Differential Distribution of Trypanosoma vivax and Trypanosoma theileri in Cattle from Distinct Agroecological Regions of Central Argentina
Abstract
1. Introduction
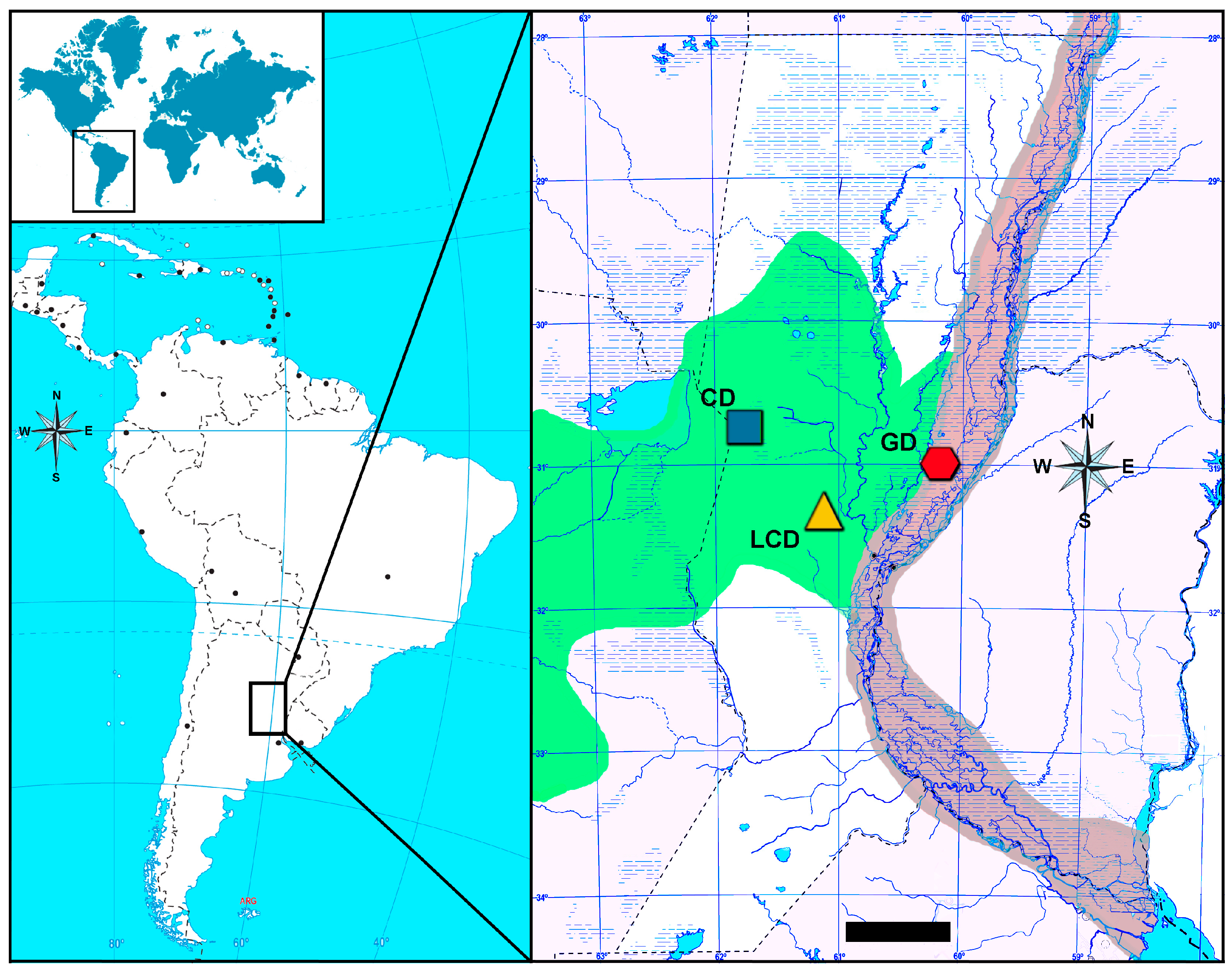
2. Materials and Methods
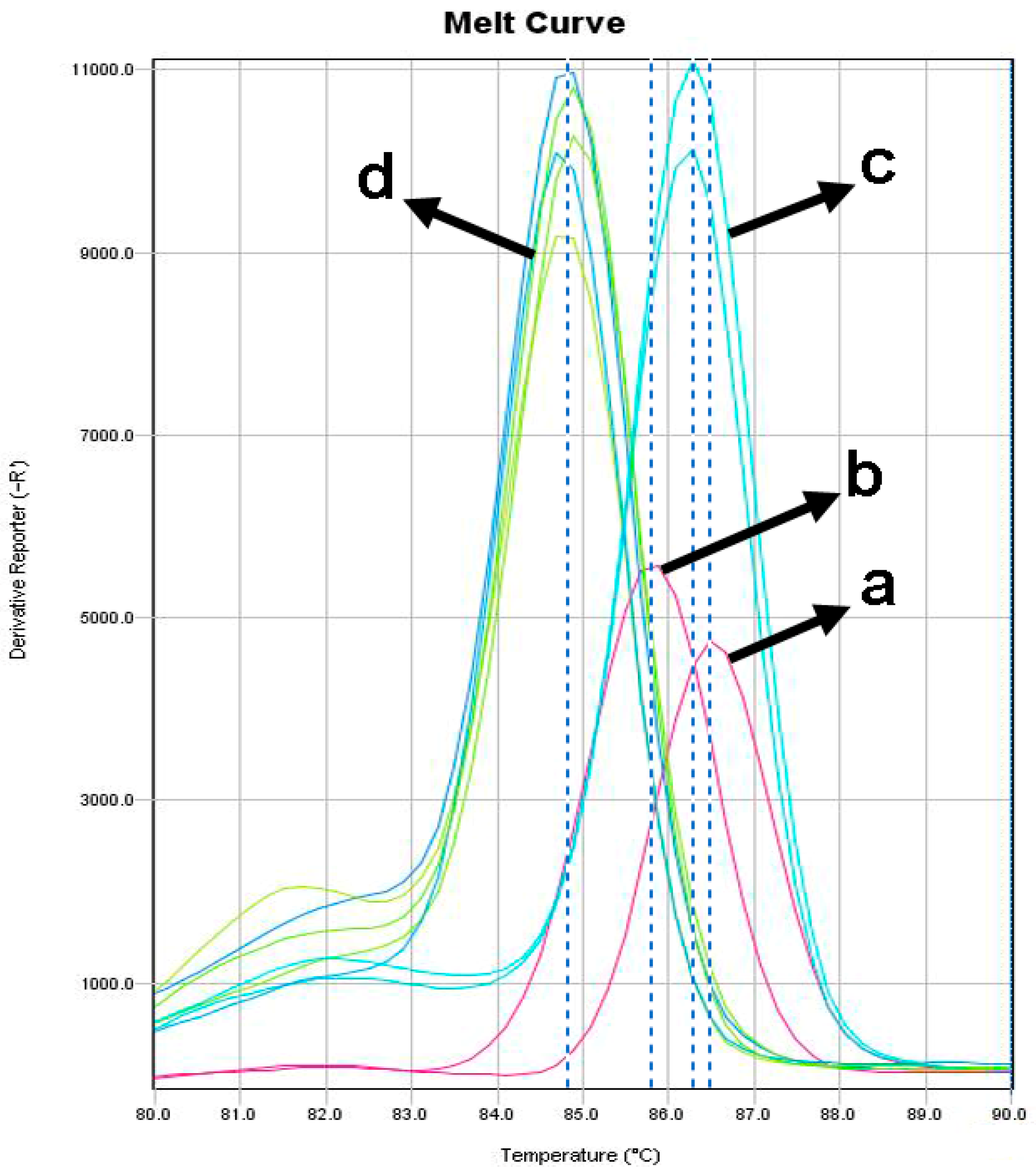
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CD | Castellanos department |
| LCD | Las Colonias department |
| GD | Garay department |
| PCR | Polymerase chain reaction |
| DNA | Deoxyribonucleic acid |
| rRNA | Ribosomal ribonucleic acid |
References
- Magez, S.; Radwanska, M. Trypanosomes and Trypanosomiasis; Springer: Vienna, Austria, 2014; ISBN 978-3-7091-1555-8. [Google Scholar]
- Muhanguzi, D.; Mugenyi, A.; Bigirwa, G.; Kamusiime, M.; Kitibwa, A.; Akurut, G.G.; Ochwo, S.; Amanyire, W.; Okech, S.G.; Hattendorf, J.; et al. African animal Trypanosomiasis as a constraint to livestock health and production in Karamoja region: A detailed qualitative and quantitative assessment. BMC Vet. Res. 2017, 13, 355. [Google Scholar] [CrossRef] [PubMed]
- Hounyèmè, R.E.; Kaboré, J.; Gimonneau, G.; Somda, M.B.; Salou, E.; Missihoun, A.A.; Bengaly, Z.; Jamonneau, V.; Boulangé, A. Molecular epidemiology of animal African Trypanosomosis in southwest Burkina Faso. PLoS Negl. Trop. Dis. 2022, 16, e0010106. [Google Scholar] [CrossRef]
- Bontempi, I.A.; Arias, D.G.; Castro, G.V.; Peverengo, L.M.; Díaz, G.F.; Allassia, M.; Greif, G.; Marcipar, I. Improved serodiagnosis of Trypanosoma vivax infections in cattle reveals high infection rates in the livestock regions of Argentina. PLoS Negl. Trop. Dis. 2024, 18, e0012020. [Google Scholar] [CrossRef]
- Cadioli, F.A.; Barnabé, P.D.A.; Machado, R.Z.; Teixeira, M.C.A.; André, M.R.; Sampaio, P.H.; Fidélis Junior, O.L.; Teixeira, M.M.G.; Marques, L.C. First report of Trypanosoma vivax outbreak in dairy cattle in São Paulo state, Brazil. Rev. Bras. Parasitol. Vet. 2012, 21, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Fritzen, A.; De Vitt, M.G.; Deolindo, G.L.; Signor, M.H.; Correa, N.G.; Ribeiro, B.G.; Marques, J.; Das Neves, G.B.; Miletti, L.C.; Da Silva, A.S. Outbreak of Trypanosoma vivax in dairy cows: Hematologic, immunological and antioxidant responses before and after treatment with isometamidium chloride. Pathogens 2025, 14, 143. [Google Scholar] [CrossRef]
