Abstract
Abscisic acid (ABA) is recognized as the key hormonal regulator of plant stress physiology. This phytohormone is also involved in plant growth and development under normal conditions. Over the last 50 years the components of ABA machinery have been well characterized, from synthesis to molecular perception and signaling; knowledge about the fine regulation of these ABA machinery components is starting to increase. In this article, we review a particular regulation of the ABA machinery that comes from the plant circadian system and extends to multiple levels. The circadian clock is a self-sustained molecular oscillator that perceives external changes and prepares plants to respond to them in advance. The circadian system constitutes the most important predictive homeostasis mechanism in living beings. Moreover, the circadian clock has several output pathways that control molecular, cellular and physiological downstream processes, such as hormonal response and transcriptional activity. One of these outputs involves the ABA machinery. The circadian oscillator components regulate expression and post-translational modification of ABA machinery elements, from synthesis to perception and signaling response. The circadian clock establishes a gating in the ABA response during the day, which fine tunes stomatal closure and plant growth response.
1. Introduction
Abscisic acid (ABA) is a phytohormone involved in the plant stress response as well as plant growth and development. ABA plays a role in different physiological processes, i.e., seed germination and early seedling growth, shoot and root growth and development, stomata closure, senescence, fruit ripening, fruit and leaf abscission, and bud dormancy, and is also a key player in the induction of tolerance to both abiotic and biotic stress [1]. ABA was initially described as a growth-inhibitor [2,3], and later as a regulator of cotton fruit abscission [4]. Nevertheless, in the last decade the most studied role of ABA has been the induction of drought resistance, among other biotic and abiotic stresses, in model plants and crops [5,6].
After half a century studying the field of ABA, we know the components and dynamics of its machinery in considerable detail, from synthesis to molecular perception and signaling [5], and the details of regulation of the ABA machinery are beginning to emerge [5,7,8]. In this article, we consider a particular regulation of the ABA machinery that comes from the plant circadian system and extends to multiple levels. The circadian clock is a self-sustained molecular oscillator present in each plant cell [9,10,11,12], although hierarchical in nature at tissue level [13]. The circadian system has both input and output pathways. The inputs allow clock synchronization to the environment or external cues, e.g., photoperiod and temperature, among others [14]. For instance, circadian oscillations are efficiently entrained by daily light/dark changes. This environmental photic diurnal cycle acts as a ‘zeitgeber’, or time giver, and induces the clock’s diurnal synchrony to 24 h [14]. On the other hand, output pathways control molecular, cellular and physiological downstream processes, such as hormonal responses, transcriptional activity, the cell cycle and growth, among others [14,15,16,17].
2. Plant Circadian System
For plants, fluctuating external stimuli challenge their survival. The prediction and anticipatory response to these threatening stimuli represent a clear adaptive advantage [18]. The circadian system supports the predictive homeostasis mechanism in almost all living organisms [19,20,21,22]. Predictive homeostasis is an anticipatory response to an expected environmental challenge in the future, such as the diurnal light-dark cycles or temperature changes [18]. The plant circadian clock allows prediction of the appearance of an environmental stimulus, and anticipation of a proper response to any disturbance that threatens a plant’s survival. Furthermore, the circadian clock contributes to oscillatory resonance with the environment and fitness for optimum plant growth [23,24].
The molecular clock mechanism (Figure 1) for self-sustained oscillation relies on regulatory loops of transcription factors, through both protein to protein and protein to promoter interactions [14]. The pseudo response regulator1/timing of cab expression1 (PRR1/TOC1) is one of the key components of the Arabidopsis circadian system [25,26]. TOC1 constitutive expression slows down the pace of the clock under diurnal conditions and leads to complete rhythm abolishment under constant light conditions [23,26]. However, the clock speeds up in toc1 reduce-of-function plant lines [23,25,27]. The morning-expressed clock genes, circadian clock-associated 1 (CCA1) and late elongated hypocotyl (LHY), participate in feedback transcriptional repression between the afternoon expressed pseudo-response regulator (PRRs: PRR5, PRR7 and PRR9) genes, TOC1, and members of the Evening Complex (EC: early flowering 3/4 “ELF3”, “ELF4”, and lux arrhythmo “LUX”). Moreover, a secondary transcriptional loop involves the mid-day expressed reveille (RVEs: RVE4, RVE6, and RVE8) genes, the PRRs, and the EC, and contributes to clock oscillatory robustness [14]. Other components, such as gigantea (GI), zeitlupe (ZTL), and night light-inducible and clock-regulated gene 1 (LNKs) fine tune oscillation properties. Furthermore, core circadian components are involved in the regulation of different outputs or key cellular events of the plant cell. Transcriptome profiling experiments in A. thaliana show that nearly a third of expressed genes are under circadian clock regulation [28,29,30]. In fact, there is a close relationship between the circadian system and hormonal circuits [17]. We focus this review on the regulation of ABA machinery by the plant circadian clock.
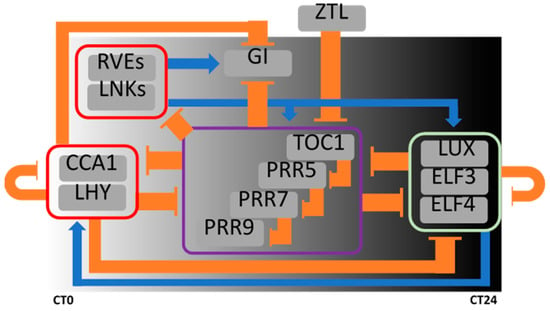
Figure 1.
Plant circadian clock. Core components of the circadian oscillator in A. thaliana and its regulatory loops network. The position in the chart of each component is relative to the time of maximal expression/activity along the day. CT: circadian time (CT0 is defined as the light on-set). Blue lines indicate activation and orange lines indicate repression. Purple box: PRRs components (PRR9, PRR7, PRR5 and TOC1). Green box: evening complex “EC” members (LUX, ELF3 and ELF4). Red box morning elements: CCA1/LHY and RVEs/LNKs. This is a simplified scheme and it does not include all of the oscillator components. LHY: LATE ELONGATED HYPOCOTYL; CCA1: CIRCADIAN CLOCK ASSOCIATED1; RVE: REVEILLE; LNK: NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED; PPR: PSEUDO RESPONSE REGULATOR; TOC1: TIMING OF CAB EXPRESSION1; ZTL: ZEITLUPE; GI: GIGANTEA; ELF3/4: EARLY FLOWERING 3/4; LUX: LUX ARRHYTHMO.
