Hirsutonosema embarrassi n. gen. n. sp. (Phylum Microsporidia) in the Ovary of Mucket (Actinonaias ligamentina), Plain Pocketbook (Lampsilis cardium), and Fatmucket (Lampsilis siliquoidea) (Unionidae) from the Embarrass River, Wisconsin, USA †
Abstract
1. Introduction
2. Results
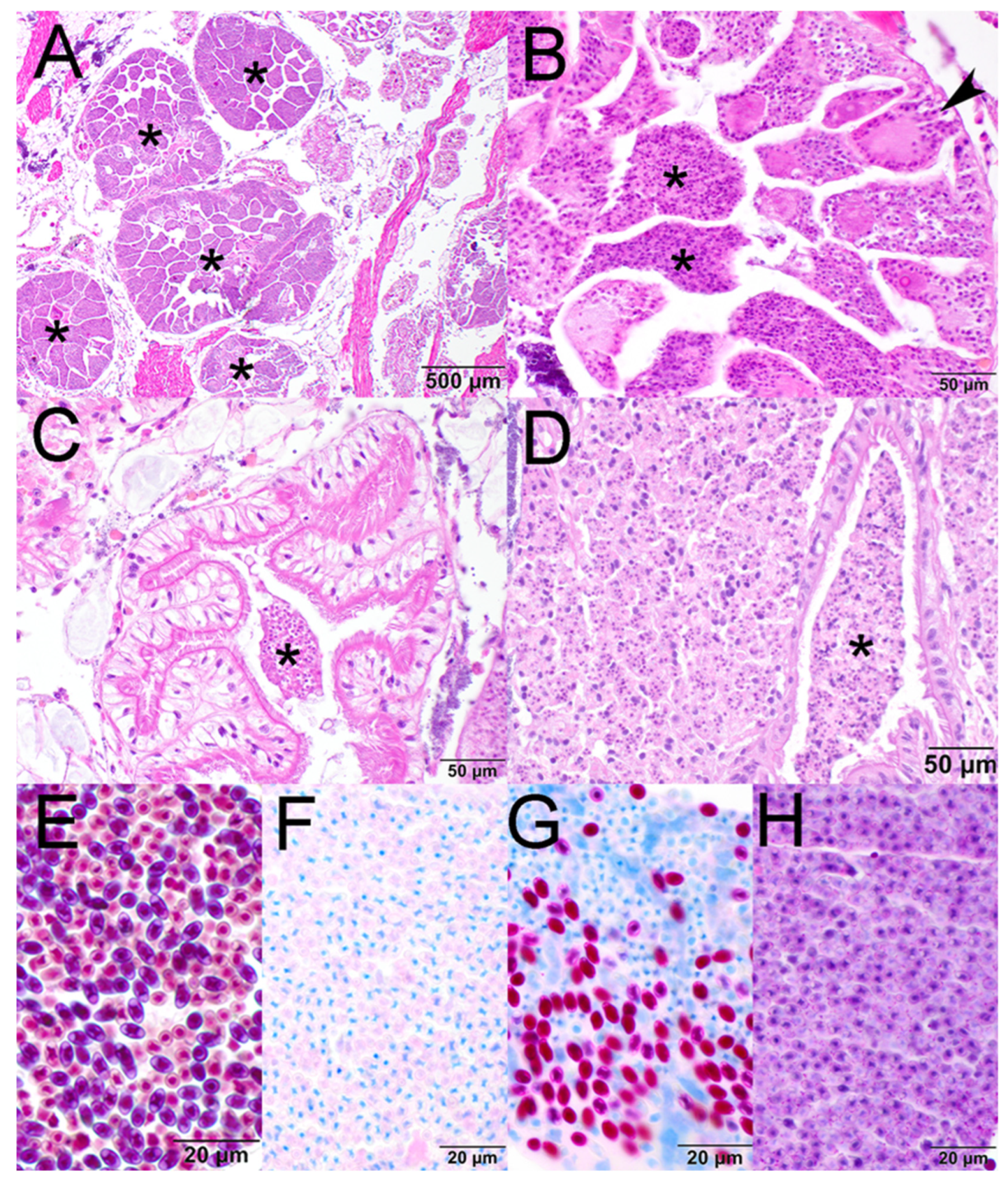
2.1. Histopathology
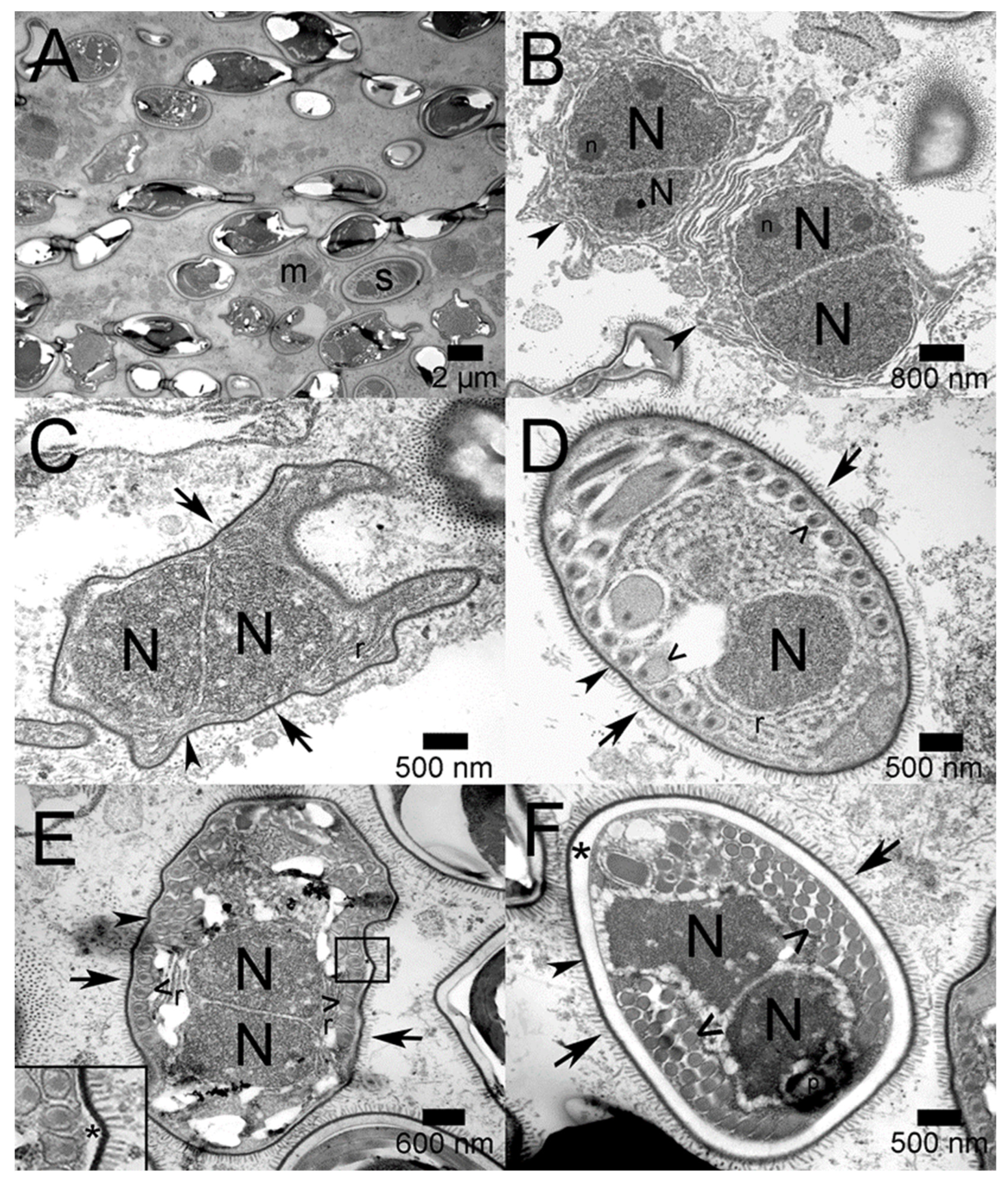
2.2. Transmission Electron Microscopy
2.3. Molecular Identification
2.4. qPCR
2.5. Taxonomic Description
2.5.1. Higher Taxonomy
2.5.2. Taxonomic Summary
- Type host: A. ligamentina (Mucket).
- Other hosts: L. siliquoidea (Fat Mucket); L. cardium (Plain Pocketbook).
- Site of infection: Cytoplasm of oocytes.
- Type locality: Embarrass River, Wisconsin, USA (44.739844° N; 88.798594° W).
- Prevalence in type host: three of five females.
- Specimens deposited: Hapantotype slides of stained gonadal tissue were
- deposited in the Smithsonian National Museum of Natural History (Washington, DC, USA) under accessions USNM 1607135 (host: A. ligamentina), USNM 1607136 (host: L. cardium), and USNM 1607137 (host: L. siliquoidea).
- Etymology: This genus name is in regard to morphology as the Latin word hirsute means “hairy”, which describes the appearance of the exospore coat. The species name is in reference to the type locality.
- DNA sequences: Partial SSU rRNA genes were submitted to GenBank under Accessions PP777264 (host: A. ligamentina; 1356 bp) and PP777265 (host: L. cardium; 1206 bp).
