholar] [CrossRef]
- Niranjan, A.; Prakash, D. Chemical constituents and biological activities of turmeric (Curcuma longa L.)—A review. J. Food Sci. Technol. 2008, 45, 109. [Google Scholar]
- Gupta, S.C.; Patchva, S.; Aggarwal, B.B. Therapeutic roles of curcumin: Lessons learned from clinical trials. AAPS J. 2013, 15, 195–218. [Google Scholar] [CrossRef] [PubMed]
- Zorofchian Moghadamtousi, S.; Abdul Kadir, H.; Hassandarvish, P.; Tajik, H.; Abubakar, S.; Zandi, K. A review on antibacterial, antiviral, and antifungal activity of curcumin. BioMed Res. Int. 2014, 2014, 186864. [Google Scholar] [CrossRef]
- Chen, T.Y.; Chen, D.Y.; Wen, H.W.; Ou, J.L.; Chiou, S.S.; Chen, J.M.; Wong, M.L.; Hsu, W.L. Inhibition of enveloped viruses infectivity by curcumin. PLoS ONE 2013, 8, e62482. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Tewari, M.; Shukla, H.S.; Roy, B.K. In silico profiling of the potentiality of curcumin and conventional drugs for CagA oncoprotein inactivation. Arch. Pharm. 2015, 348, 548–555. [Google Scholar] [CrossRef]
- Ortega, J.T.; Serrano, M.L.; Pujol, F.H.; Rangel, H.R. Unrevealing sequence and structural features of novel coronavirus using in silico approaches: The main protease as molecular target. EXCLI J. 2020, 19, 400. [Google Scholar]
- Hwang, W.C.; Lin, Y.; Santelli, E.; Sui, J.; Jaroszewski, L.; Stec, B.; Farzan, M.; Marasco, W.A.; Liddington, R.C. Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J. Biol. Chem. 2006, 281, 34610–34616. [Google Scholar] [CrossRef]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Baindara, P.; Chowdhury, T.; Roy, D.; Mandal, M.; Mandal, S.M. Surfactin-like lipopeptides from Bacillus clausii efficiently bind to spike glycoprotein of SARS-CoV-2. J. Biomol. Struct. Dyn. 2023, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. Chem. Med. Chem. 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019, 47, W357–W364. [Google Scholar] [CrossRef] [PubMed]
- Dosztányi, Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018, 27, 331–340. [Google Scholar] [CrossRef]
- Mészáros, B.; Erdős, G.; Dosztányi, Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- Mohankumar, T.; Chandramohan, V.; Lalithamba, H.S.; Jayaraj, R.L.; Kumaradhas, P.; Sivanandam, M.; Hunday, G.; Vijayakumar, R.; Balakrishnan, R.; Manimaran, D.; et al. Design and molecular dynamic investigations of 7, 8-dihydroxyflavone derivatives as potential neuroprotective agents against alpha-synuclein. Sci. Rep. 2020, 10, 599. [Google Scholar] [CrossRef]
- Mengist, H.M.; Dilnessa, T.; Jin, T. Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease. Front. Chem. 2021, 9, 622898. [Google Scholar] [CrossRef]
- Kgatle, M.M.; Lawal, I.O.; Mashabela, G.; Boshomane, T.M.G.; Koatale, P.C.; Mahasha, P.W.; Ndlovu, H.; Vorster, M.; Rodrigues, H.G.; Zeevaart, J.R.; et al. COVID-19 is a multi-organ aggressor: Epigenetic and clinical marks. Front. Immunol. 2021, 12, 752380. [Google Scholar] [CrossRef]
- Eren, T.; Baysal, G.; Doğan, F. Biocidal Activity of Bone Cements Containing Curcumin and Pegylated Quaternary Polyethylenimine. J. Poly. Environ. 2020, 28, 2469–2480. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–227. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Singh, D.; Roy, B.K. Structural interactions of curcumin biotransformed molecules with the N-terminal residues of cytotoxic-associated gene a protein provide insights into suppression of oncogenic activities. Interdiscip. Sci. Comput. Life Sci. 2017, 9, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.; Bhattacharjee, P. Promising role of curcumin against viral diseases emphasizing COVID-19 management: A review on the mechanistic insights with reference to host-pathogen interaction and immunomodulation. J. Funct. Foods 2021, 82, 104503. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jiang, H.; Shi, Y. Upregulation of heme oxygenase-1 expression by curcumin conferring protection from hydrogen peroxide-induced apoptosis in H9c2 cardiomyoblasts. Cell Biosci. 2017, 7, 20. [Google Scholar] [CrossRef]
- Vajragupta, O.; Boonchoong, P.; Morris, G.M.; Olson, A.J. Active site binding modes of curcumin in HIV-1 protease and integrase. Bioorg. Med. Chem. Lett. 2005, 15, 3364–3368. [Google Scholar] [CrossRef] [PubMed]
- Jennings, M.R.; Parks, R.J. Curcumin as an Antiviral Agent. Viruses 2020, 12, 1242. [Google Scholar] [CrossRef]
- Mounce, B.C.; Cesaro, T.; Carrau, L.; Vallet, T.; Vignuzzi, M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antivir. Res 2017, 142, 148–157. [Google Scholar] [CrossRef]