3. Abscisic Acid (ABA) Machinery and Its Regulation by the Circadian Clock
3.1. ABA Synthesis
Plants under normal or non-stress conditions synthesize basal levels of ABA, which promote optimal growth. Upon abiotic stress arrival (i.e., drought or salinity), ABA level increases notably, and activates different stress responses. The sesquiterpenoid ABA contains 15 carbon atoms in its skeleton and its synthesis in plants (Figure 2) starts from the precursor C40 β-carotene, following the carotenoid pathway [31,32]. The first steps, from C40 β-carotene to C15 xanthoxin are carried out in plastids by a battery of enzymes, well described in A. thaliana: viviparous (VPs), zeaxanthin epoxidase (ZEP/ABA1), abscisic aciid (ABA)-deficient 4 (ABA4) and nine-cis-epoxycarotenoid dioxygenase (NCEDs). Later, in the cytosol, ABA2 and ABA3 turn C15 xanthoxin into C15 abscisic acid (Figure 2). The rate-limiting step for ABA synthesis is the cleavage of 9-cis-epoxycarotenoid into xanthoxin by the NCEDs enzymes [33]. Moreover, transgenic plants with constitutive expression of NCEDs have high levels of ABA [34].
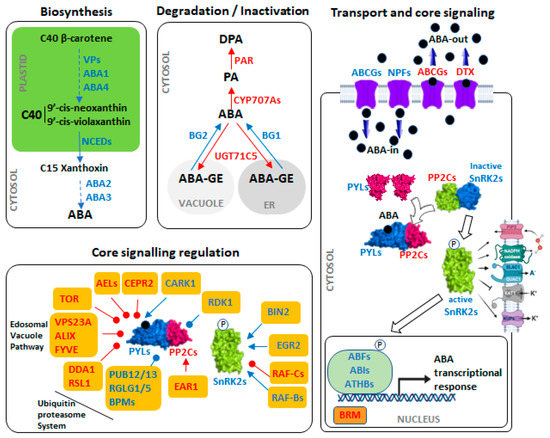
Figure 2.
Abscisic acid (ABA) machinery. Biosynthesis: ABA is synthesized from β-carotene (C40) through the carotenoid pathway. Steps from β-carotene (C40) to xanthoxin (C15) take place in plastids. In the cytoplasm, ABA2 and ABA3 convert xanthoxin into active ABA. Conversion of 9′-cis-neoxanthin and 9′-cis-violaxanthin to xanthoxin by NCEDs is a rate-limiting step in ABA biosynthesis. Degradation/Inactivation: active ABA level is regulated by catabolism through both ABA conjugation and catalytic hydroxylation. ABA can be glycosylated into inactive ABA-GE by UGT71C5. On the other hand, AtBG1 and AtBG2 can modify ABA-GE to produce active ABA. Further, ABA can be catalyzed to phaseic acid (PA) by CYP707As, which in turn is catalyzed to dihydrophaseic acid (DPA) by PA reductase (PAR). Transport and Core Signaling: ABA transport is carried out by ABCGs, NPFs and DTX transporters. Inside the cell, ABA is perceived by PYR/PYL/RCAR receptors, either at cytosol or nucleus, forming a stable ternary complex with clade A PP2Cs. The formation of this complex allows the activation of SnRK2s that leads to ABA-induced regulation of plasma membrane and nuclear targets (i.e., NADPH oxidase, KAT1, SLAC1, and ABFs/AREBs). Core Signaling Regulation: PYLs receptors, PP2Cs and SnRK2s are regulated by several protein kinases. The degradation of ABA signaling core components is regulated both by the ubiquitin-proteasome system, and through the endosomal-vacuole pathways. Components and arrows in blue mean positive function on the final ABA response, while red components and red arrows mean a negative effect. Pointed arrow: activation; rounded arrow: inactivation. (For more information about these components see Table S1).
Analyzing data from DIURNAL (http://diurnal.mocklerlab.org/, accessed on 21 December 2021; DIURNAL is a web-based tool for accessing the diurnal and circadian genome-wide expression results of genes from several array experiments conducted on common model plants), we can see that NCED2-6 have a rhythmic diurnal oscillation (under light/dark cycles of 12:12 h; Table S1), with a maximum peak or “acrophase” of expression around mid-day (ZT4-7; ZT: zeitgeber time; ZT0 is defined as the light on-set; Table S1). Moreover, NCED4 maintains its oscillation under “free-running” conditions (constant light) peaking at CT6 (CT: circadian time; CT0 is defined as the light on-set; Figure 3B). These observations indicate a possible regulation of ABA synthesis by the circadian clock. In fact, CHIPseq data obtained for PPR5, CCA1, LUX and ELF3 showed a direct regulation of NCEDs promoters by these circadian clock components (Figure 3A; Table S1) [35,36,37]. In agreement with NCEDs oscillatory patterns, a diurnal rhythm in ABA levels was reported with a maximum during mid-day (between ZT6-ZT12) [38]. Moreover, it was shown that the biosynthetic pathway of carotenoid and ABA is under the control of PRR9/7/5 [39]. In this study, the authors showed elevated ABA levels in prr9-7-5 triple mutant lines with respect to wt, suggesting that PRRs may negatively regulate ABA accumulation. Nevertheless, rhythmic oscillations in ABA levels seem also to be dependent on photoperiod and watering status, given that well-watered plants growing under long day conditions (light/dark cycles of 16:8 h) did not shown a clear oscillation [40]. Furthermore, diurnal changes in ABA level also appear to be species specific given that in A. unedo ABA oscillations with two maximum peaks were reported, one of them in the early morning, and the other at midday [41]; in contrast, in two different conifers, S. minimus and C. rhomboidea, ABA rhythms peak in early and mid-morning respectively [42]. Conversely, in N. tobacum three different peaks in ABA levels were observed throughout the 24 h day, two minor peaks at ZT1 and ZT10, and a higher peak at ZT18 or early night [43].
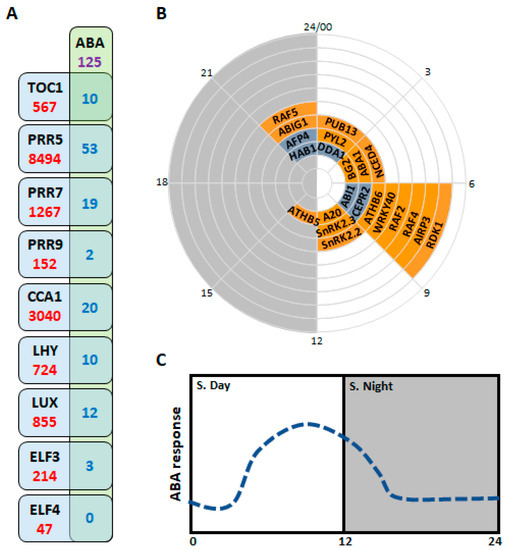
Figure 3.