2.5.3. Remarks upon the Taxonomic Summary
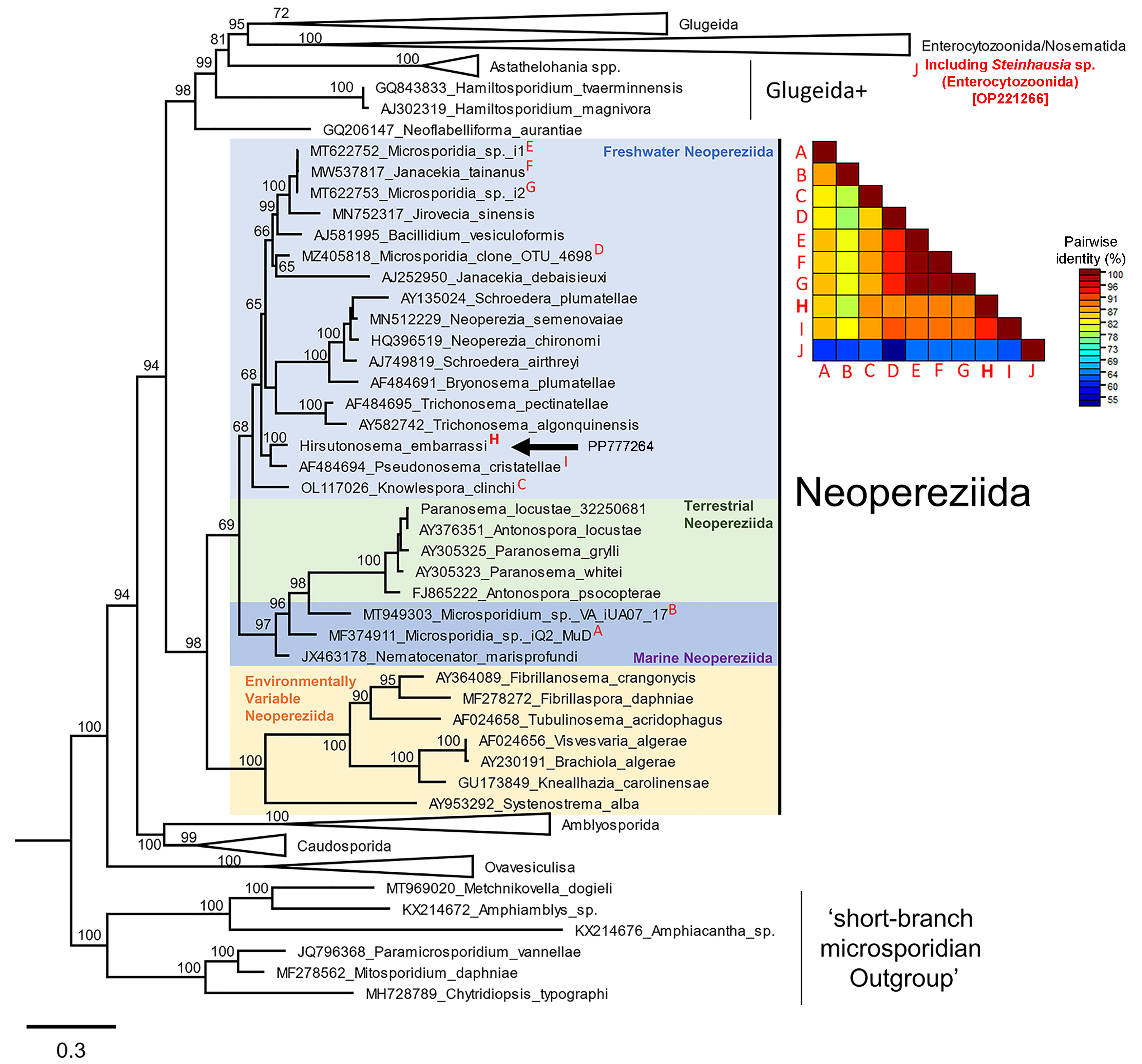
2.6. Phylogeny
3. Discussion
4. Materials and Methods
4.1. Experimental Design and Field Sampling
4.2. Tissue Processing
4.3. Transmission Electron Microscopy
4.4. Molecular Identification
4.5. qPCR Development
4.6. Phylogenetics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Graf, D.L.; Cummings, K.S. A ‘big data’ approach to global freshwater mussel diversity (Bivalvia: Unionoida), with an updated checklist of genera and species. J. Molluscan Stud. 2021, 87, eyaa034. [Google Scholar] [CrossRef]
- Lopes-Lima, M.; Burlakova, L.E.; Karatayev, A.Y.; Mehler, K.; Seddon, M.; Sousa, R. Conservation of freshwater bivalves at the global scale: Diversity, threats and research needs. Hydrobiologia 2018, 810, 1–14. [Google Scholar] [CrossRef]
- Downing, J.A.; Van Meter, P.; Woolnough, D.A. Suspects and evidence: A review of the causes of extirpation and decline in freshwater mussels. Anim. Biodivers. Conserv. 2010, 33, 151–185. [Google Scholar] [CrossRef]
- Haag, W.R. Reassessing enigmatic mussel declines in the United States. Freshw. Mollusk Biol. Conserv. 2019, 22, 43–60. [Google Scholar] [CrossRef]
- Wisconsin Mussel Species. Available online: https://wiatri.net/inventory/mussels/About/speciesList.cfm?theOrder=C (accessed on 14 August 2023).
