Nanoimprinted Hierarchical Micro-/Nanostructured Substrates for the Growth of Cardiomyocyte Fibers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Structure Design
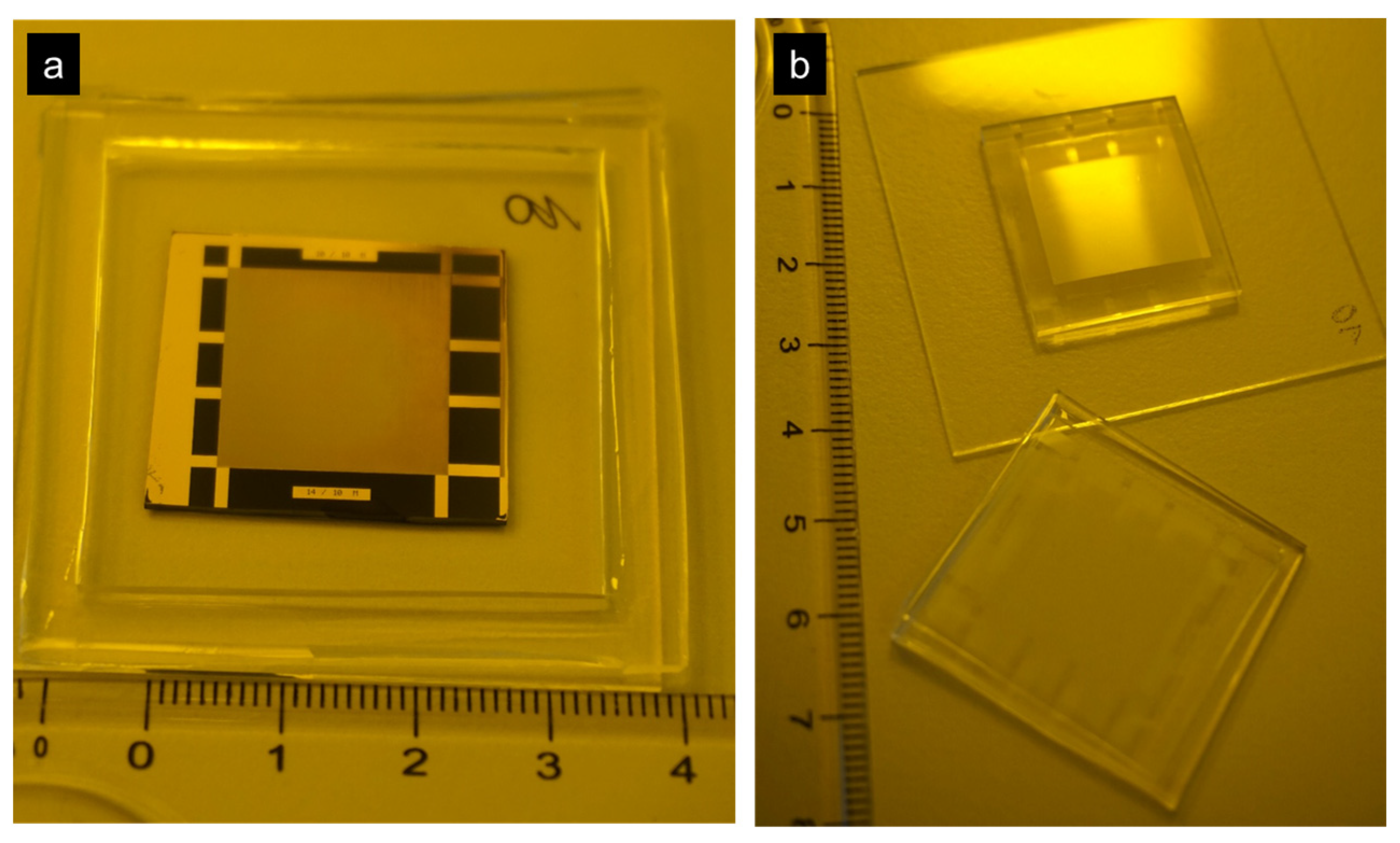
2.2. Nanoimprint Master Fabrication
2.3. Nanoimprinting
2.3.1. Basics
2.3.2. Stamps
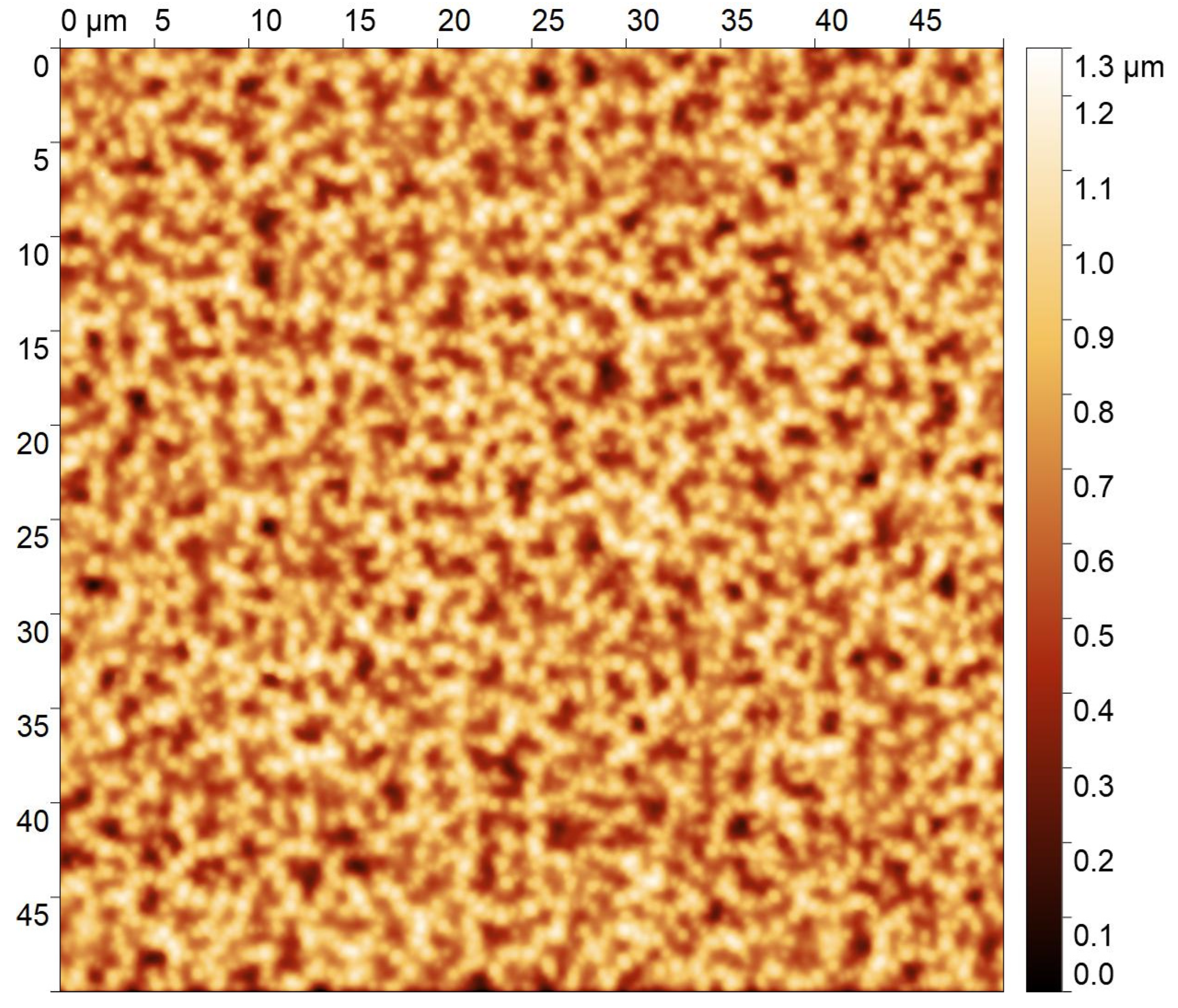
2.3.3. Imprinting Process, and Roughness on the Top or on the Bottom
2.3.4. Roll-to-Plate Nanoimprinting
2.4. Cells
3. Results
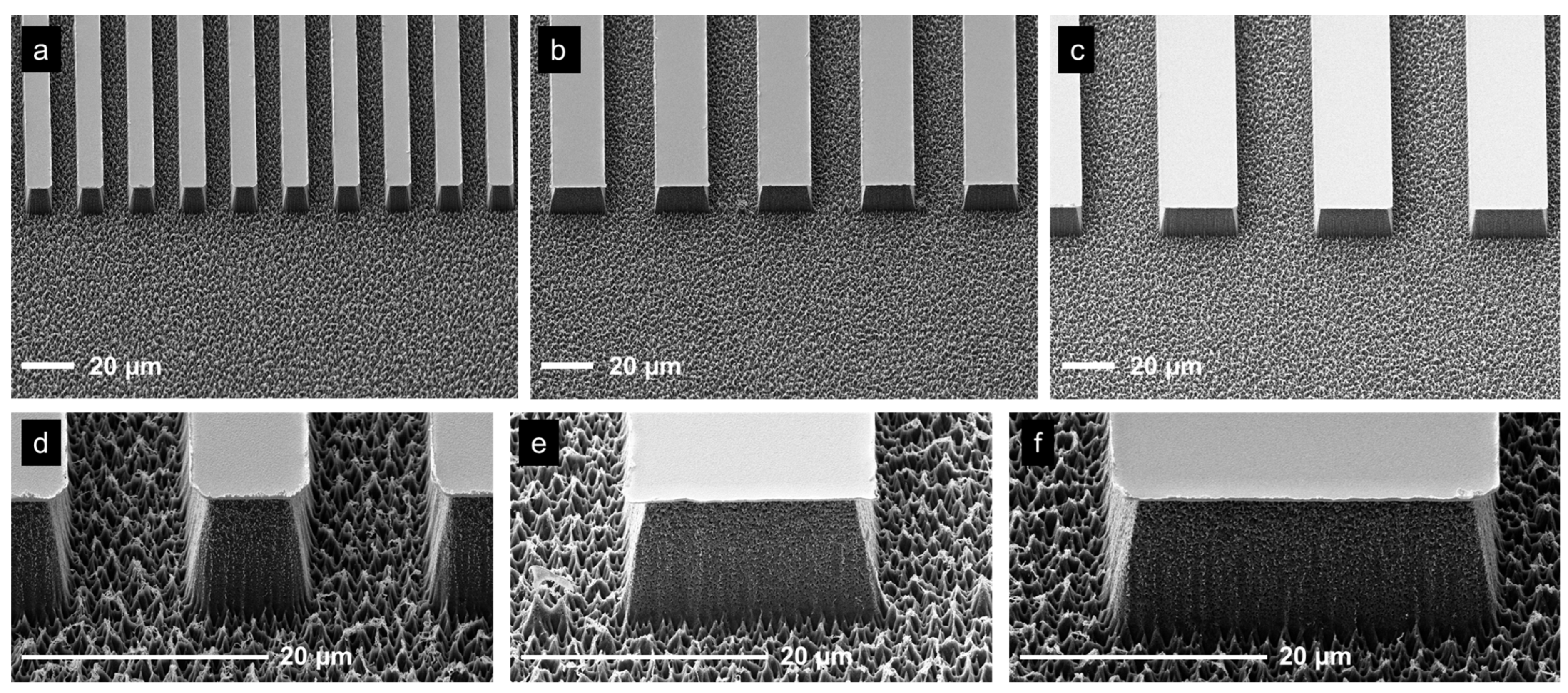
3.1. Different Sample Geometries
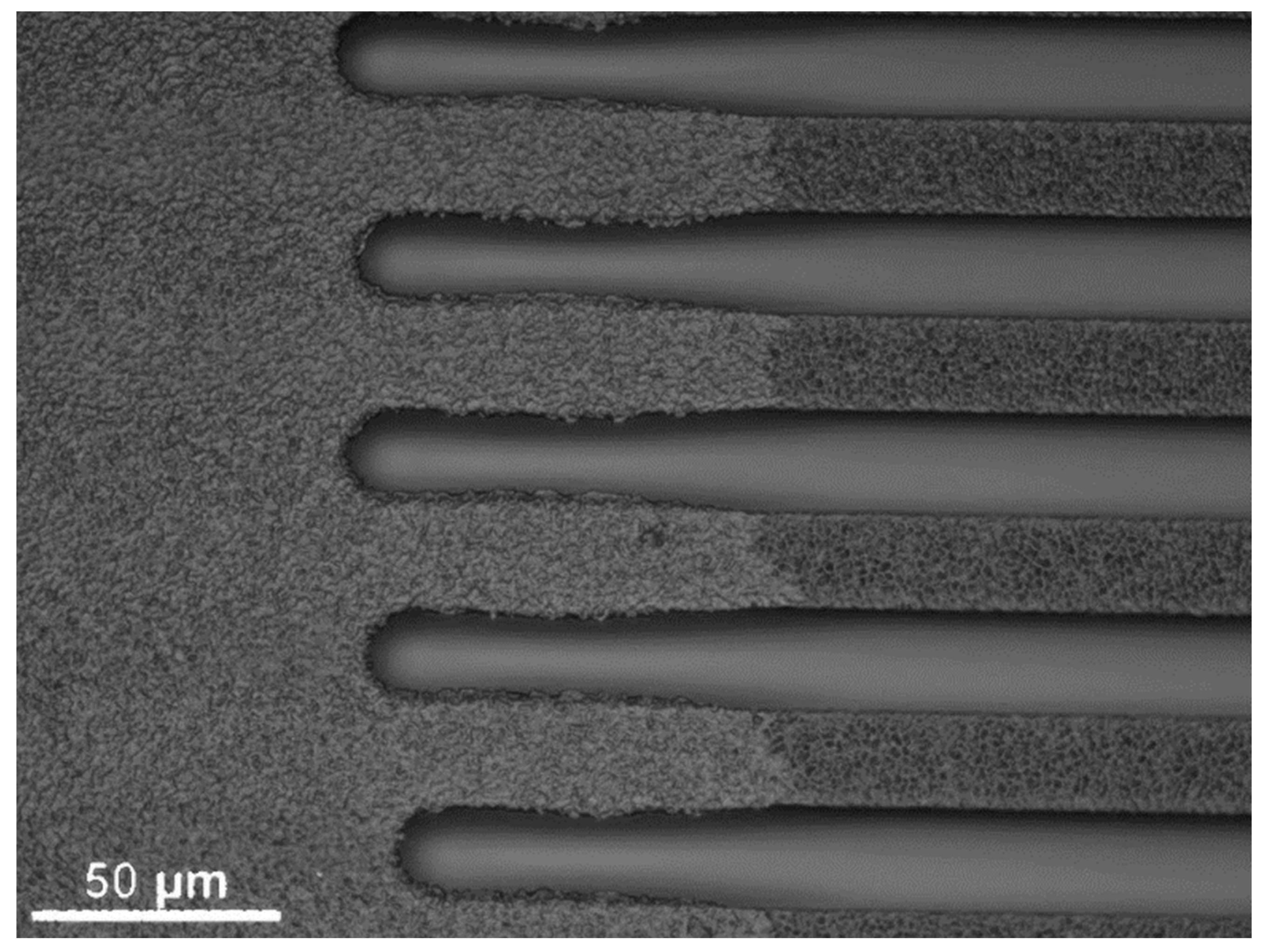
3.1.1. Open and Closed Channels
3.1.2. Residual-Layer-Free Samples
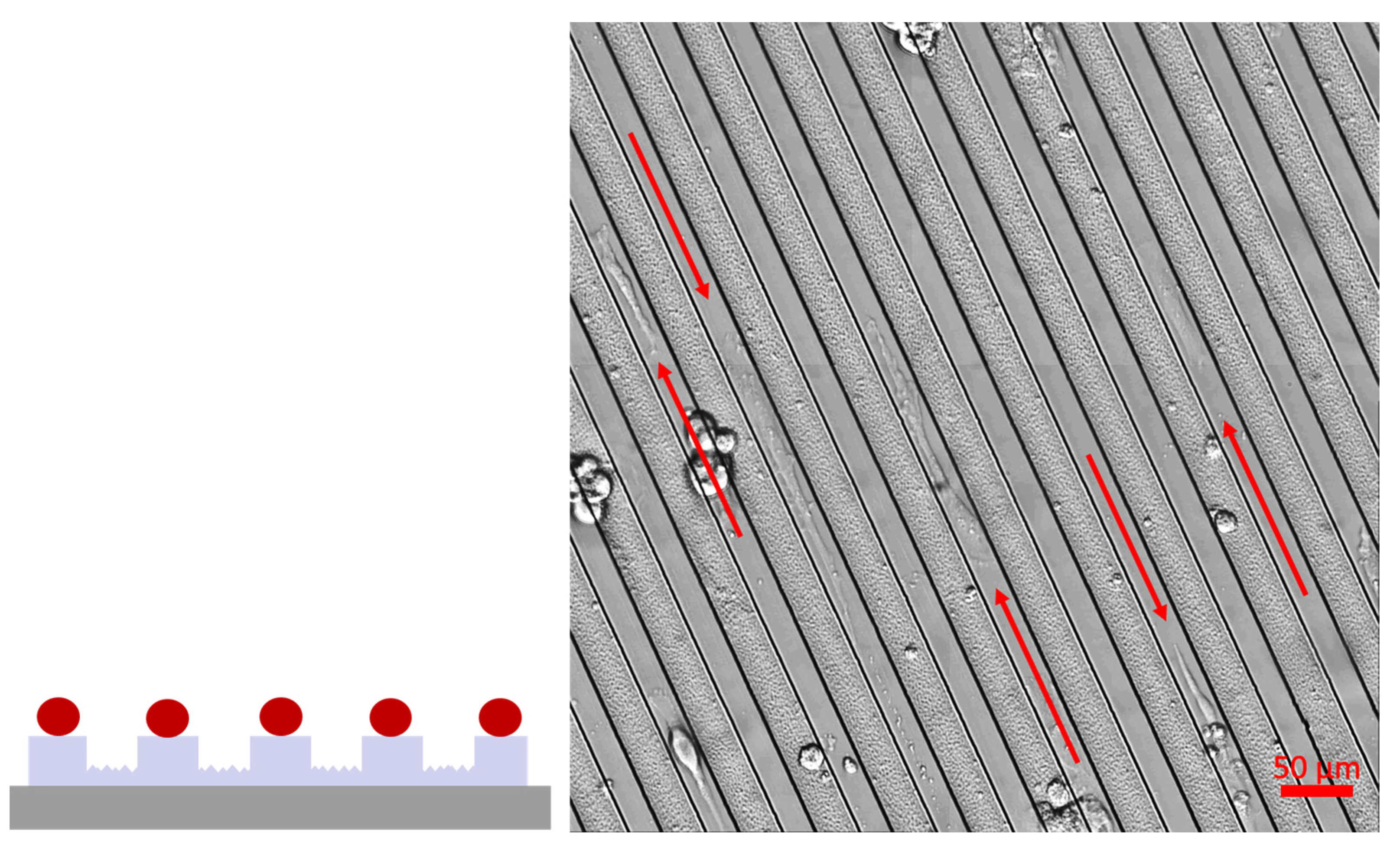
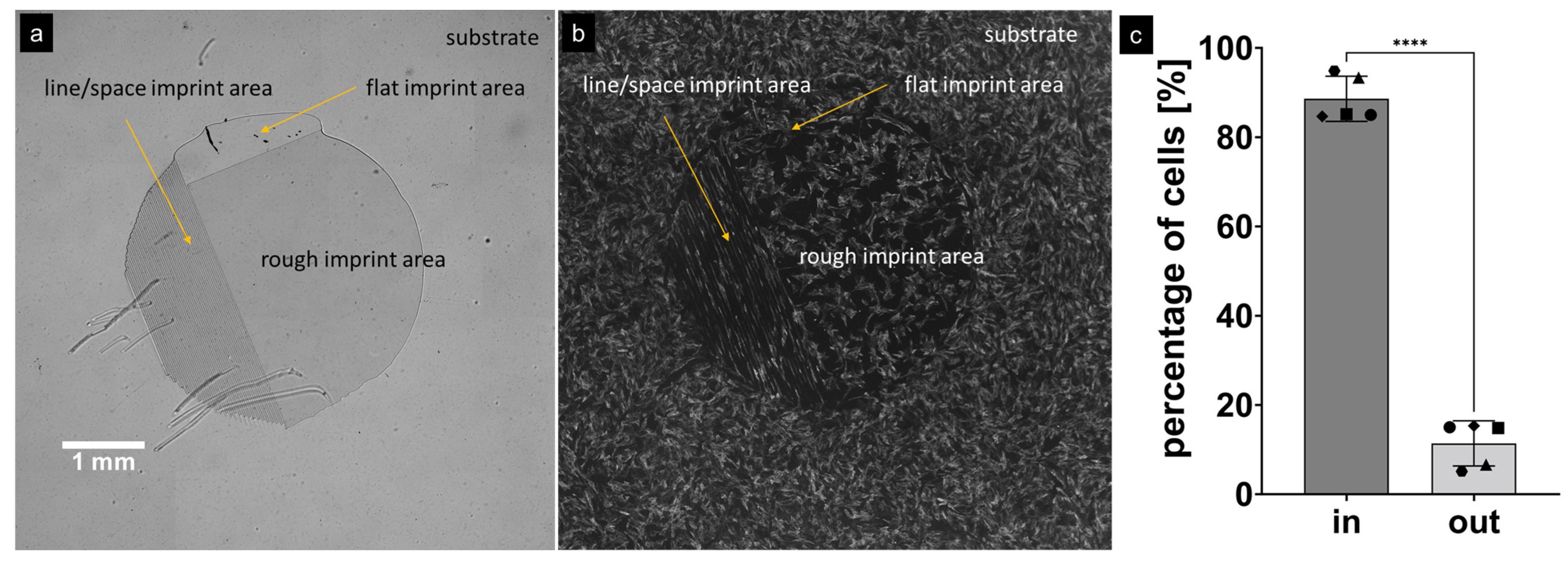
3.2. Cell Growth
4. Discussion
4.1. Nanoimprinting
4.2. Cell Growth
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
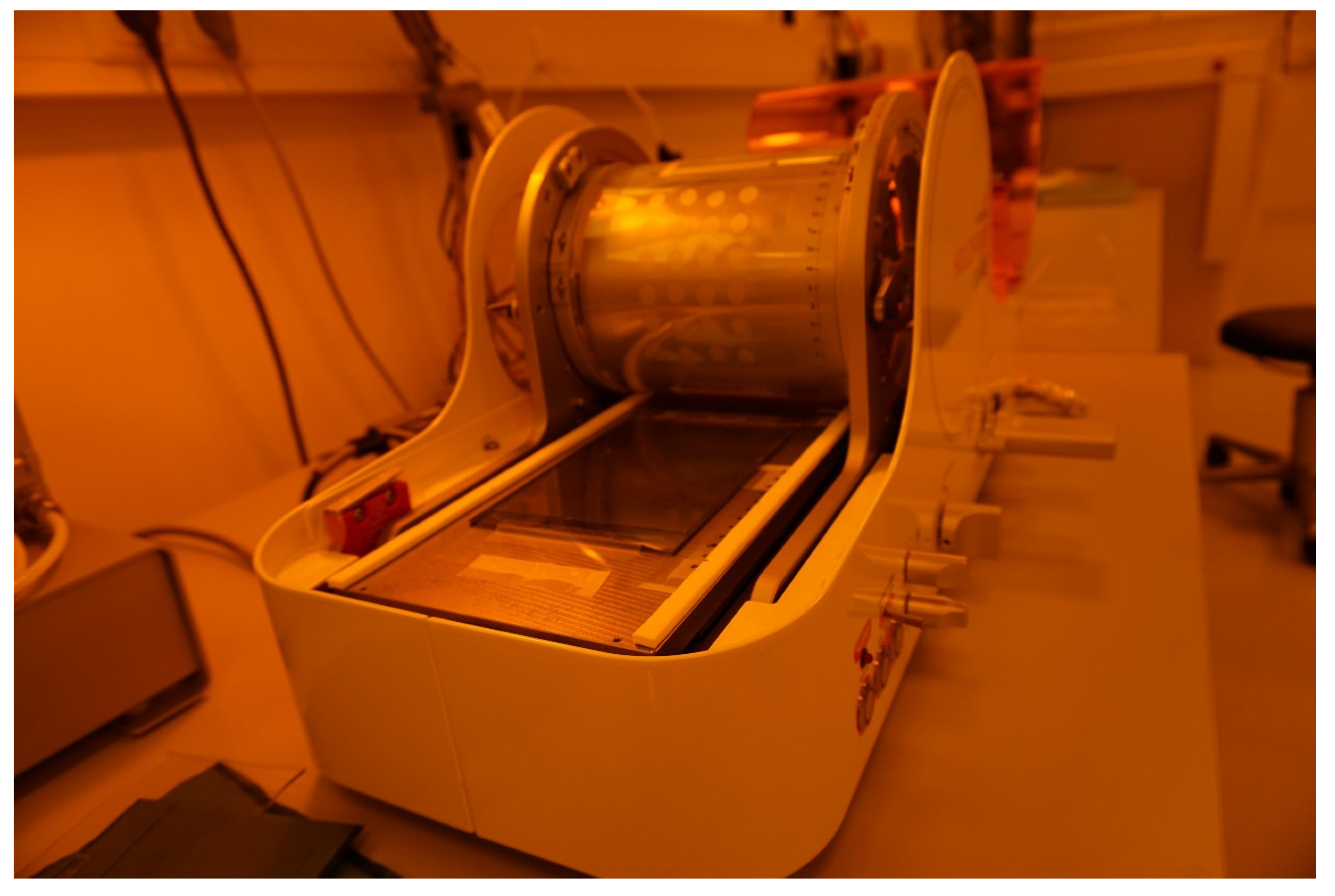
Appendix A. Roll-to-Plate Nanoimprinting

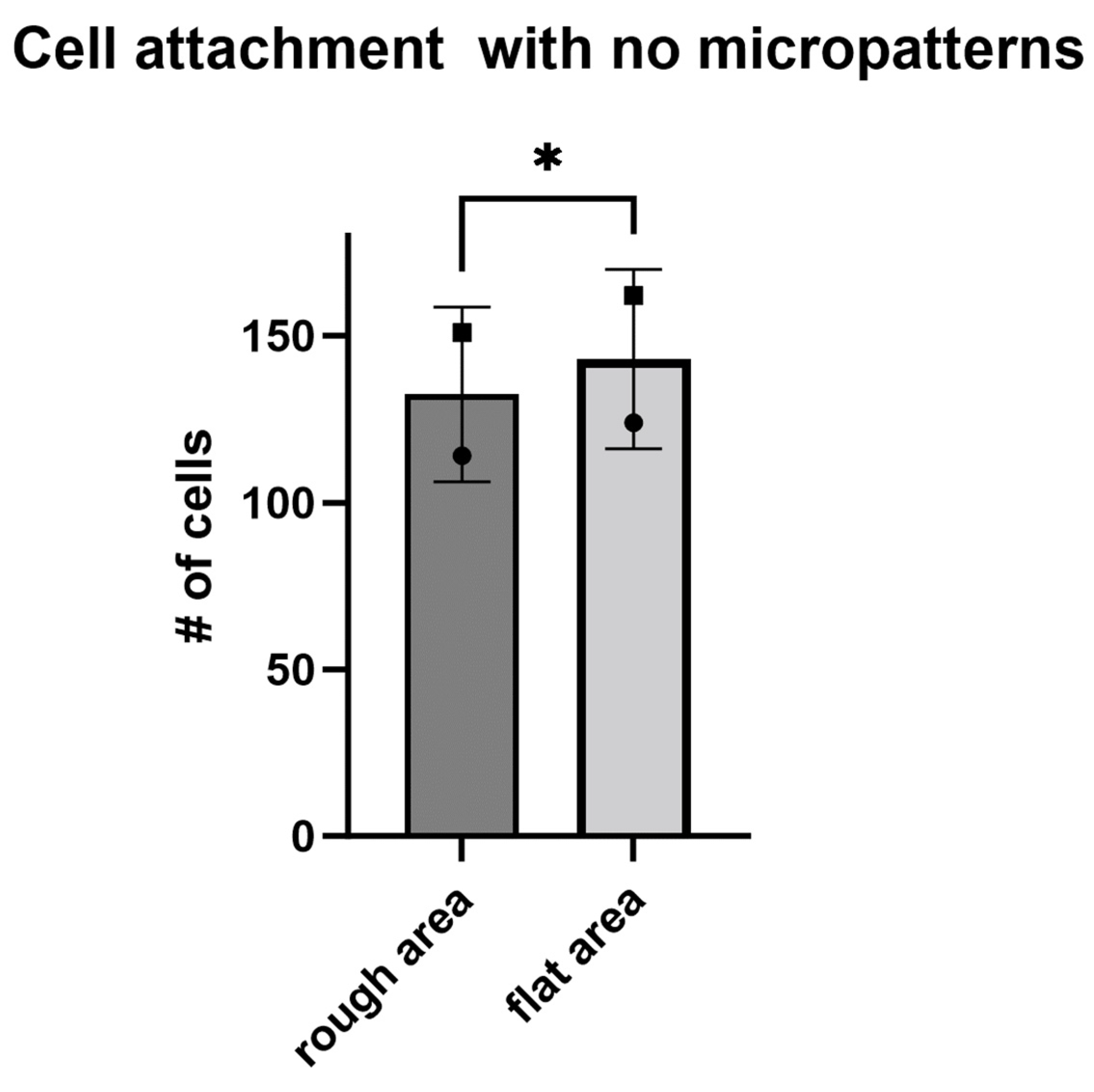
Appendix B. Cell Growth on Substrates without Micro-Channels
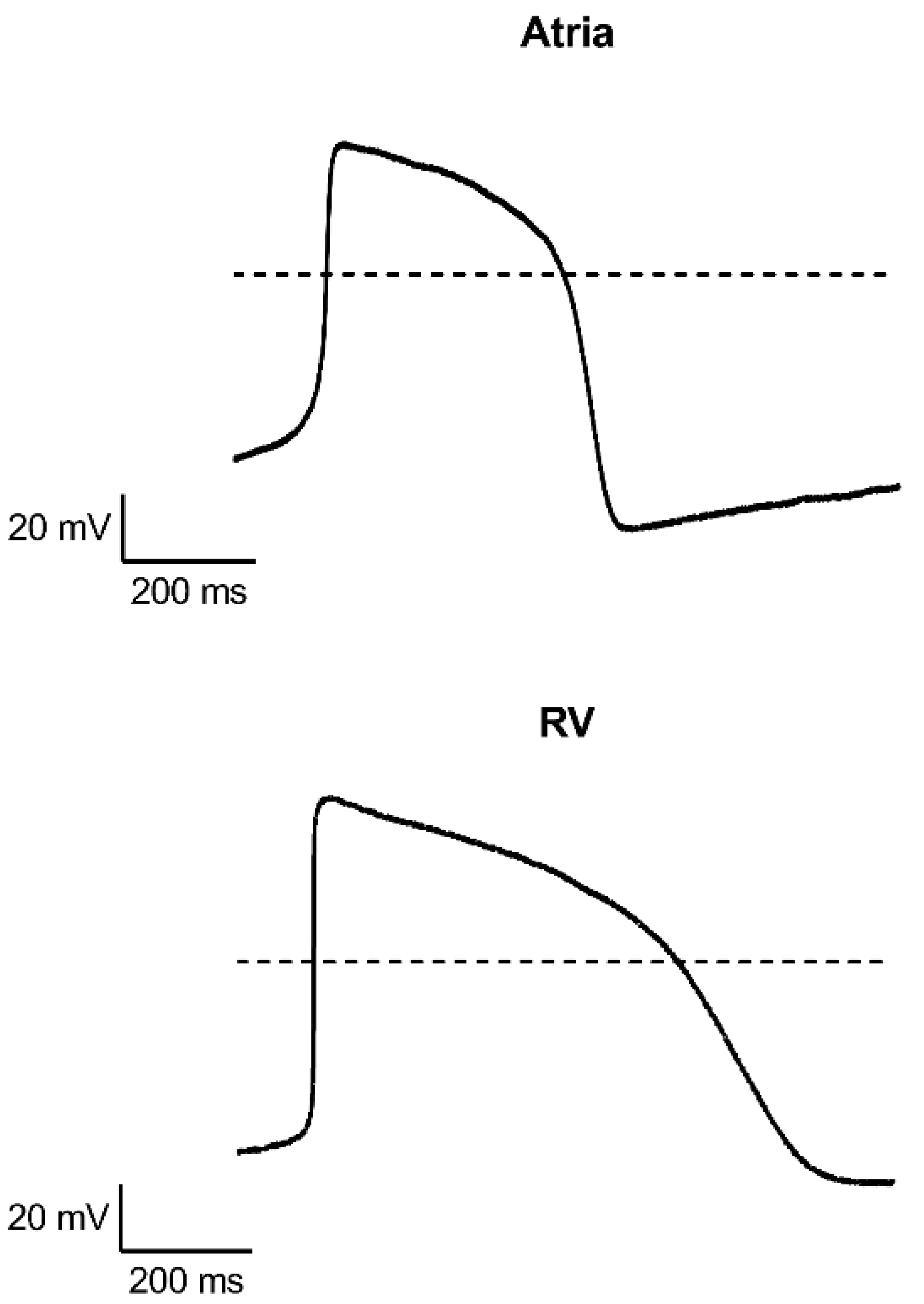
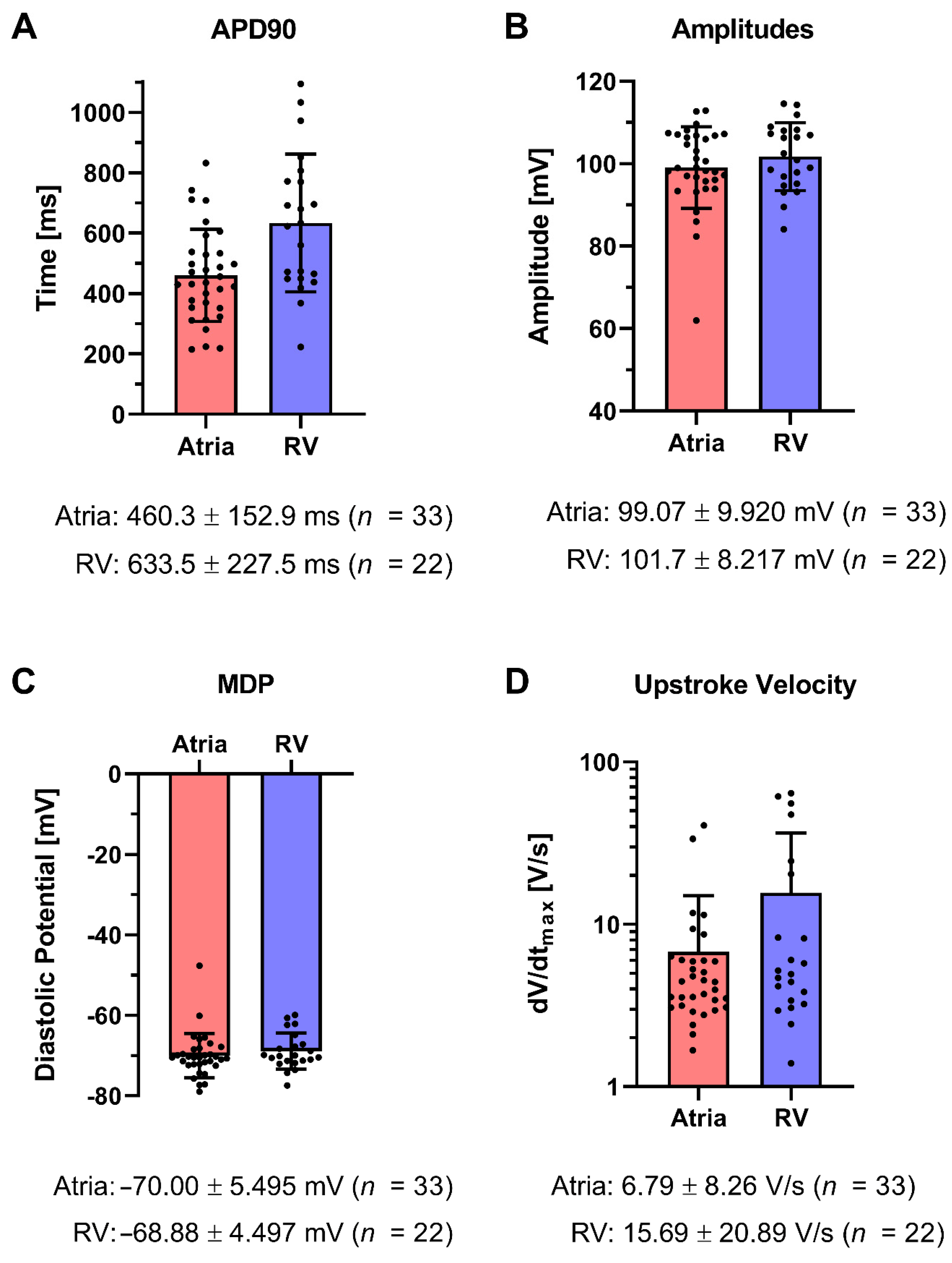
Appendix C. Measurements on Single Cardiomyocytes
Patch Clamp Recordings of Single Cardiomyocytes
References
- Ghane-Motlagh, B.; Sawan, M. A Review of Microelectrode Array Technologies: Design and Implementation Challenges. In Proceedings of the 2013 2nd International Conference on Advances in Biomedical Engineering, Tripoli, Lebanon, 11–13 September 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 38–41. [Google Scholar]
- Chen, P.; Zhang, W.; Zhou, J.; Wang, P.; Xiao, L.; Yang, M. Development of Planar Patch Clamp Technology and Its Application in the Analysis of Cellular Electrophysiology. Prog. Nat. Sci. 2009, 19, 153–160. [Google Scholar] [CrossRef]