Connections between Circadian Clock and ABA machinery. (A) Circadian clock components interact with the promoter of different ABA machinery genes. Data extracted from RNAseq experiments found in literature from A. thaliana. In purple: ABA machinery components analyzed in this review (for more information see Table S1); in blue: number of ABA machinery genes regulated by each circadian transcription factor; in red: number of genes potentially regulated by each transcription factor. (B) Phase plot or wind-rose of gene expression time of ABA machinery components. Components with circadian oscillation only plotted. The name of each component appears in the graph at the time of acrophase (maximum of gene expression). Expression data was extracted from DIURNAL web (http://diurnal.mocklerlab.org/, accessed on 21 December 2021) and obtained from plants (A. thaliana) under constant light conditions (free running). White background, from 0 to 12 h, represents the circadian subjective day, and grey background, from 12 to 24 h, the circadian subjective night. (C) Putative circadian regulated ABA response gating in plants. S. Day: circadian subjective day; S. night: circadian subjective night. 0, 12 and 24: circadian time.
On the other hand, it was shown that NCED3 expression was repressed in transgenic lines with constitutive expression of LHY, suggesting that LHY may also negatively regulate ABA accumulation [40]. Indeed, this LHY-ox line showed low ABA levels with respect to wt. By contrast, the loss-of-function lhy-11 line showed higher ABA levels in comparison to wt lines. Moreover, in wt plants, ABA accumulation was rhythmic under drought conditions, peaking in the evening, at ZT18 [40]. The phase of this rhythm was advanced in lhy-11 mutant lines. However, ABA levels were reduced and arrhythmic in LHY-ox plants under drought stress. These results are in agreement with the idea that the circadian clock regulates ABA accumulation under drought conditions. The model proposed implies direct inhibition of NCED gene expression by LHY, which in turn reduces ABA accumulation in the early morning [40].
Another circadian clock component related to the control of ABA synthesis through NCED3 regulation is gigantea (GI) [44]. Specifically, it was shown that gi-enhanced em level complex (GI-EEL) enhances drought tolerance via up-regulation of ABA levels. NCED3 expression was reduced in the eel, gi-1, and eel/gi-1 mutants under normal growth conditions. Furthermore, these mutants were also hypersensitive to drought stress due to stomatal closure inhibition and the concomitant uncontrolled water loss [44]. Moreover, it was demonstrated that GI-EEl complex binds directly to the ABA-responsive element motif in the NCED3 promoter. These results suggest that the EEL-GI complex positively regulates diurnal ABA levels, contributing to the drought tolerance of Arabidopsis [44]. Indeed, NCED3 expression resembles the GI oscillatory patterns of protein amounts [45], suggesting that GI activity regulates the circadian oscillation in NCED3 expression and ABA oscillations. Is interesting to note that, despite the fact that GI has a positive role in the tolerance response of different abiotic stress, i.e., cold, salt and drought stress [46,47,48,49,50], it also has a negative effect on water use efficiency (WUE) [51]. This apparent contradiction is in agreement with the idea that WUE, under normal conditions, follows different mechanisms than drought stress and does not always correlate reliably with drought resistance [52].
3.2. ABA Inactivation and Conjugation
Net ABA levels are not only set by synthesis, but also by degradation and inactivation. For instance, after stress conditions and the concomitant hormonal induction, ABA levels need to diminish to normal or non-stress values to resume optimal growth and development. In this process ABA catabolism can follow two different pathways: reversible conjugation or irreversible hydroxylation (Figure 2) [31]. In turn, in A. thaliana, ABA can be glycosylated by UGT71C5 to form ABA-glucose ester (ABA-GE), which is stored in vacuoles or the endoplasmic reticulum. ABA-GE is inactive but can be deconjugated to restore the active ABA by β-glycosidases, such as BG1 and BG2 [38,53,54]. The conjugation/deconjugation mechanism, played by UGT71C5 and BGs, allows plants to rapidly adapt to their changing environment through ABA-mediated responses. On the other hand, ABA can be irreversible converted to a partially active form as phaseic acid (PA) by CYP707As [55] and later to a fully inactive form as dihydrophaseic acid (DPA) by ABH2 [56].
Analyzing expression data for ABA inactivation and conjugation/deconjugation components (from DIURNAL data base), we can see that BG2 shows circadian oscillations, with high levels during the mid-subjective day (acrophase at CT6 under constant light conditions; Table S1). Moreover, the BG1 expression pattern shows diurnal rhythms (acrophase at ZT6 under light/dark 12:12 conditions). These oscillatory expression patterns in BGs are in agreement with the diurnal rhythm in ABA levels with a peak between ZT6/ZT12 [36]. Interestingly, in bg1 mutant lines this oscillation in ABA levels disappears [36]. By contrast, the CYP707A family is also diurnally regulated under light/dark cycles (12:12 h DIURNAL web tool) with maximum expression levels during the day between ZT3 to ZT7 (Table S1). Nevertheless, it seems that the diurnal regulation of ABA levels is more associated with conjugation/deconjugation than irreversible degradation, given that the diurnal fluctuations in biologically active ABA levels correlate with BG1/BG2 expression levels [36].
3.3. ABA Transport
ABA synthesis is carried out mainly in vascular parenchyma cells [57]. By contrast ABA effects are systemic. Given the anionic nature of ABA under alkaline environments, this limits its passive diffusion through the plasma membrane, so plants rely on ABA transporters to help in its uptake and efflux from apoplast into the cell and vice versa [58]. In A. thaliana, the transporters involved in ABA translocation are: “(i) detoxification efflux carrier 50 (DTX50) [59]; (ii) nitrate or di/tri-peptide transporter 4.6 (NPF4.6) [60,61]; and (iii) ABC ATP-binding cassette (ABCGs) transporters [62]. In particular, DTX50 and ABCG25/31 are involved in ABA efflux, and NPF4.5 and ABCG20/30/40 in ABA influx (Figure 2).