- Desquesnes, M. Livestock Trypanosomoses and Their Vectors in Latin America; OIE: Paris, France, 2004; ISBN 978-92-9044-634-7. [Google Scholar]
- USDA. Dairy Heifer Raiser Health and Management Practices on U.S. Dairy Operations, 2014. Available online: https://www.aphis.usda.gov/sites/default/files/dairy14_dr_partiii.pdf (accessed on 27 March 2025).
- De Melo Junior, R.D.; Azeredo Bastos, T.S.; Heller, L.M.; Couto, L.F.M.; Zapa, D.M.B.; De Assis Cavalcante, A.S.; Cruvinel, L.B.; Nicaretta, J.E.; Iuasse, H.V.; Ferreira, L.L.; et al. How many cattle can be infected by Trypanosoma vivax by reusing the same needle and syringe, and what is the viability time of this protozoan in injectable veterinary products? Parasitology 2022, 149, 270–282. [Google Scholar] [CrossRef]
- Bastos, T.S.A.; Faria, A.M.; Madrid, D.M.D.C.; Bessa, L.C.D.; Linhares, G.F.C.; Fidelis Junior, O.L.; Sampaio, P.H.; Cruz, B.C.; Cruvinel, L.B.; Nicaretta, J.E.; et al. First outbreak and subsequent cases of Trypanosoma vivax in the state of Goiás, Brazil. Rev. Bras. Parasitol. Vet. 2017, 26, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Monzón, C.M.; Mancebo, O.A.; Giménez, J.M.; Russo, A.M. Evolución de la Trypanosomosis bovina por Trypanosoma vivax en Formosa (Argentina). Años 2007-2012 y su potencial dispersión en el país. Rev. Ibero-Latinoam. de Parasitol. 2013, 72, 38–44. [Google Scholar]
- Abdala, A.A.; Larriestra, A.J.; Signorini, M. Estimación de pérdidas económicas causadas por Trypanosoma vivax en un rodeo lechero de Argentina. Rev. Vet. 2020, 31, 115. [Google Scholar] [CrossRef]
- Florentin, A.S.; Garcia Perez, H.A.; Rodrigues, C.M.F.; Dubois, E.F.; Monzón, C.M.; Teixeira, M.M.G. Molecular epidemiological insights into Trypanosoma vivax in Argentina: From the endemic Gran Chaco to outbreaks in the Pampas. Transbounding Emerg. Dis. 2022, 69, 1364–1374. [Google Scholar] [CrossRef]
- Garcia, H.A.; Blanco, P.A.; Rodrigues, A.C.; Rodrigues, C.M.F.; Takata, C.S.A.; Campaner, M.; Camargo, E.P.; Teixeira, M.M.G. Pan-American Trypanosoma (Megatrypanum) trinaperronei n. sp. in the white-tailed deer Odocoileus virginianus Zimmermann and its deer ked Lipoptena mazamae Rondani, 1878: Morphological, developmental and phylogeographical characterisation. Parasites Vectors 2020, 13, 308. [Google Scholar] [CrossRef]