- Bébarová, M. Advances in Patch Clamp Technique: Towards Higher Quality and Quantity. Gen. Physiol. Biophys. 2012, 31, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Heist, E.K.; Ruskin, J.N. Drug-Induced Arrhythmia. Circulation 2010, 122, 1426–1435. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, J.E.; Chung, M.K.; Campbell, K.B.; Hammadah, M.; Joglar, J.A.; Leclerc, J.; Rajagopalan, B.; On behalf of the American Heart Association Clinical Pharmacology Committee of the Council on Clinical Cardiology and Council on Cardiovascular and Stroke Nursing. Drug-Induced Arrhythmias: A Scientific Statement From the American Heart Association. Circulation 2020, 142, e214–e233. [Google Scholar] [CrossRef]
- Hofbauer, P.; Jahnel, S.M.; Papai, N.; Giesshammer, M.; Deyett, A.; Schmidt, C.; Penc, M.; Tavernini, K.; Grdseloff, N.; Meledeth, C.; et al. Cardioids Reveal Self-Organizing Principles of Human Cardiogenesis. Cell 2021, 184, 3299–3317.e22. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Kehat, I.; Kenyagin-Karsenti, D.; Snir, M.; Segev, H.; Amit, M.; Gepstein, A.; Livne, E.; Binah, O.; Itskovitz-Eldor, J.; Gepstein, L. Human Embryonic Stem Cells Can Differentiate into Myocytes with Structural and Functional Properties of Cardiomyocytes. J. Clin. Investig. 2001, 108, 407–414. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Burridge, P.W.; Matsa, E.; Shukla, P.; Lin, Z.C.; Churko, J.M.; Ebert, A.D.; Lan, F.; Diecke, S.; Huber, B.; Mordwinkin, N.M.; et al. Chemically Defined Generation of Human Cardiomyocytes. Nat. Methods 2014, 11, 855–860. [Google Scholar] [CrossRef]
- Curtis, A.S.G.; Gadegaard, N.; Dalby, M.J.; Riehle, M.O.; Wilkinson, C.D.W.; Aitchison, G. Cells React to Nanoscale Order and Symmetry in Their Surroundings. IEEE Trans. Nanobiosci. 2004, 3, 61–65. [Google Scholar] [CrossRef]