CHIPseq data from the circadian clock component shows that CC1 and PRR5 are able to interact with the promoter of the ABA efflux transporter ABCG25 and with the ABA influx transporter NPF4.6 [35,36]. Nonetheless, the expression levels of these transporters do not show a circadian expression pattern (DIURNAL web tool data). Possibly, other signals could be contributing to the final modulation of ABA transporter expression. However, the ABA influx transporter ABCG20 has a circadian oscillatory pattern in expression, with acrophase at CT10 or late subjective day (DIURNAL data base; Table S1).
3.4. ABA Core Signaling
ABA is perceived inside the cell through soluble receptors (Figure 2) called pyrabactin resistance1 (PYR1)/PYR1-like (PYL)/regulatory components of ABA receptors (RCAR) [63,64,65]. This ABA perception leads to interaction-inactivation of clade A protein phosphatase type 2Cs (PP2Cs). The formation of the ternary complex ABA-PYL-PP2CA relieves the inhibition on three ABA-activated subclass III SNF1-related protein kinases 2 (SnRK2s), in A. thaliana SnRK2.2/SnRK2D, 2.3/I, and 2.6/E/OST1 [66,67]. Then, activated SnRK2s induce the activation of a battery of ABA effectors from transmembrane channels (SLAC, KUP and KAT, among others) to transcription factors (i.e., ABFs, ABIs and ATHBs). Estimations in A. thaliana indicate that ABA controls nearly two thirds of water stress-responsive genes [68,69].
Published CHIPseq data sets from several circadian clock components i.e., TOC1, PRR5, PRR7, PRR9, CCA1, LHY, LUX and ELF3 [35,37,40,54,70,71] show extensive control over the ABA core signaling pathway, from ABA receptors to ABA-related transcription factors (Table S1). Moreover, the ABA receptor PYL2, the phosphatases ABI1 and HAB1, the kinases SnRK2.2 and 2.3, and the transcription factors ATHB5, ATHB6, ABIG1 and WRKY40 show a circadian expression pattern (DIURNAL web data; Figure 3B; Table S1). Furthermore, the circadian regulation of the ABA core signaling pathway also operates at post-translational levels since protein-protein interactions of TOC1-ABI3 and PRR5-ABI5 were reported [72,73]. Interestingly, phosphorylation and activation of Snrk2.2, SnRK2.3 and ABF2 also show a circadian oscillatory pattern, with maximum levels at the middle and end of the subjective day [74]. This data is in agreement with the fact that in soybean GmLCLs/RVEs negatively regulate ABA perception and signaling [75]. In A. thaliana, RVE expression decays from the middle to the end of the subjective day.
Another layer of regulation of ABA signaling is composed of ABI five binding proteins (AFPs). These proteins play a negative role in ABA signaling by interacting with and inhibiting ABI5 activity [76,77,78]. Moreover, transcription of AFPs is activated by ABA, indicating a negative feedback function of these proteins. In particular, AFP4 shows a circadian rhythm in its expression pattern (DIURNAL data; Table S1), with a maximum level at the end of the subjective night (CT23). This negative feedback operates in an antiphase respect to the activity of positive components of ABA signaling, such as SnRK2.2/2.3 or ABF2. Furthermore, AFP4 repression could be involved with TOC1, PPR5, PPR7 and LUX, as CHIPseq data showed a possible connection [35,37,70,71].
The Arabidopsis SWI/SNF chromatin-remodeling ATPase BRAHMA (BRM) modulates ABA response by preventing activation of stress response pathways during germination [79]. Moreover, core ABA signaling pathway components interact with BRM and switch its activity. For instance, SnRK2-dependent phosphorylation of BRM leads to its inhibition, and PP2CA-mediated dephosphorylation of BRM restores its activity. BRM has a negative role repressing ABA response. Interestingly, the circadian clock component LUX is able to interact with BRM [80]. However, it is necessary to characterize the function of this LUX-BRM complex, to further elucidate the real impact on the ABA signaling response.
3.5. ABA Core Signallling Regulation
Post-translational modifications play a key role in regulating protein function and stability. In particular, protein phosphorylation is a fundamental mechanism through which protein function is regulated in response to extracellular stimuli [81]. ABA core signaling components are phosphorylated by multiple protein kinases (Figure 2), including in A. thaliana: target of rapamycin (TOR), Arabidopsis early flowering 1 (EL1)-like casein kinase (AEL), C-terminally encoded peptide receptor 2 (CEPR2), cytosolic ABA receptor kinase 1 (CARK1), receptor dead kinase 1 (RDK1), enhancer of ABA coreceptor (EAR1), brassino-steroid insensitive 2 (BIN2), and mitogen activated kinase kinase kinase RAFs [7,82,83,84,85,86,87,88,89,90]. Some of these kinases have a positive effect on ABA response, i.e., BIN2, RAF-Bs, RDKs and CARK1. Moreover, DIURNAL expression data shows circadian oscillation in RAF2, RAF4 and RAF5 and RDK1, with acrophases from early to mid-subjective day (Figure 3B; Table S1). In addition, PRR5 CHIPseq data [35] show a possible regulation of RAF2 and RDK1, among other kinases (Table S1). Otherwise, kinases, such as TOR, CEPR2, AEL, RAF-Cs and EAR1 have a negative effect on ABA signaling. In particular, CEPR2 shows a circadian oscillatory pattern in gene expression, with a peak at CT9 (Table S1). Moreover, CEPR2 promoter is possibly regulated by several circadian clock components (i.e., PRR5, CCA1, LHY and LUX), as can be inferred from CHIPseq data [35,36,37,40].
On the other hand, protein ubiquitination is a reversible modification involved in a myriad of processes in plant biology. ABA signaling relies on a fine-tuned protein turnover [8]; specifically, the half-life of its components is regulated both through the ubiquitin-26S proteasome system and the endocytic/vacuolar degradation pathway (Figure 2). E3 Ub ligases have been reported that target different ABA signaling core components, i.e., ABA receptors, PP2Cs, SnRK2s, and ABFs/ABI5 transcription factors (Figure 2). The degradation of PYLs is regulated by both the ubiquitin ligase substrate adaptor DDA1 and RING-type E3 ligase RSL1 via the ubiquitin-proteasome system, and by the ESCRT-I components VPS23A and ALIX through the endosomal-vacuole pathways [91,92,93,94]. PP2Cs are also degraded by the 26S proteasome pathway through the PUB12/13 U-box, RGLG1/5 RING-type and (CUL3)-RING based E3 ligases by interacting with the adaptor BTB/POZ and math domain proteins (BPMs) [95,96,97,98]. Circadian clock regulation of these protein degradation systems is less apparent. From CHIPseq data, we only can attribute some regulatory function to PRR5 and PRR7 (Table S1) [35,71]. Moreover, only PUB13, AIRP3 and DDA1 show a circadian expression pattern (Figure 3B; Table S1).
