The Antarctic Scallop Adamussium colbecki Is Unable to Transcriptomically Respond to Captivity and Moderate Thermal Stress
Abstract
1. Introduction
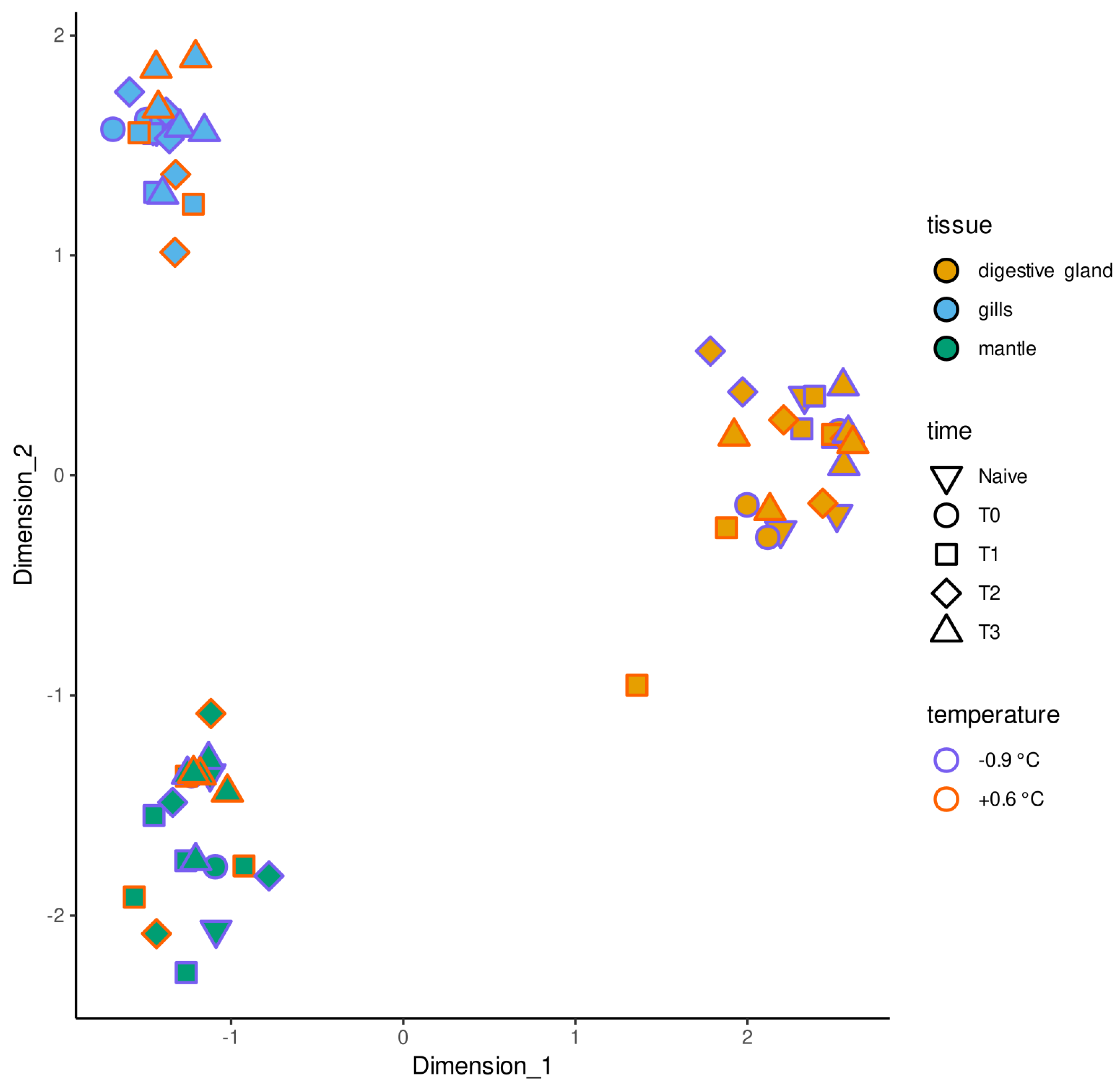
2. Results
2.1. Transcriptome Assembly and Refinement
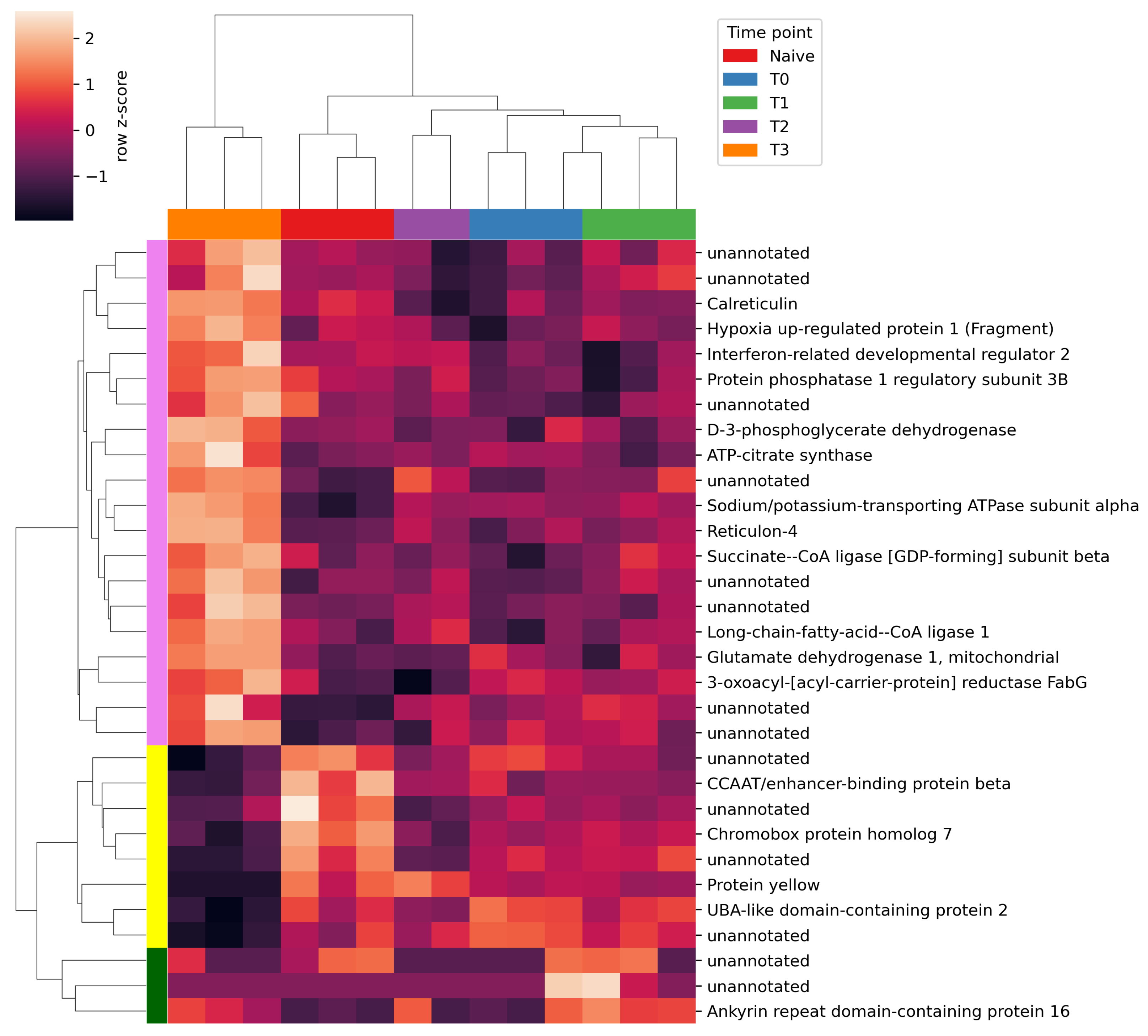
2.2. Differential Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Sampling and Experimental Setup
4.2. RNA Extraction and Library Preparation
4.3. Transcriptome Assembly and Refinement
4.4. Gene Expression Quantification and Pre-Processing
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stockton, W. The Biology and Ecology of the Epifaunal Scallop Adamussium colbecki on the West Side of McMurdo Sound, Antarctica. Mar. Biol. 1984, 78, 171–178. [Google Scholar] [CrossRef]
- Schiaparelli, S.; Linse, K. A Reassessment of the Distribution of the Common Antarctic Scallop Adamussium colbecki (Smith, 1902). Deep. Sea Res. Part II Top. Stud. Oceanogr. 2006, 53, 912–920. [Google Scholar] [CrossRef]
- Berkman, P.A. The Population Biology of the Antarctic Scallop, Adamussium colbecki (Smith 1902) at New Harbor, Ross Sea. In Antarctic Ecosystems; Kerry, K.R., Hempel, G., Eds.; Springer: Berlin/Heidelberg, Germany, 1990; pp. 281–288. [Google Scholar] [CrossRef]
- Chiantore, M.; Cattaneo-Vietti, R.; Povero, P.; Albertelli, G. The Population Structure and Ecology of the Antarctic Scallop Adamussium colbecki in Terra Nova Bay. In Ross Sea Ecology; Springer: Berlin/Heidelberg, Germany, 2000; pp. 563–573. [Google Scholar]
- Beu, A.; Taviani, M. Early Miocene Mollusca from McMurdo Sound, Antarctica (ANDRILL 2A Drill Core), with a Review of Antarctic Oligocene and Neogene Pectinidae (Bivalvia). Palaeontology 2014, 57, 299–342. [Google Scholar] [CrossRef]
- Quaglio, F.; Warren, L.V.; Anelli, L.E.; Dos Santos, P.R.; Rocha-Campos, A.C.; Gaździcki, A.; Strikis, P.C.; Ghilardi, R.P.; Tiossi, A.B.; Simões, M.G. Shell Beds from the Low Head Member (Polonez Cove Formation, Early Oligocene) at King George Island, West Antarctica: New Insights on Facies Analysis, Taphonomy and Environmental Significance. Antarct. Sci. 2014, 26, 400–412. [Google Scholar] [CrossRef]
- Egorova, E.N. Fossil Bivalves in Marine Sediments of Vestfold Hills in Eastern Antarctica. Biol. Bull. 2016, 43, 619–627. [Google Scholar] [CrossRef]
- Moro, G.; Buonocore, F.; Barucca, M.; Spazzali, F.; Canapa, A.; Pallavicini, A.; Scapigliati, G.; Gerdol, M. The First Transcriptomic Resource for the Antarctic Scallop Adamussium colbecki. Mar. Genom. 2019, 44, 61–64. [Google Scholar] [CrossRef]