- Dalby, M.J.; Gadegaard, N.; Oreffo, R.O.C. Harnessing Nanotopography and Integrin–Matrix Interactions to Influence Stem Cell Fate. Nat. Mater. 2014, 13, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Carthew, J.; Abdelmaksoud, H.H.; Hodgson-Garms, M.; Aslanoglou, S.; Ghavamian, S.; Elnathan, R.; Spatz, J.P.; Brugger, J.; Thissen, H.; Voelcker, N.H.; et al. Precision Surface Microtopography Regulates Cell Fate via Changes to Actomyosin Contractility and Nuclear Architecture. Adv. Sci. 2021, 8, 2003186. [Google Scholar] [CrossRef] [PubMed]
- Han, E.D.; Kim, B.H.; Seo, Y.H. Anti-Cell Adhesion Characteristics of Nanotextured Surface for Implantable Biomedical Device. Int. J. Precis. Eng. Manuf. 2017, 18, 239–244. [Google Scholar] [CrossRef]
- Mobini, S.; Kuliasha, C.A.; Siders, Z.A.; Bohmann, N.A.; Jamal, S.-M.; Judy, J.W.; Schmidt, C.E.; Brennan, A.B. Microtopographical Patterns Promote Different Responses in Fibroblasts and Schwann Cells: A Possible Feature for Neural Implants. J. Biomed. Mater. Res. Part A 2021, 109, 64–76. [Google Scholar] [CrossRef]
- Batalov, I. Engineering Aligned Human Cardiac Muscle Using Developmentally Inspired Fibronectin Micropatterns. Sci. Rep. 2021, 11, 11502. [Google Scholar] [CrossRef]
- Zhang, F.; Low, H.Y. Ordered Three-Dimensional Hierarchical Nanostructures by Nanoimprint Lithography. Nanotechnology 2006, 17, 1884–1890. [Google Scholar] [CrossRef]
- Durret, J.; Szkutnik, P.-D.; Frolet, N.; Labau, S.; Gourgon, C. Superhydrophobic Polymeric Films with Hierarchical Structures Produced by Nanoimprint (NIL) and Plasma Roughening. Appl. Surf. Sci. 2018, 445, 97–106. [Google Scholar] [CrossRef]
- Choo, S.; Choi, H.-J.; Lee, H. Replication of Rose-Petal Surface Structure Using UV-Nanoimprint Lithography. Mater. Lett. 2014, 121, 170–173. [Google Scholar] [CrossRef]
- Latthe, S.; Terashima, C.; Nakata, K.; Fujishima, A. Superhydrophobic Surfaces Developed by Mimicking Hierarchical Surface Morphology of Lotus Leaf. Molecules 2014, 19, 4256–4283. [Google Scholar] [CrossRef]