4. Conclusions and Remarks
ABA plays an important role in the response to environmental challenges, including water deficit, salinity, freezing, wounding, and pathogen attack. Moreover, it participates in several developmental processes, i.e., seed germination, early seedling development, and reproduction [1,68,99,100,101,102]. Elucidating the regulation mechanisms on the ABA machinery is vital to further understand its dynamics. On the other hand, the circadian clock is a self-sustained molecular oscillator, with several output pathways that control processes, such as hormonal response and transcriptional activity. One of these outputs involves ABA machinery regulation. The circadian clock anticipates external changes and prepares plants to respond to them in advance, contributing to the plant predictive homeostasis. Moreover, the circadian system also couples plant physiology to external environmental cycles to maximize metabolic efficiency. This constitutes an efficient way to increase stress tolerance without substantially decreasing plant growth [103].
The linkage of circadian clock and ABA machinery possibly establishes a circadian gating of ABA responses. Apparently, this gating maximizes ABA response during the mid-late day and minimizes it during late night and early morning (Figure 3C). This gating in ABA response is directly related to stomatal closure and plant growth. The regulation of stomatal closure by ABA is well established [104]. Moreover, in C3 and C4 plants stomata are open during the day. It is useful to have a system ready to operate during light hours, when it is more required, given that plants at this time of day are more susceptible to water loss. Interestingly, plants also adjust growth to night-light cycles. For instance, root growth occurs mostly at night [105,106], and hypocotyl growth rate is maximal at the end of the night [103,107,108]. Given that ABA plays a negative role in plant growth [109], it is consistent that during the night hours ABA response is low. An interesting way to understand circadian clock regulated ABA gating could involve the use of reporter lines with GFP or luciferase driven by promoters carrying ABA response cis-acting elements. This kind of approach might specifically answer the time and degree of ABA response during the day.
It is worth noting that light-dark cycles must play an important role in regulating ABA machinery and the gating in ABA response, bypassing the circadian system. A high number of ABA machinery components (75 out of 124; 60%) show oscillations in gene expression under diurnal cycles (12:12 h of light:dark; Table S1), but do not under circadian conditions (i.e., constant light), when the clock is in free running mode, without external time cues.
Interactions between ABA signaling and the circadian clock are not unidirectional. There is evidence for the reciprocal impact of ABA on the amplitude, period and phase of the circadian clock. Application of ABA lengthened the period of CCA1 and of the marker gene for clock output chlorophyll A/B-binding protein2 (CAB2) by ~2 h in free-running conditions (constant light) compared with mock treatments [110]. Furthermore, ABA also induces the expression of TOC1 [111] and represses glycine rich protein 7 (AtGRP7) expression [112]. Moreover, CCA1 expression is induced by dry after-ripening; after-ripening modifies the transcriptional amplitude of clock gene oscillations [47]. In addition, there is evidence demonstrating that ABA regulates the GI pathway and activates florigen genes under drought conditions [113]. Furthermore, it was reported that an ABA-inducible R2R3-type MYB transcription factor (MYB96) binds directly to the TOC1 promoter to activate its expression. In turn, TOC1 indirectly regulates MYB96 expression, through CCA1 activity [114]. This feedback network directly links ABA responses with the circadian clock and provides a mechanism whereby the ABA gating is controlled to optimize plant fitness.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/stresses2010006/s1, Table S1: ABA machinery components analyzed in this review.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemberg, T. Growth-Inhibiting Substances in Terminal Buds of Fraxinus. Physiol. Plant. 1949, 2, 37–44. [Google Scholar] [CrossRef]
- Hemberg, T. Significance of Growth-Inhibiting Substances and Auxins for the Rest-Period of the Potato Tuber. Physiol. Plant. 1949, 2, 24–36. [Google Scholar] [CrossRef]
- Ohkuma, K.; Lyon, J.L.; Addicott, F.T.; Smith, O.E. Abscisin II, an Abscission-Accelerating Substance from Young Cotton Fruit. Science 1963, 142, 1592–1593. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Partida, R.; Rosario, S.; Lozano-Juste, J. An Update on Crop ABA Receptors. Plants 2021, 10, 1087. [Google Scholar] [CrossRef]
- Lozano-Juste, J.; Alrefaei, A.F.; Rodriguez, P.L. Plant Osmotic Stress Signaling: MAPKKKs Meet SnRK2s. Trends Plant Sci. 2020, 25, 1179–1182. [Google Scholar] [CrossRef] [PubMed]
- Coego, A.; Julian, J.; Lozano-Juste, J.; Pizzio, G.; Alrefaei, A.; Rodriguez, P. Ubiquitylation of ABA Receptors and Protein Phosphatase 2C Coreceptors to Modulate ABA Signaling and Stress Response. Int. J. Mol. Sci. 2021, 22, 7103. [Google Scholar] [CrossRef]
- Yakir, E.; Hassidim, M.; Melamed-Book, N.; Hilman, D.; Kron, I.; Green, R.M. Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant J. 2011, 68, 520–531. [Google Scholar] [CrossRef]
- Wenden, B.; Toner, D.L.K.; Hodge, S.K.; Grima, R.; Millar, A.J. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proc. Natl. Acad. Sci. USA 2012, 109, 6757–6762. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Shimizu, H.; Nohales, M.A.; Araki, T.; Kay, S.A. Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 2014, 515, 419–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordage, S.; Sullivan, S.; Laird, J.; Millar, A.J.; Nimmo, H.G. Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol. 2016, 212, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Hirata, Y.; Aihara, K.; Mas, P. A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell 2015, 163, 148–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nohales, M.A.; Kay, S.A. Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 2016, 23, 1061–1069. [Google Scholar] [CrossRef]