- Rodrigues, A.C.; Campaner, M.; Takata, C.S.A.; Dell’ Porto, A.; Milder, R.V.; Takeda, G.F.; Teixeira, M.M.G. Brazilian isolates of Trypanosoma (Megatrypanum) theileri: Diagnosis and differentiation of isolates from cattle and water buffalo based on biological characteristics and randomly amplified DNA sequences. Vet. Parasitol. 2003, 116, 185–207. [Google Scholar] [CrossRef]
- Kostygov, A.Y.; Frolov, A.O.; Malysheva, M.N.; Ganyukova, A.I.; Drachko, D.; Yurchenko, V.; Agasoi, V.V. Development of two species of the Trypanosoma theileri complex in Tabanids. Parasites Vectors 2022, 15, 95. [Google Scholar] [CrossRef]
- Brotánková, A.; Fialová, M.; Čepička, I.; Brzoňová, J.; Svobodová, M. Trypanosomes of the Trypanosoma theileri group: Phylogeny and new potential vectors. Microorganisms 2022, 10, 294. [Google Scholar] [CrossRef]
- Calzolari, M.; Rugna, G.; Clementi, E.; Carra, E.; Pinna, M.; Bergamini, F.; Fabbi, M.; Dottori, M.; Sacchi, L.; Votýpka, J. Isolation of a trypanosome related to Trypanosoma theileri (Kinetoplastea: Trypanosomatidae) from Phlebotomus perfiliewi (Diptera: Psychodidae). BioMed Res. Int. 2018, 2018, 2597074. [Google Scholar] [CrossRef]
- Votýpka, J.; Rádrová, J.; Skalický, T.; Jirků, M.; Jirsová, D.; Mihalca, A.D.; D’Amico, G.; Petrželková, K.J.; Modrý, D.; Lukeš, J. A tsetse and Tabanid fly survey of African great apes habitats reveals the presence of a novel trypanosome lineage but the absence of Trypanosoma brucei. Int. J. Parasitol. 2015, 45, 741–748. [Google Scholar] [CrossRef]
- Latif, A.A.; Bakheit, M.A.; Mohamed, A.E.E.; Zweygarth, E. High infection rates of the tick Hyalomma anatolicum anatolicum with Trypanosoma theileri. Onderstepoort J. Vet. Res. 2004, 71, 251–256. [Google Scholar] [CrossRef]
- Martins, J.R.; Leite, R.C.; Doyle, R.L. Tripanosomatides like Trypanosoma theileri in the cattle tick Boophilus microplus. Rev. Bras. Parasitol. Vet. 2008, 17, 113–114. [Google Scholar] [CrossRef]
- Braun, U.; Rogg, E.; Walser, M.; Nehrbass, D.; Guscetti, F.; Mathis, A.; Deplazes, P. Trypanosoma theileri in the cerebrospinal fluid and brain of a heifer with suppurative meningoencephalitis. Vet. Rec. 2002, 150, 18–19. [Google Scholar] [CrossRef]