- Schift, H. Nanoimprint Lithography: An Old Story in Modern Times? A Review. J. Vac. Sci. Technol. B Microelectron. Nanometer Struct. 2008, 26, 458. [Google Scholar] [CrossRef]
- Chou, S.Y. Nanoimprint Lithography. J. Vac. Sci. Technol. B 1996, 14, 4129. [Google Scholar] [CrossRef]
- Haisma, J. Mold-Assisted Nanolithography: A Process for Reliable Pattern Replication. J. Vac. Sci. Technol. B Microelectron. Nanometer Struct. 1996, 14, 4124. [Google Scholar] [CrossRef]
- Mühlberger, M. Nanoimprint Lithography Technology and Applications; Nanomaterials; MDPI: Basel, Switzerland, 2022; ISBN 978-3-0365-4482-3. [Google Scholar]
- Xia, Y.; Whitesides, G.M. Soft Lithography. Annu. Rev. Mater. Sci. 1998, 28, 153–184. [Google Scholar] [CrossRef]
- Muehlberger, M. Nanoimprinting of Biomimetic Nanostructures. Nanomanufacturing 2022, 2, 17–40. [Google Scholar] [CrossRef]
- Muehlberger, M.; Ruttloff, S.; Nees, D.; Moharana, A.; Belegratis, M.R.; Taus, P.; Kopp, S.; Wanzenboeck, H.D.; Prinz, A.; Fechtig, D. Nanoimprint Replication of Biomimetic, Multilevel Undercut Nanostructures. Nanomaterials 2021, 11, 1051. [Google Scholar] [CrossRef] [PubMed]
- Pribyl, M.; Taus, P.; Prado-López, S.; Dozio, S.M.; Schrenk, W.; Haslinger, M.J.; Kopp, S.; Mühlberger, M.; Wanzenboeck, H.D. Dense High Aspect Ratio Nanostructures for Cell Chip Applications—Fabrication, Replication, and Cell Interactions. Micro Nano Eng. 2022, 15, 100121. [Google Scholar] [CrossRef]
- Wanzenboeck, H.D.; Waid, S.; Bertagnolli, E.; Muehlberger, M.; Bergmair, I.; Schoeftner, R. Nanoimprint Lithography Stamp Modification Utilizing Focused Ion Beams. J. Vac. Sci. Technol. B 2009, 27, 7. [Google Scholar] [CrossRef]
- Schizas, C.; Karalekas, D. Mechanical Characteristics of an Ormocomp® Biocompatible Hybrid Photopolymer. J. Mech. Behav. Biomed. Mater. 2011, 4, 99–106. [Google Scholar] [CrossRef]