- Fung-Uceda, J.; Lee, K.; Seo, P.J.; Polyn, S.; De Veylder, L.; Mas, P. The Circadian Clock Sets the Time of DNA Replication Licensing to Regulate Growth in Arabidopsis. Dev. Cell 2018, 45, 101–113.e4. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Gil, S.; Grasser, K.D.; Mas, P. Targeted Recruitment of the Basal Transcriptional Machinery by LNK Clock Components Controls the Circadian Rhythms of Nascent RNAs in Arabidopsis. Plant Cell 2018, 30, 907–924. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Mas, P. A Functional Connection between the Circadian Clock and Hormonal Timing in Arabidopsis. Genes 2018, 9, 567. [Google Scholar] [CrossRef] [Green Version]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef]
- Hurley, J.M.; Dasgupta, A.; Emerson, J.M.; Zhou, X.; Ringelberg, C.S.; Knabe, N.; Lipzen, A.M.; Lindquist, E.A.; Daum, C.G.; Barry, K.W.; et al. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proc. Natl. Acad. Sci. USA 2014, 111, 16995–17002. [Google Scholar] [CrossRef] [Green Version]
- McClung, C.R. The Plant Circadian Oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Welkie, D.; Rubin, B.E.; Diamond, S.; Hood, R.D.; Savage, D.F.; Golden, S.S. A Hard Day’s Night: Cyanobacteria in Diel Cycles. Trends Microbiol. 2019, 27, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Koronowski, K.B.; Sassone-Corsi, P. Communicating clocks shape circadian homeostasis. Science 2021, 371, eabd0951. [Google Scholar] [CrossRef] [PubMed]
- Mas, P.; Alabadí, D.; Yanovsky, M.J.; Oyama, T.; Kay, S.A. Dual Role of TOC1 in the Control of Circadian and Photomorphogenic Responses in Arabidopsis. Plant Cell 2003, 15, 223–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashino, T.; Ito, S.; Niwa, Y.; Kunihiro, A.; Nakamichi, N.; Mizuno, T. Involvement of Arabidopsis Clock-Associated Pseudo-Response Regulators in Diurnal Oscillations of Gene Expression in the Presence of Environmental Time Cues. Plant Cell Physiol. 2008, 49, 1839–1850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strayer, C.; Oyama, T.; Schultz, T.F.; Raman, R.; Somers, D.E.; Más, P.; Panda, S.; Kreps, J.A.; Kay, S.A. Cloning of the Arabidopsis Clock Gene TOC1, an Autoregulatory Response Regulator Homolog. Science 2000, 289, 768–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, S.; Matsushika, A.; Kojima, M.; Yamashino, T.; Mizuno, T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002, 43, 58–69. [Google Scholar] [CrossRef] [Green Version]
- Somers, D.E.; Webb, A.A.; Pearson, M.; Kay, S.A. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 1998, 125, 485–494. [Google Scholar] [CrossRef]
- Covington, M.F.; Harmer, S.L. The Circadian Clock Regulates Auxin Signaling and Responses in Arabidopsis. PLoS Biol. 2007, 5, e222. [Google Scholar] [CrossRef] [Green Version]
- Michael, T.P.; Breton, G.; Hazen, S.P.; Priest, H.; Mockler, T.C.; Kay, S.A.; Chory, J. A Morning-Specific Phytohormone Gene Expression Program underlying Rhythmic Plant Growth. PLoS Biol. 2008, 6, e225. [Google Scholar] [CrossRef] [Green Version]
- Michael, T.P.; Mockler, T.C.; Breton, G.; McEntee, C.; Byer, A.; Trout, J.D.; Hazen, S.P.; Shen, R.; Priest, H.D.; Sullivan, C.M.; et al. Network Discovery Pipeline Elucidates Conserved Time-of-Day–Specific cis-Regulatory Modules. PLoS Genet. 2008, 4, e14. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, X.; Zeevaart, J.A.D. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc. Natl. Acad. Sci. USA 1999, 96, 15354–15361. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Zeevaart, J.A. Overexpression of a 9-cis-Epoxycarotenoid Dioxygenase Gene in Nicotiana plumbaginifolia Increases Abscisic Acid and Phaseic Acid Levels and Enhances Drought Tolerance. Plant Physiol. 2002, 128, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef] [Green Version]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.; Ecker, J.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef] [Green Version]
- Ezer, D.; Jung, J.-H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stöckle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of Glucosidase via Stress-Induced Polymerization Rapidly Increases Active Pools of Abscisic Acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [Green Version]
- Fukushima, A.; Kusano, M.; Nakamichi, N.; Kobayashi, M.; Hayashi, N.; Sakakibara, H.; Mizuno, T.; Saito, K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 2009, 106, 7251–7256. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef]
- Burschka, C.; Tenhunen, J.D.; Hartung, W. Diurnal variations in abscisic acid content and stomatal response to applied abscisic acid in leaves of irrigated and non-irrigated Arbutus unedo plants under naturally fluctuating environmental conditions. Oecologia 1983, 58, 128–131. [Google Scholar] [CrossRef] [PubMed]
- McAdam, S.A.M.; Brodribb, T.J.; Ross, J.J.; Jordan, G.J. Augmentation of abscisic acid (ABA) levels by drought does not induce short-term stomatal sensitivity to CO2 in two divergent conifer species. J. Exp. Bot. 2010, 62, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nováková, M.; Motyka, V.; Dobrev, P.I.; Malbeck, J.; Gaudinová, A.; Vanková, R. Diurnal variation of cytokinin, auxin and abscisic acid levels in tobacco leaves. J. Exp. Bot. 2005, 56, 2877–2883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, D.; Kim, W.-Y.; Cha, J.-Y.; Park, H.J.; Shin, G.; Park, J.; Lim, C.J.; Chun, H.J.; Li, N.; Kim, D.H.; et al. The Gigantea-Enhanced em level Complex Enhances Drought Tolerance via Regulation of Abscisic Acid Synthesis. Plant Physiol. 2020, 184, 443–458. [Google Scholar] [CrossRef]
- Yu, J.-W.; Rubio, V.; Lee, N.-Y.; Bai, S.; Lee, S.-Y.; Kim, S.-S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 Control Circadian Function and Photoperiodic Flowering by Regulating GI Stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Ye, M.; Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005, 24, 683–690. [Google Scholar] [CrossRef]