- Amato, B.; Mira, F.; Di Marco Lo Presti, V.; Guercio, A.; Russotto, L.; Gucciardi, F.; Vitale, M.; Lena, A.; Loria, G.R.; Puleio, R.; et al. A case of bovine trypanosomiasis caused by Trypanosoma theileri in Sicily, Italy. Parasitol. Res. 2019, 118, 2723–2727. [Google Scholar] [CrossRef]
- Suganuma, K.; Kayano, M.; Kida, K.; Gröhn, Y.T.; Miura, R.; Ohari, Y.; Mizushima, D.; Inoue, N. Genetic and seasonal variations of Trypanosoma theileri and the association of Trypanosoma theileri infection with dairy cattle productivity in northern Japan. Parasitol. Int. 2022, 86, 102476. [Google Scholar] [CrossRef]
- Morello, J.H.; Matteucci, S.D.; Rodriguez, A.F.; Silva, M.E. Ecorregiones y Complejos Ecosistémicos Argentinos, Primera edición; Facultad de Arquitectura, Diseño y Urbanismo, GEPAMA Grupo de Ecología del Paisaje y Medio Ambiente, Universidad de Buenos Aires: Buenos Aires, Argentina, 2012; ISBN 978-987-1922-00-0. [Google Scholar]
- Eberhardt, A.T.; Monje, L.D.; Zurvera, D.A.; Beldomenico, P.M. Detection of Trypanosoma evansi infection in wild capybaras from Argentina using smear mcroscopy and real-time PCR assays. Vet. Parasitol. 2014, 202, 226–233. [Google Scholar] [CrossRef]
- Monje, L.D.; Costa, F.B.; Colombo, V.C.; Labruna, M.B.; Antoniazzi, L.R.; Gamietea, I.; Nava, S.; Beldomenico, P.M. Dynamics of exposure to Rickettsia parkeri in cattle in the Paraná River Delta, Argentina. J. Med. Entomol. 2016, 53, 660–665. [Google Scholar] [CrossRef]
- Keatley, S.; Botero, A.; Fosu-Nyarko, J.; Pallant, L.; Northover, A.; Thompson, R.C.A. Species-level identification of trypanosomes infecting Australian wildlife by high-resolution melting—real time quantitative polymerase chain reaction (HRM-qPCR). Int. J. Parasitol. Parasites Wildl. 2020, 13, 261–268. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- De Souza Pimentel, D.; Do Nascimento Ramos, C.A.; Ramos, R.A.D.N.; De Araújo, F.R.; Borba, M.L.; Da Gloria Faustino, M.A.; Alves, L.C. First report and molecular characterization of Trypanosoma vivax in cattle from state of Pernambuco, Brazil. Vet. Parasitol. 2012, 185, 286–289. [Google Scholar] [CrossRef]