- Listed Species with Spatial Current Range Believed to or Known to Occur in Wisconsin. Available online: https://ecos.fws.gov/ecp/report/species-listings-by-state?stateAbbrev=WI&stateName=Wisconsin&statusCategory=Listed (accessed on 14 August 2023).
- Wisconsin Endangered and Threatened Species Laws and List. Available online: https://p.widencdn.net/byxof6/ER001 (accessed on 14 August 2023).
- Wisconsin Natural Heritage Working List. Available online: https://dnr.wisconsin.gov/sites/default/files/topic/NHI/NHIWorkingList.pdf (accessed on 14 August 2023).
- The Clam Chronicle 2019 Spring. Available online: https://wiatri.net/inventory/mussels/news/pdf/2019Spring_ClamChronicle.pdf (accessed on 14 August 2023).
- Bojko, J.; Reinke, A.W.; Stentiford, G.D.; Williams, B.; Rogers, M.S.J.; Bass, D. Microsporidia: A new taxonomic, evolutionary, and ecological synthesis. Trends Parasitol. 2022, 38, 642–659. [Google Scholar] [CrossRef]
- Han, B.; Takvorian, P.M.; Weiss, L.M. Invasion of host cells by microsporidia. Front. Microbiol. 2020, 11, 172. [Google Scholar] [CrossRef]
- Dunn, A.; Smith, J. Microsporidian life cycles and diversity: The relationship between virulence and transmission. Microbes Infect. 2001, 3, 381–388. [Google Scholar] [CrossRef]
- Canning, E.U. Microsporidia. In Parasitic Protozoa, 2nd ed.; Kreier, J.P., Ed.; Academic Press, Inc.: San Diego, CA, USA, 1993; Volume 6, pp. 299–370. [Google Scholar]
- Vávra, J.; Ronny Larsson, J.I. Structure of Microsporidia. In Microsporidia: Pathogens of Opportunity; Weiss, L.M., Becnel, J.J., Eds.; John Wiley & Sons, Inc.: Chichester, UK, 2014; pp. 1–70. [Google Scholar]
- Cali, A.; Becnel, J.J.; Takvorian, P.M. Microsporidia. In Handbook of the Protists, 2nd ed.; Archibald, J.M., Simpson, A.G.B., Slamovits, C.H., Eds.; Springer International Publishing AG: Cham, Switzerland, 2017; pp. 1559–1618. [Google Scholar]
- Canning, E.U.; Refardt, D.; Vossbrinck, C.R.; Okamura, B.; Curry, A. New diplokaryotic microsporidia (Phylum Microsporidia) from freshwater bryozoans (Bryozoa, Phylactolaemata). Eur. J. Protistol. 2002, 38, 247–265. [Google Scholar] [CrossRef]
- Canning, E.U.; Okamura, B.; Curry, A. A new microsporidium, Nosema cristatellae n. sp. in the Bryozoan Cristatella mucedo (Bryozoa, Phylactolaemata). J. Invertebr. Pathol. 1997, 70, 177–183. [Google Scholar] [CrossRef]
- Weng, M.; Zhang, J.; Li, A.; Zhang, Q.; Sato, H. Morphological and molecular characterization of a new microsporidium, Janacekia tainanus n. sp. from the adipose tissue of Kiefferulus tainanus (Diptera: Chironomidae) in China. J. Invertebr. Pathol. 2021, 182, 107578. [Google Scholar] [CrossRef]
- Liu, X.; Ren, S.; Chen, Z.; Yin, Q.; Xiang, J.; Yu, J.; Li, D.; Zhang, J. Jirovecia branchilis n. sp. (Microsporidia) from glands of Branchiura sowerbyi (Oligochaeta: Tubificidae) in China. Eur. J. Protistol. 2023, 88, 125972. [Google Scholar] [CrossRef]
- Morris, D.J.; Terry, R.S.; Ferguson, K.B.; Smith, J.E.; Adams, A. Ultrastructural and molecular characterization of Bacillidium vesiculoformis n. sp. (Microspora: Mrazekiidae) in the freshwater oligochaete Nais simplex (Oligochaeta: Naididae). Parasitology 2005, 130, 31–40. [Google Scholar] [CrossRef]