- Denny, M.; Miller, L. Jet Propulsion in the Cold: Mechanics of Swimming in the Antarctic Scallop Adamussiumcolbecki. J. Exp. Biol. 2006, 209, 4503–4514. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.S.; Hauer, L.; Cziko, P.A.; Meister, K. Cryofouling Avoidance in the Antarctic Scallop Adamussium colbecki. Commun. Biol. 2022, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Heilmayer, O.; Brey, T. Saving by Freezing? Metabolic Rates of Adamussium colbecki in a Latitudinal Context. Mar. Biol. 2003, 143, 477–484. [Google Scholar] [CrossRef]
- Peck, L.S.; Powell, D.K.; Tyler, P.A. Very Slow Development in Two Antarctic Bivalve Molluscs, the Infaunal Clam Laternula Elliptica and the Scallop Adamussium colbecki. Mar. Biol. 2007, 150, 1191–1197. [Google Scholar] [CrossRef]
- Regoli, F.; Nigro, M.; Bertoli, E.; Principato, G.; Orlando, E. Defenses against Oxidative Stress in the Antarctic Scallop Adamussium colbecki and Effects of Acute Exposure to Metals. In Interactions and Adaptation Strategies of Marine Organisms; Springer: Berlin/Heidelberg, Germany, 1997; pp. 139–144. [Google Scholar]
- Storch, D.; Heilmayer, O.; Hardewig, I.; Portner, H.O. In Vitro Protein Synthesis Capacities in a Cold Stenothermal and a Temperate Eurythermal Pectinid. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 2003, 173, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Greco, S.; Gaetano, A.; Voltarel, G.; Pallavicini, A.; Giulianini, P.; Gerdol, M. Comparative Transcriptomic Analysis Reveals Adaptive Traits in Antarctic Scallop Adamussium colbecki. Fishes 2023. under review. [Google Scholar] [CrossRef]
- Peck, L.S.; Webb, K.E.; Bailey, D.M. Extreme Sensitivity of Biological Function to Temperature in Antarctic Marine Species. Funct. Ecol. 2004, 18, 625–630. [Google Scholar] [CrossRef]
- Bailey, D.M. The Thermal Dependence of Swimming and Muscle Physiology in Temperate and Antarctic Scallops. Ph.D. Thesis, University of St Andrews, St Andrews, UK, 2001. [Google Scholar]
- Bailey, D.M.; Peck, L.S.; Bock, C.; Pörtner, H.O. High-Energy Phosphate Metabolism during Exercise and Recovery in Temperate and Antarctic Scallops: An In Vivo 31P-NMR Study. Physiol. Biochem. Zool. 2003, 76, 622–633. [Google Scholar] [CrossRef]
- Quaglio, F.; Whittle, R.J.; Gaździcki, A.; Guimarães, S.M. A new fossil Adamussium (Bivalvia: Pectinidae) from Antarctica. Pol. Polar Res. 2010, 31, 291–302. [Google Scholar] [CrossRef]