- Böse, R.; Friedhoff, K.T.; Olbrich, S. Transmission of Megatrypanum trypanosomes to Cervus dama by Tabanidae. J. Protozool. 1987, 34, 110–113. [Google Scholar] [CrossRef]
- Quintana, M.G.; Fernández, M.S.; Salomón, O.D. Distribution and abundance of Phlebotominae, vectors of Leishmaniasis, in Argentina: Spatial and temporal analysis at different scales. J. Trop. Med. 2012, 2012, 652803. [Google Scholar] [CrossRef]
- Lucas, M.; Krolow, T.K.; Riet-Correa, F.; Barros, A.T.M.; Krüger, R.F.; Saravia, A.; Miraballes, C. Diversity and seasonality of horse flies (Diptera: Tabanidae) in Uruguay. Sci. Rep. 2020, 10, 401. [Google Scholar] [CrossRef]
- Maraghi, S.; Molyneux, D.H. Studies on cross-immunity in Herpetosoma trypanosomes of Microtus, Clethrionomys and Apodemus. Parasitol. Res. 1989, 75, 175–177. [Google Scholar] [CrossRef]
- Welburn, S.; Picozzi, K.; Coleman, P.G.; Packer, C. Patterns in age-seroprevalence consistent with acquired immunity against Trypanosoma brucei in Serengeti lions. PLoS Negl. Trop. Dis. 2008, 2, e347. [Google Scholar] [CrossRef] [PubMed]
- Doherty, M.L.; Windle, H.; Voorheis, H.P.; Larkin, H.; Casey, M.; Clery, D.; Murray, M. Clinical disease associated with Trypanosoma theileri infection in a calf in Ireland. Vet. Rec. 1993, 132, 653–656. [Google Scholar] [CrossRef]
- Hajihassani, A.; Maroufi, S.; Esmaeilnejad, B.; Khorram, H.; Tavassoli, M.; Dalir-Naghadeh, B.; Samiei, A. Hemolytic anemia associated with Trypanosoma theileri in a cow from Kurdistan province, west of Iran. Vet. Res. Forum. 2020, 11, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Sood, N.K.; Singla, L.D.; Singh, R.S.; Uppal, S.K. Association of Trypanosoma theileri with peritonitis in a pregnant cross-bred cow: A case report. Vet. Med. 2011, 56, 82–84. [Google Scholar] [CrossRef]

| Trypanosoma Prevalence by Breed | Trypanosoma Prevalence by Site | Trypanosoma spp. Prevalence | ||||
|---|---|---|---|---|---|---|
| Dairy Breed (Holando Argentino) | Beef Breed (Braford) | Delta and Islands of Paraná | Espinal | |||
| Garay Department | Castellanos Department | Las Colonias Department | ||||
| Trypanosoma spp. | 8.27 ± 4.68% (11/133) | 21.8 ± 8.68% (19/87) | 8.63 ± 3.71% (19/220) | 0.91 ± 1.25% (2/220) | 4.09 ± 2.62% (9/220) | 13.63 ± 4.53% (30/220) |