- Knowles, S.; Leis, E.M.; Richard, J.C.; Cole, R.; Agbalog, R.E.; Putnam, J.G.; Goldberg, T.L.; Waller, D.L. A novel gonadotropic microsporidian parasite (Microsporidium clinchi n. sp.) infecting a declining population of pheasantshell mussels (Actinonaias pectorosa) (Unioinidae) from the Clinch River, USA. Parasitologia 2022, 2, 1–12. [Google Scholar] [CrossRef]
- Bojko, J. Systematic identity and phylogenetic analysis of Knowlespora clinchi (Knowles et al. 2022) gen. et comb. nov. from pheasantshell mussels (Actinonaiais pectorosa). J. Invertebr. Pathol. 2022, 194, 107817. [Google Scholar] [CrossRef]
- Liu, X.; Ren, S.; Chen, Z.; Yuan, X.; Song, R.; Yu, J.; Li, D.; Xiang, J.; Zhang, J. Two new species of Bacillidium (Microsporidia) from coelomocytes of Branchiura sowerbyi (Oligochaeta: Naididae) in China. J. Invertebr. Pathol. 2022, 192, 107785. [Google Scholar] [CrossRef]
- Sapir, A.; Dillman, A.R.; Connon, S.A.; Grupe, B.M.; Ingels, J.; Mundo-Ocampo, M.; Levin, L.A.; Baldwin, J.G.; Orphan, V.J.; Sternberg, P.W. Microsporidia-nematode associations in methane seeps reveal basal fungal parasitism in the deep sea. Front. Microbiol. 2014, 5, 43. [Google Scholar] [CrossRef]
- Liu, X.; Wen, M.; Zhao, Y.; Li, A.; Zhang, J. Morphological and molecular characterization of a new freshwater microsporidium, Jirovecia sinensis sp. n. (Microsporidia) infecting the coelomocytes of Branchiura sowerbyi (Oligochaeta: Naididae) in China. J. Invertebr. Pathol. 2020, 173, 107368. [Google Scholar] [CrossRef]
- Morris, D.J.; Terry, R.S.; Adams, A. Development and molecular characterisation of the microsporidian Schroedera airthreyi n. sp. in a freshwater bryozoan Plumatella sp. (Bryozoa: Phylactolaemata). J. Eukaryot. Microbiol. 2005, 52, 31–37. [Google Scholar] [CrossRef]
- Issi, I.V.; Tokarev, Y.S.; Seliverstova, E.V.; Voronin, V.N. Taxonomy of Neoperezia chironomi and Neoperezia semenovaiae comb. nov. (Microsporidia, Aquasporidia): Lessons from ultrastructure and ribosomal DNA sequence data. Eur. J. Protistol. 2012, 48, 17–29. [Google Scholar] [CrossRef]
- Desser, S.S.; Koehler, A.; Barta, J.R.; Kamyab, J.; Ringuette, M.J. Trichonosema algonquinensis n. sp. (Phylum microsporidia) in Pectinatella magnifica (Bryozoa: Phylactolaemata) from Algonquin Park, Ontario, Canada. J. Eukaryot. Microbiol. 2004, 51, 389–393. [Google Scholar] [CrossRef]
- Morris, D.J.; Adams, A. Development of Schroedera plumatellae gen. n., sp. n. (Microsporidia) in Plumatella fungosa (Bryozoa: Phylactolaemata). Acta Protozool. 2002, 41, 383–396. [Google Scholar]
- Vossbrinck, C.R.; Andreadis, T.G.; Vavra, J.; Becnel, J.J. Molecular phylogeny and evolution of mosquito parasitic Microsporidia (Microsporidia: Amblyosporidae). J. Eukaryot. Microbiol. 2004, 51, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Cheney, S.A.; Lafranchi-Tristem, N.J.; Canning, E.U. Phylogenetic relationships of Pleistophora-like microsporidia based on small subunit ribosomal DNA sequences and implications for the source of Trachipleistophora hominis infections. J. Eukaryot. Microbiol. 2000, 47, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, F. Small subunit rDNA phylogeny of Bacillidium sp. (Microspora, Mrazekiidae) infecting oligochaets. Parasitology 1999, 118 Pt 6, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Karpov, S.A.; Mamkaeva, M.A.; Aleoshin, V.V.; Nassonova, E.; Lilje, O.; Gleason, F.H. Morphology, phylogeny, and ecology of the aphelids (Aphelidea, Opisthokonta) and proposal for the new superphylum Opisthosporidia. Front. Microbiol. 2014, 5, 112. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Balbiani, E. Sur les microsporidies ou psorospermies des articulés. C. R. Acad. Sci. 1882, 95, 1168–1171. [Google Scholar]