- Benedetti, M.; Lanzoni, I.; Nardi, A.; d’Errico, G.; Di Carlo, M.; Fattorini, D.; Nigro, M.; Regoli, F. Oxidative Responsiveness to Multiple Stressors in the Key Antarctic Species, Adamussium colbecki: Interactions between Temperature, Acidification and Cadmium Exposure. Mar. Environ. Res. 2016, 121, 20–30. [Google Scholar] [CrossRef]
- Truebano, M.; Diz, A.P.; Thorne, M.A.; Clark, M.S.; Skibinski, D.O. Proteome Response to Heat Stress in the Antarctic Clam Laternula elliptica. J. Integr. OMICS 2013, 3, 34–43. [Google Scholar] [CrossRef]
- Bonacci, S.; Browne, M.A.; Dissanayake, A.; Hagger, J.A.; Corsi, I.; Focardi, S.; Galloway, T.S. Esterase Activities in the Bivalve Mollusc Adamussium colbecki as a Biomarker for Pollution Monitoring in the Antarctic Marine Environment. Mar. Pollut. Bull. 2004, 49, 445–455. [Google Scholar] [CrossRef]
- Regoli, F.; Nigro, M.; Chiantore, M.; Winston, G. Seasonal Variations of Susceptibility to Oxidative Stress in Adamussium colbecki, a Key Bioindicator Species for the Antarctic Marine Environment. Sci. Total. Environ. 2002, 289, 205–211. [Google Scholar] [CrossRef]
- Cronin, K.E.; Walker, S.E.; Mann, R.; Chute, A.S.; Long, M.C.; Bowser, S.S. Growth and Longevity of the Antarctic Scallop AdamussiumColbecki under Annual and Multiannual Sea Ice. Antarct. Sci. 2020, 32, 466–475. [Google Scholar] [CrossRef]
- Heilmayer, O.; Brey, T.; Chiantore, M.; Cattaneo-Vietti, R.; Arntz, W.E. Age and Productivity of the Antarctic Scallop, Adamussium colbecki, in Terra Nova Bay (Ross Sea, Antarctica). J. Exp. Mar. Biol. Ecol. 2003, 288, 239–256. [Google Scholar] [CrossRef]
- Masson-Delmotte, V.; Zhai, P.; Pörtner, H.O.; Roberts, D.; Skea, J.; Shukla, P.R.; Pirani, A.; Moufouma-Okia, W.; Péan, C.; Pidcock, R.; et al. Policymakers. In Global Warming of 1.5 C. An IPCC Special Report on the Impacts of Global Warming of 1.5 C above Pre-Industrial Levels and Related Global Greenhouse Gas Emission Pathways, in the Context of Strengthening the Global Response to the Threat of Climate Change, Sustainable Development, and Efforts to Eradicate Poverty; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2018; pp. 3–24. [Google Scholar] [CrossRef]
- Greco, S.; Gaetano, A.S.; Furlanis, G.; Capanni, F.; Manfrin, C.; Giulianini, P.G.; Santovito, G.; Edomi, P.; Pallavicini, A.; Gerdol, M. Gene Expression Profiling of Trematomus bernacchii in Response to Thermal and Stabling Stress. Fishes 2022, 7, 387. [Google Scholar] [CrossRef]