- Stewart, M.D.; Willson, C.G. Imprint Materials for Nanoscale Devices. Mrs. Bull. 2005, 30, 947. [Google Scholar] [CrossRef]
- Vig, A.L.; Mäkelä, T.; Majander, P.; Lambertini, V.; Ahopelto, J.; Kristensen, A. Roll-to-Roll Fabricated Lab-on-a-Chip Devices. J. Micromech. Microeng. 2011, 21, 035006. [Google Scholar] [CrossRef]
- Smolka, M.; Haase, A.; Palfinger, U.; Nees, D.; Kuna, L.; Hesse, J.; Stadlober, B.; Geidel, S.; Nestler, J.; Ladenhauf, N.; et al. Roll-to-Roll Pilot Line for Large-Scale Manufacturing of Microfluidic Devices. In Proceedings of the Single-use Technologies II: Bridging Polymer Science to Biotechnology Applications, Tomar, Portugal, 7–10 May 2017; p. 23. [Google Scholar]
- Smolka, M.; Ruttloff, S.; Nees, D.; Prietl, C.; Satzinger, V.; Lamprecht, B.; Hütter, P.; Hesse, J.; Kokkinis, G.; Kriechhammer, G.; et al. High Throughput Roll-to-Roll Production of Microfluidic Chips. Proceedings 2018, 2, 1054. [Google Scholar] [CrossRef]
- Nguyen, V.T.H.; Jensen, F.; Hübner, J.; Leussink, P.; Jansen, H. On the Formation of Black Silicon in SF6-O2 Plasma: The Clear, Oxidize, Remove, and Etch (CORE) Sequence and Black Silicon on Demand. J. Vac. Sci. Technol. A 2020, 38, 043004. [Google Scholar] [CrossRef]
- Zhu, F.; Zhang, X.; Zhang, H. Formation Mechanism of Multi-Functional Black Silicon Based on Optimized Deep Reactive Ion Etching Technique with SF6/C4F8. Sci. China Technol. Sci. 2015, 58, 381–389. [Google Scholar] [CrossRef]
- Silvestre, C.M.; Nguyen, V.; Jansen, H.; Hansen, O. Deep Reactive Ion Etching of ‘Grass-Free’ Widely-Spaced Periodic 2D Arrays, Using Sacrificial Structures. Microelectron. Eng. 2020, 223, 111228. [Google Scholar] [CrossRef]
- Kroll, M.; Käsebier, T.; Otto, M.; Salzer, R.; Wehrspohn, R.; Kley, E.-B.; Tünnermann, A.; Pertsch, T. Optical Modeling of Needle like Silicon Surfaces Produced by an ICP-RIE Process. In Proceedings of the fo SPIE Photonics Europe, Brussels, Belgium, 12–16 April 2010; Wehrspohn, R.B., Gombert, A., Eds.; SPIE: Brussels, Belgium, 2010; p. 772505. [Google Scholar]