- Vossbrinck, C.R.; Debrunner-Vossbrinck, B.; Weiss, L. Phylogeny of the microsporidia. In Microsporidia: Pathogens of Opportunity, 1st ed.; Weiss, L.M., Becnel, J.J., Eds.; John Wiley & Sons, Inc.: Chinchester, UK, 2014; pp. 203–220. [Google Scholar]
- Voronin, V. On a macrotaxonomy of the phylum Microsporidia. Parazitologiia 2001, 35, 35–44. [Google Scholar] [PubMed]
- Carella, F.; De Vico, G. Pathology, epidemiology, and phylogeny of mussel egg disease due to the microsporidian Steinhausia mytilovum (Field, 1924) in the Mediterranean mussel (Mytilus galloprovincialis). J. Invertebr. Pathol. 2023, 198, 107927. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Field, I.A. Biology and economic value of the sea mussel Mytilus edulis. Bull. US Bur. Fish. 1923, 38, 127–259. [Google Scholar]
- Léger, L.; Hollande, A. Sur un nouveau protiste à facies de Chytridiopsis, parasite des ovules de l’huit. CR Séanc. Soc. Biol. 1917, 80, 61–64. [Google Scholar]
- Farley, C.A. Neoplasms in estuarine mollusks and approaches ot ascertain causes. Ann. N. Y. Acad. Sci. 1977, 298, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Villalba, A.; Carballal, M.J.; López, M.C. Pathologic Conditions of Three Carpet Shell Clam Species of Galicia (NW. of Spain), Proceedings of World Aquaculture ’93—From discovery to commercialization, Torremolinos, Spain, 26–28 May 1993; Carrillo, M., Ed.; European Aquaculture Society: Oostende, Belgium, 1993; p. 85. [Google Scholar]
- Anderson, T.; Hine, P.; Lester, R. A Steinhausia-like infection in the ovocytes of Sydney rock oysters Saccostrea commercialis. Dis. Aquat. Organ. 1995, 22, 143–146. [Google Scholar] [CrossRef]
- Carballal, M.J.; Iglesias, D.; Santamarina, J.; Ferro-Soto, B.; Villalba, A. Parasites and pathologic conditions of the cockle Cerastoderma edule populations of the coast of Galicia (NW Spain). J. Invertebr. Pathol. 2001, 78, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Comtet, T.; Garcia, C.; Le Coguic, Y.; Joly, J.P. Infection of the cockle Cerastoderma edule in the Baie des Veys (France) by the microsporidian parasite Steinhausia sp. Dis. Aquat. Organ. 2003, 57, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.M.D.; Scardua, M.P.; Vieira, C.B.; Alves, A.C.; Dungan, C.F. Survey of pathologies in Crassostrea gasar (Adanson, 1757) oysters from cultured and wild populations in the São Francisco Estuary, Sergipe, Northeast Brazil. J. Shellfish. Res. 2015, 34, 289–296. [Google Scholar] [CrossRef]
- Wadi, L.; Reinke, A.W. Evolution of microsporidia: An extremely successful group of eukaryotic intracellular parasites. PLoS Pathog. 2020, 16, e1008276. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.L.; Lawrence, C.; Nichols, D.K.; Brubaker, J.F.; Peterson, T.S.; Murray, K.N.; Kent, M.L. Pleistophora hyphessobryconis (Microsporidia) infecting zebrafish Danio rerio in research facilities. Dis. Aquat. Organ. 2010, 91, 47–56. [Google Scholar] [CrossRef]
- Geneious. Available online: www.geneious.com (accessed on 28 March 2024).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- National Library of Medicine National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 28 March 2024).