| Trypanosoma vivax | 4.51 ± 3.53% (6/133) | ND | ND | 0.91 ± 1.25% (2/220) | 1.82 ± 1.77% (4/220) | 2.73 ± 2.15% (6/220) |
| Trypanosoma theileri | 3.75 ± 3.23% (5/133) | 21.8 ± 8.68% (19/87) | 8.63 ± 3.71% (19/220) | ND | 2.27 ± 1.97% (5/220) | 10.91 ± 4.12% (24/220) |
| Prevalence Comparison | Observed Trend | p-Value |
|---|---|---|
| Trypanosoma spp. prevalence by site | LCD = CD < GD | 0.0069 *,a |
| Trypanosoma theileri prevalence by site | LCD = CD < GD | 7.29 × 10−5 *,a |
| Trypanosoma vivax prevalence by site | LCD = CD = GD | 0.087 b |
| Trypanosoma spp. prevalence by cattle type | Dairy < Beef | 0.0197 *,b |
| Trypanosoma theileri prevalence by cattle type | Dairy < Beef | 0.0001 *,b |
| Trypanosoma vivax prevalence by cattle type | Dairy > Beef | 0.027 *,b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Facelli Fernández, M.C.; Barolin, J.; Allassia, M.; Gonzalez, J.H.; Beldomenico, P.M.; Monje, L.D. Differential Distribution of Trypanosoma vivax and Trypanosoma theileri in Cattle from Distinct Agroecological Regions of Central Argentina. Parasitologia 2025, 5, 27. https://doi.org/10.3390/parasitologia5020027
Facelli Fernández MC, Barolin J, Allassia M, Gonzalez JH, Beldomenico PM, Monje LD. Differential Distribution of Trypanosoma vivax and Trypanosoma theileri in Cattle from Distinct Agroecological Regions of Central Argentina. Parasitologia. 2025; 5(2):27. https://doi.org/10.3390/parasitologia5020027
Chicago/Turabian StyleFacelli Fernández, Maria Celeste, Johann Barolin, Martin Allassia, Javier Hernan Gonzalez, Pablo Martin Beldomenico, and Lucas Daniel Monje. 2025. "Differential Distribution of Trypanosoma vivax and Trypanosoma theileri in Cattle from Distinct Agroecological Regions of Central Argentina" Parasitologia 5, no. 2: 27. https://doi.org/10.3390/parasitologia5020027
APA StyleFacelli Fernández, M. C., Barolin, J., Allassia, M., Gonzalez, J. H., Beldomenico, P. M., & Monje, L. D. (2025). Differential Distribution of Trypanosoma vivax and Trypanosoma theileri in Cattle from Distinct Agroecological Regions of Central Argentina. Parasitologia, 5(2), 27. https://doi.org/10.3390/parasitologia5020027