- Ansaloni, F.; Gerdol, M.; Torboli, V.; Fornaini, N.R.; Greco, S.; Giulianini, P.G.; Coscia, M.R.; Miccoli, A.; Santovito, G.; Buonocore, F.; et al. Cold Adaptation in Antarctic Notothenioids: Comparative Transcriptomics Reveals Novel Insights in the Peculiar Role of Gills and Highlights Signatures of Cobalamin Deficiency. Int. J. Mol. Sci. 2021, 22, 1812. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, K. Bottom macroalgae of the Admiralty Bay (King George Island, South Shetlands, Antarctica). Pol. Polar Res. 1990, 11, 95–131. [Google Scholar]
- Brand, A.R. Chapter 11—Scallop Ecology: Distributions and Behaviour. In Scallops; Shumway, S.E., Parsons, G.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; Volume 40, pp. 469–533. [Google Scholar] [CrossRef]
- Mehta, M.B.; Shewale, S.V.; Sequeira, R.N.; Millar, J.S.; Hand, N.J.; Rader, D.J. Hepatic Protein Phosphatase 1 Regulatory Subunit 3B (Ppp1r3b) Promotes Hepatic Glycogen Synthesis and Thereby Regulates Fasting Energy Homeostasis. J. Biol. Chem. 2017, 292, 10444–10454. [Google Scholar] [CrossRef] [PubMed]
- Axelsen, K.B.; Palmgren, M.G. Evolution of Substrate Specificities in the P-Type ATPase Superfamily. J. Mol. Evol. 1998, 46, 84–101. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Strittmatter, S. The Reticulons: A Family of Proteins with Diverse Functions. Genome Biol. 2007, 8, 234. [Google Scholar] [CrossRef]
- Vietor, I.; Huber, L.A. Role of TIS7 Family of Transcriptional Regulators in Differentiation and Regeneration. Differ. Res. Biol. Divers. 2007, 75, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Li, J.; Wang, J.; Li, J.; Ge, H.; Zhai, Q. Transcriptome Analysis of the Hepatopancreas in Exopalaemon carinicauda Infected with an AHPND-causing Strain of Vibrio parahaemolyticus. Fish Shellfish. Immunol. 2017, 67, 620–633. [Google Scholar] [CrossRef]
- Wang, W.; Xia, X.; Mao, L.; Wang, S. The CCAAT/Enhancer-Binding Protein Family: Its Roles in MDSC Expansion and Function. Front. Immunol. 2019, 10, 1804. [Google Scholar] [CrossRef]
- Maertens, G.; El, M.A.S.; Racek, T.; Stock, J.; Nicholls, J.; Rodriguez-Niedenführ, M.; Gil, J.; Peters, G. Several Distinct Polycomb Complexes Regulate and Co-Localize on the INK4a Tumor Suppressor Locus. PLoS ONE 2009, 4, e6380. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, X.; Lu, Z.; Zhang, B.; Guan, Z.; Liu, Z.; Zhong, Q.; Gu, L.; Zhou, J.; Zhu, B.; et al. Polycomb CBX7 Directly Controls Trimethylation of Histone H3 at Lysine 9 at the P16 Locus. PLoS ONE 2010, 5, e13732. [Google Scholar] [CrossRef]