- Knowles, S.; Leis, E.M.; Richard, J.C.; Standish, I.F.; Bojko, J.; Weinzinger, J.; Waller, D.L. 2018 Embarrass River Microsporidia: U.S. Geological Survey Data Release; USGS Data Release Products: Madison, WI, USA, 2024. [CrossRef]
| Species | Number of Males (M), Females (F), Undetermined (U) | Infected | Prevalence in Females | Gravid Females | Prevalence in Gravid Females |
|---|---|---|---|---|---|
| Actinonaias ligamentina | 4M 5F | 3/9 | 60% (3/5) | 4/5 | 50% (2/4) |
| Lampsilis cardium | 4F | 4/4 | 100% (4/4) | 3/4 | 100% (3/3) |
| Potamilus fragilis | 1F 1U | 0/2 | 0% (0/1) | 0/1 | NA |
| Lampsilis siliquoidea | 1M 2F | 1/3 | 50% (1/2) | 2/2 | 50% (1/2) |
| Potamilus alatus | 4F | 0/4 | 0% (0/4) | 1/4 | 0% (0/1) |
| Truncilla truncata | 5M 2F | 0/7 | 0% (0/2) | 1/2 | 0% (0/1) |
| Totals | 10M 18F 1U | 8/29 | 44% (8/18) | 11/18 | 55% 6/11 |
| Species/GenBank Accession Number | Percent Identity | Host (Phylum) | Tissue Infected | Spore Shape and Size in µm | Polar Filament Coils and Rows | Exospore Layers | Publication |
|---|---|---|---|---|---|---|---|
| Hirsutonosema embarrassi | 100 | Actinonaias ligamentina, Lampsilis cardium, Lampsilis siliquoidea (Mollusca) | Ovary | Round-to-oval 4.1 × 2.88 | 27–28 1–3 | 1 with a spiky “hairy” coat | This paper |
| Pseudonosema cristatellae AF484694.1 | 92.36 | Cristatella mucedo (Bryozoa) | Epithelial cells of body wall | Broadly pyriform 7.3 × 5.1 |
22–32 1–4 | 1 | [16,17] |
| Microsporidia sp. MT622752.1, MT622753.1 |
88.79 88.72 |
Kiefferulus barbatitarsis (Arthopoda) | Adipose tissue | NA | NA | NA | NA |
| Janacekia tainanus MW537817.1 | 88.72 |
Kiefferulus tainanus
(Arthropoda) | Adipose tissue | Oval 6.1 × 3.7 |
13–17 1 | 1 with tubular secretions | [18] |
| Jirovecia branchilis OP234512.1, OP234513.1 |
88.49 88.42 |
Branchiura sowerbyi
(Annelida) | Glands | Rod-like 26.6 × 1.8 |
Manubrium NA | 1 in a polar sac | [19] |
| Bacillidium vesiculoformis AJ581995.1 | 87.85 |
Nais simplex
(Annelida) | Intestinal hemocytes | Cylindrical 12.2 × 1.6 |
Manubrium NA | 1 covered in spherical granules | [20] |
| Knowlespora clinchi OL117026.1 | 87.03 |
Actinonaias pectorosa (Mollusca) | Gonad | Round-to-oval 4.6 × 3.7 |
~13–14 1–2 | 1 | [21,22] |
| Bacillidium sinensis ON054957.1 | 87.18 |
Branchiura sowerbyi
(Annelida) | Coelomocytes | Rod-like 15.9 × 2.5 |
Manubrium NA | 16 | [23] |
| Nematocenator marisprofundi JX463178.1 | 84.63 |
Desmodora marci (Nematoda) | Muscle | Pyriform ~8.5 × 2.7 * |
3–5 1 | 1 | [24] |
| Jirovecia sinensis MN752318.1, MN752317.1 |
88.48 88.36 |
Branchiura sowerbyi
(Annelida) | Coelom | Rod-like 17 × 2 |
Manubrium NA | 1 in a polar sac/s | [25] |
| Bacillidium branchilis ON054958.1, ON054959.1 |
87.35 86.98 |
Branchiura sowerbyi
(Annelida) | Coelomocytes | Rod-like 9.8 × 1.7 |
Manubrium NA | 2 | [23] |
| Microsporidium sp. BPAR4 FJ756101.1 | 86.38 |
Dorogostaiskia parasitica
(Arthropoda) | NA | NA | NA | NA | NA |
| Schroedera airthreyi AJ749819.1 | 87.39 |
Plumatella
sp. (Bryozoa) | Cells of body wall | Ovoid 8.7 × 5.8 |
37–38 1–2 | 2 | [26] |
| Neoperezia chironomid HQ396519.1 | 86.98 |
Chironomus plumosus
(Arthropoda) | Adipose tissue | Wide oval 6.1 × 3.4 |
24–27 1–3 | 1 | [27] |
| Trichonosema pectinatellae AF484695.1 | 86.89 |
Pectinatella magnifica
(Bryozoa) | Coelomic cell | Elongated pyriform 9.5 × 4.5 |
25 1–3 | 1 with surface spikes | [16] |
| Neoperezia semenovaiae HQ396520.1 | 86.27 |
Chironomus plumosus
(Arthropoda) | Adipose tissue | Wide oval 6.0 × 3.5 |
17–32 1–4 | 2 | [27] |
| Bryonosema plumatellae AF484692.1, AF484691.1 |
86.29 85.62 |
Plumatella nitens
(Bryozoa) | Coelomic cells | Broadly pyriform 8.4 × 5.8 |