- Jansen, H.; de Boer, M.; Legtenberg, R.; Elwenspoek, M. The Black Silicon Method: A Universal Method for Determining the Parameter Setting of a Fluorine-Based Reactive Ion Etcher in Deep Silicon Trench Etching with Profile Control. J. Micromech. Microeng. 1995, 5, 115. [Google Scholar] [CrossRef]
- Holland-Moritz, H.; Ilinov, A.; Djurabekova, F.; Nordlund, K.; Ronning, C. Sputtering and Redeposition of Ion Irradiated Au Nanoparticle Arrays: Direct Comparison of Simulations to Experiments. New J. Phys. 2017, 19, 013023. [Google Scholar] [CrossRef]
- Lugstein, A.; Basnar, B.; Smoliner, J.; Bertagnolli, E. FIB Processing of Silicon in the Nanoscale Regime. Appl. Phys. A Mater. Sci. Process. 2003, 76, 545–548. [Google Scholar] [CrossRef]
- Lindsey, S.; Waid, S.; Hobler, G.; Wanzenböck, H.D.; Bertagnolli, E. Inverse Modeling of FIB Milling by Dose Profile Optimization. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. At. 2014, 341, 77–83. [Google Scholar] [CrossRef]
- Kim, H.-B.; Hobler, G.; Steiger, A.; Lugstein, A.; Bertagnolli, E. Full Three-Dimensional Simulation of Focused Ion Beam Micro/Nanofabrication. Nanotechnology 2007, 18, 245303. [Google Scholar] [CrossRef]
- Rumyantsev, A.V.; Borgardt, N.I.; Volkov, R.L. Simulation of Redeposited Silicon Sputtering under Focused Ion Beam Irradiation. J. Surf. Investig. 2018, 12, 607–612. [Google Scholar] [CrossRef]
- Gassend, B.L.P.; Velasquez-Garcia, L.F.; Akinwande, A.I. Design and Fabrication of DRIE-Patterned Complex Needlelike Silicon Structures. J. Microelectromech. Syst. 2010, 19, 589–598. [Google Scholar] [CrossRef]
- Grigoropoulos, S. Highly Anisotropic Silicon Reactive Ion Etching for Nanofabrication Using Mixtures OfSF[Sub 6]/CHF[Sub 3] Gases. J. Vac. Sci. Technol. B 1997, 15, 640. [Google Scholar] [CrossRef]