- Mueller, T.; Feigon, J. Solution Structures of UBA Domains Reveal a Conserved Hydrophobic Surface for Protein-Protein Interactions. J. Mol. Biol. 2002, 319, 1243–1255. [Google Scholar] [CrossRef]
- Gerdol, M.; Moreira, R.; Cruz, F.; Gómez-Garrido, J.; Vlasova, A.; Rosani, U.; Venier, P.; Naranjo-Ortiz, M.; Murgarella, M.; Greco, S.; et al. Massive Gene Presence-Absence Variation Shapes an Open Pan-Genome in the Mediterranean Mussel. Genome Biol. 2020, 21, 275. [Google Scholar] [CrossRef]
- Ansell, A.D.; Ansell, A.D.; Cattaneo-Vietti, R.; Chiantore, M. Swimming in the Antarctic Scallop Adamussium colbecki: Analysis of in Situ Video Recordings. Antarct. Sci. 1998, 10, 369–375. [Google Scholar] [CrossRef]
- Chiantore, M.; Cattaneo-Vietti, R.; Heilmayer, O. Antarctic Scallop (Adamussium colbecki) Annual Growth Rate at Terra Nova Bay. Polar Biol. 2003, 26, 416–419. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- MacManes, M. The Oyster River Protocol: A Multi-Assembler and Kmer Approach for de Novo Transcriptome Assembly. PeerJ 2018, 6, e5428. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.; Seppey, M.; Simão, F.; Zdobnov, E. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Kriventseva, E.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.; Zdobnov, E. OrthoDB V10: Sampling the Diversity of Animal, Plant, Fungal, Protist, Bacterial and Viral Genomes for Evolutionary and Functional Annotations of Orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Ono, H.; Ishii, K.; Kozaki, T.; Ogiwara, I.; Kanekatsu, M.; Yamada, T. Removal of Redundant Contigs from de Novo RNA-Seq Assemblies via Homology Search Improves Accurate Detection of Differentially Expressed Genes. BMC Genom. 2015, 16, 1031. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Liu, J.; Wang, C.; Wang, H.; Zhang, L.; Hu, J.; Bao, L.; Wang, S. High-Quality Reannotation of the King Scallop Genome Reveals No ‘gene-Rich’ Feature and Evolution of Toxin Resistance. Comput. Struct. Biotechnol. J. 2021, 19, 4954–4960. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, J.; Jiao, W.; Li, J.; Xun, X.; Sun, Y.; Guo, X.; Huan, P.; Dong, B.; Zhang, L.; et al. Scallop Genome Provides Insights into Evolution of Bilaterian Karyotype and Development. Nat. Ecol. Evol. 2017, 1, 0120. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Li, W.; Jaroszewski, L.; Godzik, A. Clustering of Highly Homologous Sequences to Reduce the Size of Large Protein Databases. Bioinformatics 2001, 17, 282–283. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Greco, S. annot.aM. Available online: https://gitlab.com/54mu/annotaM (accessed on 20 July 2022).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the edgeR Quasi-Likelihood Pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential Expression Analysis of Multifactor RNA-Seq Experiments with Respect to Biological Variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq Data Using Factor Analysis of Control Genes or Samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [PubMed]

| Parameter | Value |
|---|---|
| Total contigs | 44,475 |
| GC content | 40.76 |
| Contig N50 | 1911 |
| Median contig length | 886 |
| Mean contig length | 1329.11 |
| Total assembled bases | 591,112,155 |
| BUSCO single (%) | 76 |
| BUSCO duplicated (%) | 13.9 |
| BUSCO fragmented (%) | 4.4 |
| BUSCO missing (%) | 5.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greco, S.; Gaetano, A.S.; Manfrin, C.; Capanni, F.; Santovito, G.; Pallavicini, A.; Giulianini, P.G.; Gerdol, M. The Antarctic Scallop Adamussium colbecki Is Unable to Transcriptomically Respond to Captivity and Moderate Thermal Stress. Stresses 2023, 3, 475-487. https://doi.org/10.3390/stresses3020034
Greco S, Gaetano AS, Manfrin C, Capanni F, Santovito G, Pallavicini A, Giulianini PG, Gerdol M. The Antarctic Scallop Adamussium colbecki Is Unable to Transcriptomically Respond to Captivity and Moderate Thermal Stress. Stresses. 2023; 3(2):475-487. https://doi.org/10.3390/stresses3020034
Chicago/Turabian StyleGreco, Samuele, Anastasia Serena Gaetano, Chiara Manfrin, Francesca Capanni, Gianfranco Santovito, Alberto Pallavicini, Piero Giulio Giulianini, and Marco Gerdol. 2023. "The Antarctic Scallop Adamussium colbecki Is Unable to Transcriptomically Respond to Captivity and Moderate Thermal Stress" Stresses 3, no. 2: 475-487. https://doi.org/10.3390/stresses3020034
APA StyleGreco, S., Gaetano, A. S., Manfrin, C., Capanni, F., Santovito, G., Pallavicini, A., Giulianini, P. G., & Gerdol, M. (2023). The Antarctic Scallop Adamussium colbecki Is Unable to Transcriptomically Respond to Captivity and Moderate Thermal Stress. Stresses, 3(2), 475-487. https://doi.org/10.3390/stresses3020034