≤33 1–3 | 1 | [16] |
| Neoperezia semenovaiae MN512230.1 | 86.27 | NA | NA | NA | NA | NA | NA |
| Neoperezia semenovaiae MN512229.1 | 86.15 | NA | NA | NA | NA | NA | NA |
| Trichonosema algonquinensis AY582742.1 | 86.13 |
Pectinatella magnifica
(Bryozoa) | Epithelial cells | Ovoid 8.5 × 4.4 |
20–33 1–2 | 1 with finger-like processes | [28] |
| Schroedera plumatellae AY135024.1 | 84.50 |
Plumatella fungosa
(Bryozoa) | Testes | Oval 7.2 × 5.0 |
22–23 1–3 | 1 with bubble-like extensions near poles | [29] |
| Janacekia debaisieuxi AY090070.1 | 84.00 | Odagoamia ornuta (Arthropoda) | NA | NA | NA | NA | [30] |
| Janacekia debaisieuxi AJ252950.1 | 83.88 |
Simulium equinum
or S. ornatum (Arthropoda) | NA | NA | NA | NA | [31] |
| Bacillidium sp. AF104087.1 | 83.82 |
Lumbriculus
sp. (Annelida) | Coelomocytes | Long-slender 24–27.2 × 3.2 |
Manubrium NA | NA | [32] |
| gBlock® Copies | Cq Mean | Cq Mean SD | Cq CV (%) |
|---|---|---|---|
| Intra-assay | |||
| 10,000,000 | 17.37 | 0.28 | 1.60 |
| 1,000,000 | 20.30 | 0.13 | 0.63 |
| 100,000 | 23.73 | 0.29 | 1.21 |
| 10,000 | 26.98 | 0.36 | 1.32 |
| 1000 | 30.19 | 0.28 | 0.94 |
| 100 | 34.07 | 0.27 | 0.78 |
| 10 | 37.31 | 0.72 | 1.93 |
| 1 | 39.50 | NA | NA |
| Inter-assay | |||
| 10,000,000 | 17.16 | 0.35 | 2.02 |
| 1,000,000 | 20.17 | 0.22 | 1.08 |
| 100,000 | 23.65 | 0.31 | 1.31 |
| 10,000 | 26.83 | 0.40 | 1.49 |
| 1000 | 30.16 | 0.30 | 0.99 |
| 100 | 33.87 | 0.37 | 1.10 |
| 10 | 37.20 | 0.66 | 1.77 |
| 1 | 39.91 | 0.79 | 1.97 |
| gBlock® Copies | SQ Mean | SQ SD | SQ CV (%) |
|---|---|---|---|
| 10,000,000 | 11,935,000.00 | 948,063.82 | 7.94 |
| 1,000,000 | 1,063,125.00 | 78,387.16 | 7.37 |
| 100,000 | 80,337.50 | 11,217.60 | 13.96 |
| 10,000 | 6697.50 | 468.25 | 6.99 |
| 1000 | 530.50 | 29.02 | 5.47 |
| 100 | 44.56 | 8.81 | 19.77 |
| 10 | 26.24 | 1.92 | 7.34 |
| 1 | 1.47 | 0.51 | 34.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knowles, S.; Leis, E.M.; Richard, J.C.; Standish, I.F.; Bojko, J.; Weinzinger, J.; Waller, D.L. Hirsutonosema embarrassi n. gen. n. sp. (Phylum Microsporidia) in the Ovary of Mucket (Actinonaias ligamentina), Plain Pocketbook (Lampsilis cardium), and Fatmucket (Lampsilis siliquoidea) (Unionidae) from the Embarrass River, Wisconsin, USA. Parasitologia 2024, 4, 184-198. https://doi.org/10.3390/parasitologia4020016
Knowles S, Leis EM, Richard JC, Standish IF, Bojko J, Weinzinger J, Waller DL. Hirsutonosema embarrassi n. gen. n. sp. (Phylum Microsporidia) in the Ovary of Mucket (Actinonaias ligamentina), Plain Pocketbook (Lampsilis cardium), and Fatmucket (Lampsilis siliquoidea) (Unionidae) from the Embarrass River, Wisconsin, USA. Parasitologia. 2024; 4(2):184-198. https://doi.org/10.3390/parasitologia4020016
Chicago/Turabian StyleKnowles, Susan, Eric M. Leis, Jordan C. Richard, Isaac F. Standish, Jamie Bojko, Jesse Weinzinger, and Diane L. Waller. 2024. "Hirsutonosema embarrassi n. gen. n. sp. (Phylum Microsporidia) in the Ovary of Mucket (Actinonaias ligamentina), Plain Pocketbook (Lampsilis cardium), and Fatmucket (Lampsilis siliquoidea) (Unionidae) from the Embarrass River, Wisconsin, USA" Parasitologia 4, no. 2: 184-198. https://doi.org/10.3390/parasitologia4020016
APA StyleKnowles, S., Leis, E. M., Richard, J. C., Standish, I. F., Bojko, J., Weinzinger, J., & Waller, D. L. (2024). Hirsutonosema embarrassi n. gen. n. sp. (Phylum Microsporidia) in the Ovary of Mucket (Actinonaias ligamentina), Plain Pocketbook (Lampsilis cardium), and Fatmucket (Lampsilis siliquoidea) (Unionidae) from the Embarrass River, Wisconsin, USA. Parasitologia, 4(2), 184-198. https://doi.org/10.3390/parasitologia4020016