- Penfield, S.; Hall, A. A Role for Multiple Circadian Clock Genes in the Response to Signals That Break Seed Dormancy inArabidopsis. Plant Cell 2009, 21, 1722–1732. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.-Y.; Ali, Z.; Park, H.J.; Park, S.J.; Cha, J.-Y.; Perez-Hormaeche, J.; Quintero, F.J.; Shin, G.; Kim, M.R.; Qiang, Z.; et al. Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 2013, 4, 1352. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Zhang, X.; Wang, W.; Wang, Y.; Ming, F. The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 2013, 8, e73541. [Google Scholar] [CrossRef]
- Riboni, M.; Galbiati, M.; Tonelli, C.; Conti, L. GIGANTEA Enables Drought Escape Response via Abscisic Acid-Dependent Activation of the Florigens and SUPPRESSOR of OVEREXPRESSION of CONSTANS1. Plant Physiol. 2013, 162, 1706–1719. [Google Scholar] [CrossRef] [Green Version]
- Simon, N.M.L.; Graham, C.A.; Comben, N.E.; Hetherington, A.M.; Dodd, A.N. The Circadian Clock Influences the Long-Term Water Use Efficiency of Arabidopsis. Plant Physiol. 2020, 183, 317–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobata, T.; Okuno, T.; Yamamoto, T. Contributions of Capacity for Soil Water Extraction and Water Use Efficiency to Maintenance of Dry matter Production in Rice subjected to Drought. Jpn. J. Crop Sci. 1996, 65, 652–662. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A.; et al. A vacuolar beta-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Yan, J.-P.; Li, D.-K.; Luo, Q.; Yan, Q.; Liu, Z.-B.; Ye, L.-M.; Wang, J.-M.; Li, X.-F.; Yang, Y. UDP-Glucosyltransferase71C5, a Major Glucosyltransferase, Mediates Abscisic Acid Homeostasis in Arabidopsis. Plant Physiol. 2015, 167, 1659–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.-K.; Ye, M.; Li, B.; Noel, J.P. Co-evolution of Hormone Metabolism and Signaling Networks Expands Plant Adaptive Plasticity. Cell 2016, 166, 881–893. [Google Scholar] [CrossRef] [Green Version]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought Induction of Arabidopsis 9-cis-Epoxycarotenoid Dioxygenase Occurs in Vascular Parenchyma Cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef] [Green Version]
- Boursiac, Y.; Léran, S.; Corratgé-Faillie, C.; Gojon, A.; Krouk, G.; Lacombe, B. ABA transport and transporters. Trends Plant Sci. 2013, 18, 325–333. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, H.; Pan, Y.; Yu, Y.; Luan, S.; Li, L. A DTX/MATE-Type Transporter Facilitates Abscisic Acid Efflux and Modulates ABA Sensitivity and Drought Tolerance in Arabidopsis. Mol. Plant 2014, 7, 1522–1532. [Google Scholar] [CrossRef] [Green Version]
- Kanno, Y.; Hanada, A.; Chiba, Y.; Ichikawa, T.; Nakazawa, M.; Matsui, M.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 2012, 109, 9653–9658. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, T.; Kanno, Y.; Suzuki, H.; Watanabe, S.; Seo, M. Arabidopsis NPF4.6 and NPF5.1 Control Leaf Stomatal Aperture by Regulating Abscisic Acid Transport. Genes 2021, 12, 885. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Seo, M.; Shinozaki, K. ABA Transport and Plant Water Stress Responses. Trends Plant Sci. 2018, 23, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.-Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Márquez, J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [Green Version]
- Vlad, F.; Rubio, S.; Rodrigues, A.; Sirichandra, C.; Belin, C.; Robert, N.; Leung, J.; Rodriguez, P.L.; Laurière, C.; Merlot, S. Protein Phosphatases 2C Regulate the Activation of the Snf1-Related Kinase OST1 by Abscisic Acid in Arabidopsis. Plant Cell 2009, 21, 3170–3184. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Wu, W.; Abrams, S.R.; Cutler, A.J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 2008, 59, 2991–3007. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Huang, W.; Pérez-García, P.; Pokhilko, A.; Millar, A.J.; Antoshechkin, I.; Riechmann, J.L.; Mas, P. Mapping the Core of the Arabidopsis Circadian Clock Defines the Network Structure of the Oscillator. Science 2012, 336, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Carlsson, J.; Takeuchi, T.; Newton, L.; Farré, E.M. Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 2013, 76, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Kurup, S.; Jones, H.; Holdsworth, M.J. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 2000, 21, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Han, X.; Yang, J.; Jiang, Y.; Hu, Y. The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 2021, 33, 3022–3041. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.K.; Nomura, Y.; Wang, L.; Nakagami, H.; Somers, D.E. Quantitative Circadian Phosphoproteomic Analysis of Arabidopsis Reveals Extensive Clock Control of Key Components in Physiological, Metabolic, and Signaling Pathways. Mol. Cell. Proteom. 2015, 14, 2243–2260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.; Xie, G.Z.; Zhang, S.; Li, B.; Wang, X.; Li, Y.; Liu, T.; Xu, X. GmLCLs negatively regulate ABA perception and signalling genes in soybean leaf dehydration response. Plant Cell Environ. 2020, 44, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Molina, L.; Mongrand, S.; Kinoshita, N.; Chua, N.-H. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 2003, 17, 410–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, M.E.; Lynch, T.; Peeters, J.; Snowden, C.; Finkelstein, R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 2008, 67, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Lynch, T.J.; Erickson, B.J.; Miller, D.R.; Finkelstein, R.R. ABI5-binding proteins (AFPs) alter transcription of ABA-induced genes via a variety of interactions with chromatin modifiers. Plant Mol. Biol. 2016, 93, 403–418. [Google Scholar] [CrossRef]
- Peirats-Llobet, M.; Han, S.-K.; Gonzalez-Guzman, M.; Jeong, C.W.; Rodriguez, L.; Belda-Palazon, B.; Wagner, D.; Rodriguez, P.L. A Direct Link between Abscisic Acid Sensing and the Chromatin-Remodeling ATPase BRAHMA via Core ABA Signaling Pathway Components. Mol. Plant 2016, 9, 136–147. [Google Scholar] [CrossRef] [Green Version]