- Franz, G.; Oberhausen, W.; Meyer, R.; Amann, M.-C. Residual-Free Reactive Ion Etching of Gold Layers. AIP Adv. 2018, 8, 075026. [Google Scholar] [CrossRef]
- Micro Resist Technology OrmoComp®—Micro Resist Technology. Available online: https://www.microresist.de/produkt/ormocomp/ (accessed on 21 February 2023).
- Kidwell, D.A.; Lee, W.-K.; Perkins, K.; Gilpin, K.M.; O’Shaughnessy, T.J.; Robinson, J.T.; Sheehan, P.E.; Mulvaney, S.P. Chemistries for Making Additive Nanolithography in OrmoComp Permissive for Cell Adhesion and Growth. ACS Appl. Mater. Interfaces 2019, 11, 19793–19798. [Google Scholar] [CrossRef]
- Stensborg Welcome to Stensborg A/S. Available online: https://www.stensborg.com/ (accessed on 15 December 2022).
- Zhuang, G.; Kutter, J.P. Anti-Stiction Coating of PDMS Moulds for Rapid Microchannel Fabrication by Double Replica Moulding. J. Micromech. Microeng. 2011, 21, 105020. [Google Scholar] [CrossRef]
- Mühlberger, M.; Bergmair, I.; Klukowska, A.; Kolander, A.; Leichtfried, H.; Platzgummer, E.; Loeschner, H.; Ebm, C.h.; Grützner, G.; Schöftner, R. UV-NIL with Working Stamps Made from Ormostamp. Microelectron. Eng. 2009, 86, 691–693. [Google Scholar] [CrossRef]
- PROFACTOR BGL-GZ-83, Profactor: Steyr, Austria, 2022.
- Deckgläser, Rund. Available online: https://at.vwr.com/store/product/11973345/deckglaser-rund (accessed on 9 December 2022).
- Diener Elektronik Nano Plasmaanlage Diener Elektronik. Available online: https://www.plasma.com/produkte/plasmaanlagen/niederdruckplasma-anlagen/nano/ (accessed on 29 November 2017).
- PROFACTOR HMNP-12|Coatings|Profactor. Available online: https://www.profactor.at/en/solutions/nil-coatings/#HMNP-12 (accessed on 3 October 2023).
- Stensborg Desktop R2P NanoImprinter. Available online: https://www.stensborg.com/desktop-r2p-nanoimprinter (accessed on 15 December 2022).