- Balasubramanian, A.; Pilankatta, R.; Teramoto, T.; Sajith, A.M.; Nwulia, E.; Kulkarni, A.; Padmanabhan, R. Inhibition of dengue virus by curcuminoids. Antivir. Res. 2019, 162, 71–78. [Google Scholar] [CrossRef]
- Richart, S.M.; Li, Y.L.; Mizushina, Y.; Chang, Y.Y.; Chung, T.Y.; Chen, G.H.; Tzen, J.T.; Shia, K.S.; Hsu, W.L. Synergic effect of curcumin and its structural analogue (Monoacetylcurcumin) on anti-influenza virus infection. J. Food Drug Anal. 2018, 26, 1015–1023. [Google Scholar] [CrossRef]
- Yang, X.X.; Li, C.M.; Huang, C.Z. Curcumin modified silver nanoparticles for highly efficient inhibition of respiratory syncytial virus infection. Nanoscale 2016, 8, 3040–3048. [Google Scholar] [CrossRef]
- Zahedipour, F.; Hosseini, S.A.; Sathyapalan, T.; Majeed, M.; Jamialahmadi, T.; Al-Rasadi, K.; Banach, M.; Sahebkar, A. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother. Res. 2020, 34, 2911–2920. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kashyap, P.; Chowdhury, S.; Kumar, S.; Panwar, A.; Kumar, A. Identification of phytochemicals as potential therapeutic agents that binds to Nsp15 protein target of coronavirus (SARS-CoV-2) that are capable of inhibiting virus replication. Phytomedicine 2021, 85, 153317. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Thomas, P.D.; Dill, K.A. An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. USA 1996, 93, 11628–11633. [Google Scholar] [CrossRef] [PubMed]
- Dosztanyi, Z.; Csizmok, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, B.; Simon, I.; Dosztányi, Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009, 5, e1000376. [Google Scholar] [CrossRef]


| Compounds | Caco-2 Permeability | AMES Toxicity | Carcinogens | Rat Acute Toxicity LD50 (mol/kg) |
|---|---|---|---|---|
| HCQ | Caco2- | AMES toxic | Non-carcinogens | 2.6348 |
| FAV | Caco2- | Non-AMES toxic | Non-carcinogens | 2.1259 |
| REM | Caco2- | Non-AMES toxic | Non-carcinogens | 2.7169 |
| CUR | Caco2+ | Non-AMES toxic | Non-carcinogens | 2.5468 |
| BDMC | Caco2+ | Non-AMES toxic | Non-carcinogens | 2.2754 |
| DMC | Caco2+ | Non-AMES toxic | Non-carcinogens | 2.2792 |
| Compounds | Mpro (kcal/mol) | S1-RBD (kcal/mol) |
|---|---|---|
| HCQ | −24.58 | −35.87 |
| FAV | −22.87 | −23.77 |
| REM | −23.48 | −38.44 |
| CUR | −20.47 | −38.84 |
| BDMC | −27.47 | −28.07 |
| DMC | −20.58 | −30.29 |
| Viruses | Doses | Curcumin and Its Derivatives | Outcomes | Ref. |
|---|---|---|---|---|
| Human Immunodeficiency Virus (HIV) | 20–120 µM | Curcumin | Inhibited HIV-1 protease | [45] |
| 0.7–12 µM | Synthetic curcumin analogue lacking the β-diketone moiety named as curcumin A | Lowered late viral genome-copy levels | [46] | |
| Zika Virus (ZIKV) | 10 µM–1 mM | Curcumin, bisdemethoxycurcumin, demethoxycurcumin | Inactivated virus or hindered cell attachment | [47] |
| Dengue virus (DENV) | 36–66 µM | Curcumin, bisdemethoxycurcumin, acyclic and cyclohexanone analogues of curcumin | Inhibited the viral protease activity, downregulated acetyl-CoA carboxylase and fatty acid synthase | [48] |
| Influenza A Virus (IAV) | 25–200 µM | Curcumin | Decreased IAV neuraminidase (NA) activity, inhibitor of the PI3K/Akt signalling pathway | [49] |
| Human Respiratory Syncytial Virus (HRSV) | 5–15 µM | Curcumin-stabilized silver nanoparticles | Inhibited the viral G protein expression involved in viral attachment | [50] |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) | >10 µM | Curcumin | Suppressed the main protease and inhibited the viral non-structural protein Nsp15 | [51,52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, A.K.; Singh, D.; Yadav, P.; Singh, M.; Singh, S.K.; Kumar, A. Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach. Stresses 2023, 3, 615-628. https://doi.org/10.3390/stresses3030043
Srivastava AK, Singh D, Yadav P, Singh M, Singh SK, Kumar A. Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach. Stresses. 2023; 3(3):615-628. https://doi.org/10.3390/stresses3030043
Chicago/Turabian StyleSrivastava, Akhileshwar Kumar, Divya Singh, Priya Yadav, Monika Singh, Sandeep Kumar Singh, and Ajay Kumar. 2023. "Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach" Stresses 3, no. 3: 615-628. https://doi.org/10.3390/stresses3030043
APA StyleSrivastava, A. K., Singh, D., Yadav, P., Singh, M., Singh, S. K., & Kumar, A. (2023). Paradigm of Well-Orchestrated Pharmacokinetic Properties of Curcuminoids Relative to Conventional Drugs for the Inactivation of SARS-CoV-2 Receptors: An In Silico Approach. Stresses, 3(3), 615-628. https://doi.org/10.3390/stresses3030043