- Efroni, I.; Han, S.-K.; Kim, H.J.; Wu, M.-F.; Steiner, E.; Birnbaum, K.D.; Hong, J.C.; Eshed, Y.; Wagner, D. Regulation of Leaf Maturation by Chromatin-Mediated Modulation of Cytokinin Responses. Dev. Cell 2013, 24, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Champion, A.; Kreis, M.; Mockaitis, K.; Picaud, A.; Henry, Y. Arabidopsis kinome: After the casting. Funct. Integr. Genom. 2004, 4, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, J.; Wang, H.; Yang, C.; Chen, Y.; Li, Y.; Pan, S.; Dong, R.; Tang, G.; Barajas-Lopez, J.D.; et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 9651–9656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Kumar, R.; Baek, D.; Hyun, T.-K.; Chung, W.S.; Yun, D.-J.; Kim, J.-Y. Arabidopsis thaliana RECEPTOR DEAD KINASE1 Functions as a Positive Regulator in Plant Responses to ABA. Mol. Plant 2017, 10, 223–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; He, J.; Zhao, Y.; Wu, T.; Zhou, X.; Ding, Y.; Kong, L.; Wang, X.; Wang, Y.; Li, J.; et al. EAR1 Negatively Regulates ABA Signaling by Enhancing 2C Protein Phosphatase Activity. Plant Cell 2018, 30, 815–834. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhao, Y.; Li, Z.; Hsu, C.-C.; Liu, X.; Fu, L.; Hou, Y.-J.; Du, Y.; Xie, S.; Zhang, C.; et al. Reciprocal Regulation of the TOR Kinase and ABA Receptor Balances Plant Growth and Stress Response. Mol. Cell 2017, 69, 100–112.e6. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-H.; Qu, L.; Xu, Z.-H.; Zhu, J.-K.; Xue, H.-W. EL1-like Casein Kinases Suppress ABA Signaling and Responses by Phosphorylating and Destabilizing the ABA Receptors PYR/PYLs in Arabidopsis. Mol. Plant 2018, 11, 706–719. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, X.; Li, D.; Sun, Y.; Li, Y.; Luo, Q.; Liu, Z.; Wang, J.; Li, X.; Zhang, H.; et al. CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discov. 2018, 4, 30. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, D.; Xu, Y.; Jin, S.; Zhang, L.; Zhang, S.; Yang, G.; Huang, J.; Yan, K.; Wu, C.; et al. CEPR2 phosphorylates and accelerates the degradation of PYR/PYLs in Arabidopsis. J. Exp. Bot. 2019, 70, 5457–5469. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Kong, X.; Huang, Q.; Zhang, Q.; Ge, H.; Zhang, L.; Li, G.; Peng, L.; Liu, Z.; Wang, J.; et al. CARK1 phosphorylates subfamily III members of ABA receptors. J. Exp. Bot. 2018, 70, 519–528. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Wang, Y.; Liu, X.; Ma, L.; Zhang, Z.; Mu, C.; Zhang, Y.; Peng, L.; Xie, S.; et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021, 12, 2456. [Google Scholar] [CrossRef]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Muñoz-Bertomeu, J.; Ibañez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2014, 80, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Irigoyen, M.L.; Iniesto, E.; Rodriguez, L.; Puga, M.I.; Yanagawa, Y.; Pick, E.; Strickland, E.; Paz-Ares, J.; Wei, N.; De Jaeger, G.; et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 2014, 26, 712–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.; Lou, L.; Tian, M.; Li, Q.; Ding, Y.; Cao, X.; Wu, Y.; Belda-Palazon, B.; Rodriguez, P.L.; Yang, S.; et al. ESCRT-I Component VPS23A Affects ABA Signaling by Recognizing ABA Receptors for Endosomal Degradation. Mol. Plant 2016, 9, 1570–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-León, M.; Cuyas, L.; Abd El-Moneim, D.; Rodriguez, L.; Belda-Palazon, B.; Sánchez-Quant, E.; Fernández, Y.; Roux, B.; Zamarreño, A.M.; Garcia-Mina, J.M.; et al. Stomatal apertura and turnover of ABA receptors are regulated by Arabidopsis ALIX. Plant Cell 2019, 31, 2411–2429. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Zhang, X.; Peirats-Llobet, M.; Belda-Palazon, B.; Wang, X.; Cui, S.; Yu, X.; Rodriguez, P.L.; An, C. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 2016, 28, 2178–2196. [Google Scholar] [CrossRef] [Green Version]
- Belda-Palazon, B.; Julian, J.; Coego, A.; Wu, Q.; Zhang, X.; Batistic, O.; AlQuraishi, S.A.; Kudla, J.; An, C.; Rodriguez, P.L. ABA inhibits myristoylation and induces shuttling of the RGLG 1 E3 ligase to promote nuclear degradation of PP 2 CA. Plant J. 2019, 98, 813–825. [Google Scholar] [CrossRef]
- Julian, J.; Coego, A.; Lozano-Juste, J.; Lechner, E.; Wu, Q.; Zhang, X.; Merilo, E.; Belda-Palazon, B.; Park, S.; Cutler, S.R.; et al. The MATH-BTB BPM3 and BPM5 subunits of Cullin3-RING E3 ubiquitin ligases target PP2CA and other clade A PP2Cs for degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 15725–15734. [Google Scholar] [CrossRef] [Green Version]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 2002, 14 (Suppl. 1), S15–S45. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant–pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Toh, S.; Torii, K.U. Chemical control of stomatal function and development. Curr. Opin. Plant Biol. 2021, 60, 102010. [Google Scholar] [CrossRef] [PubMed]
- Yazdanbakhsh, N.; Sulpice, R.; Graf, A.; Stitt, M.; Fisahn, J. Circadian control of root elongation and C partitioning inArabidopsis thaliana. Plant Cell Environ. 2011, 34, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Kircher, S.; Schopfer, P. Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 11217–11221. [Google Scholar] [CrossRef] [Green Version]
- DeMontaigu, A.; Tóth, R.; Coupland, G. Plant development goes like clockwork. Trends Genet. 2010, 26, 296–306. [Google Scholar] [CrossRef]
- Yan, J.; Li, S.; Kim, Y.J.; Zeng, Q.; Radziejwoski, A.; Wang, L.; Nomura, Y.; Nakagami, H.; Somers, D.E. TOC1 clock protein phosphorylation controls complex formation with NF-YB/C to repress hypocotyl growth. EMBO J. 2021, 40, e108684. [Google Scholar] [CrossRef] [PubMed]
- Belda-Palazón, B.; Adamo, M.; Valerio, C.; Ferreira, L.J.; Confraria, A.; Reis-Barata, D.; Rodrigues, A.; Meyer, C.; Rodriguez, P.L.; Baena-González, E. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nat. Plants 2020, 6, 1345–1353. [Google Scholar] [CrossRef]
- Hanano, S.; Domagalska, M.A.; Nagy, F.; Davis, S.J. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells 2006, 11, 1381–1392. [Google Scholar] [CrossRef]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, S.; Jiang, L.; Song, S.; Jing, R.; Xu, G. AtGRP7 is involved in the regulation of abscisic acid and stress responses in arabidopsis. Cell. Mol. Biol. Lett. 2006, 11, 526–535. [Google Scholar] [CrossRef] [PubMed]
- Riboni, M.; Test, A.R.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil Lee, H.; Mas, P.; Seo, P.J. MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci. Rep. 2016, 6, 17754. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).