- Lee, H.; Im, J.S.; Choi, D.B.; An, J.; Kim, S.-B.; Yeon, S.; Yoon, S.; Woo, D.-H. Three-Dimensional Cardiac Organoid Formation Accelerates the Functional Maturation of Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Organoid 2022, 2, e14. [Google Scholar] [CrossRef]
- ICell Cardiomyocytes|FujiFilm Cellular Dynamics, Inc. Available online: https://www.fujifilmcdi.com/products/cardiac-cells/icell-cardiomyocytes (accessed on 12 December 2022).
- Kim, E.; Xia, Y.; Whitesides, G.M. Micromolding in Capillaries: Applications in Materials Science. J. Am. Chem. Soc. 1996, 118, 5722–5731. [Google Scholar] [CrossRef]
- Nečas, D.; Klapetek, P. Gwyddion: An Open-Source Software for SPM Data Analysis. Open Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]
- Smith, A.S.T.; Choi, E.; Gray, K.; Macadangdang, J.; Ahn, E.H.; Clark, E.C.; Laflamme, M.A.; Wu, J.C.; Murry, C.E.; Tung, L.; et al. NanoMEA: A Tool for High-Throughput, Electrophysiological Phenotyping of Patterned Excitable Cells. Nano Lett. 2020, 20, 1561–1570. [Google Scholar] [CrossRef]
- Verschuuren, M.A.; Megens, M.; Ni, Y.; van Sprang, H.; Polman, A. Large Area Nanoimprint by Substrate Conformal Imprint Lithography (SCIL). Adv. Opt. Technol. 2017, 6, 243–264. [Google Scholar] [CrossRef]
- Mohapatra, S.; Kumari, S.; Moirangthem, R.S. Fabrication of a Cost-Effective Polymer Nanograting as a Disposable Plasmonic Biosensor Using Nanoimprint Lithography. Mater. Res. Express 2017, 4, 076202. [Google Scholar] [CrossRef]
- Resnick, D.J.; Choi, J. A Review of Nanoimprint Lithography for High-Volume Semiconductor Device Manufacturing. Adv. Opt. Technol. 2017, 6, 229–241. [Google Scholar] [CrossRef]
- Buhl, J.; Yoo, D.; Köpke, M.; Gerken, M. Two-Dimensional Nanograting Fabrication by Multistep Nanoimprint Lithography and Ion Beam Etching. Nanomanufacturing 2021, 1, 39–48. [Google Scholar] [CrossRef]
- Oh, J.; Hoffman, J.B.; Hong, S.; Jo, K.D.; Román-Kustas, J.; Reed, J.H.; Dana, C.E.; Cropek, D.M.; Alleyne, M.; Miljkovic, N. Dissolvable Template Nanoimprint Lithography: A Facile and Versatile Nanoscale Replication Technique. Nano Lett. 2020, 20, 6989–6997. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Tzadka, S.; Yehuda, D.; Schvartzman, M. Soft Thermal Nanoimprint with a 10 Nm Feature Size. Soft Matter 2019, 15, 2897–2904. [Google Scholar] [CrossRef]
- Cox, L.M.; Martinez, A.M.; Blevins, A.K.; Sowan, N.; Ding, Y.; Bowman, C.N. Nanoimprint Lithography: Emergent Materials and Methods of Actuation. Nano Today 2020, 31, 100838. [Google Scholar] [CrossRef]
- Nowduri, B.; Schulte, S.; Decker, D.; Schäfer, K.-H.; Saumer, M. Biomimetic Nanostructures Fabricated by Nanoimprint Lithography for Improved Cell-Coupling. Adv. Funct. Mater. 2020, 30, 2004227. [Google Scholar] [CrossRef]
- Heitz, J.; Plamadeala, C.; Muck, M.; Armbruster, O.; Baumgartner, W.; Weth, A.; Steinwender, C.; Blessberger, H.; Kellermair, J.; Kirner, S.V.; et al. Femtosecond Laser-Induced Microstructures on Ti Substrates for Reduced Cell Adhesion. Appl. Phys. A 2017, 123, 734. [Google Scholar] [CrossRef]
- Fosodeder, P.; Baumgartner, W.; Steinwender, C.; Hassel, A.W.; Florian, C.; Bonse, J.; Heitz, J. Repellent Rings at Titanium Cylinders against Overgrowth by Fibroblasts. Adv. Opt. Technol. 2020, 9, 113–120. [Google Scholar] [CrossRef]
- Stensborg Soft Lithography with Desktop R2P NanoImprinter. Available online: https://www.youtube.com/watch?v=3GSQHuDmbHQ (accessed on 17 August 2023).
- Schmidt, C.; Deyett, A.; Ilmer, T.; Caballero, A.T.; Haendeler, S.; Pimpale, L.; Netzer, M.A.; Ginistrelli, L.C.; Cirigliano, M.; Mancheno, E.J.; et al. Multi-Chamber Cardioids Unravel Human Heart Development and Cardiac Defects. Biorxiv 2022. [Google Scholar] [CrossRef]
- Li, G.-R.; Feng, J.; Yue, L.; Carrier, M. Transmural Heterogeneity of Action Potentials AndI To1 in Myocytes Isolated from the Human Right Ventricle. Am. J. Physiol. Heart Circ. Physiol. 1998, 275, H369–H377. [Google Scholar] [CrossRef] [PubMed]
- Workman, A.J.; Kane, K.A.; Rankin, A.C. The Contribution of Ionic Currents to Changes in Refractoriness of Human Atrial Myocytes Associated with Chronic Atrial Fibrillation. Cardiovasc. Res. 2001, 52, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Guo, L.; Fiene, S.J.; Anson, B.D.; Thomson, J.A.; Kamp, T.J.; Kolaja, K.L.; Swanson, B.J.; January, C.T. High Purity Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes: Electrophysiological Properties of Action Potentials and Ionic Currents. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H2006–H2017. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mühlberger, M.M.; Kopp, S.; Deyett, A.A.; Pribyl, M.; Haslinger, M.J.; Siegel, A.M.; Taus, P.; Guillén, E.; Torres-Caballero, A.; Baltov, B.; et al. Nanoimprinted Hierarchical Micro-/Nanostructured Substrates for the Growth of Cardiomyocyte Fibers. Nanomanufacturing 2023, 3, 416-433. https://doi.org/10.3390/nanomanufacturing3040026
Mühlberger MM, Kopp S, Deyett AA, Pribyl M, Haslinger MJ, Siegel AM, Taus P, Guillén E, Torres-Caballero A, Baltov B, et al. Nanoimprinted Hierarchical Micro-/Nanostructured Substrates for the Growth of Cardiomyocyte Fibers. Nanomanufacturing. 2023; 3(4):416-433. https://doi.org/10.3390/nanomanufacturing3040026
Chicago/Turabian StyleMühlberger, Michael M., Sonja Kopp, Alison A. Deyett, Markus Pribyl, Michael J. Haslinger, Anica M. Siegel, Philipp Taus, Elena Guillén, Aranxa Torres-Caballero, Bozhidar Baltov, and et al. 2023. "Nanoimprinted Hierarchical Micro-/Nanostructured Substrates for the Growth of Cardiomyocyte Fibers" Nanomanufacturing 3, no. 4: 416-433. https://doi.org/10.3390/nanomanufacturing3040026
APA StyleMühlberger, M. M., Kopp, S., Deyett, A. A., Pribyl, M., Haslinger, M. J., Siegel, A. M., Taus, P., Guillén, E., Torres-Caballero, A., Baltov, B., Netzer, M. A., Prado-López, S., Yde, L., Stensborg, J., Mendjan, S., Hering, S., & Wanzenboeck, H. D. (2023). Nanoimprinted Hierarchical Micro-/Nanostructured Substrates for the Growth of Cardiomyocyte Fibers. Nanomanufacturing, 3(4), 416-433. https://doi.org/10.3390/nanomanufacturing3040026







