Abstract
Sarcocystis aucheniae is a coccidian parasite that produces macroscopic sarcocysts in South American camelid (SAC) muscles and causes a disease known as SAC sarcocystosis. This parasitosis hampers the commercialization of llama and alpaca meat, a vital economic activity in the Andean regions. No control or prevention methods are available, and diagnosis is based on postmortem visual inspection of carcasses. The aim of this study was to identify S. aucheniae B-cell epitopes suitable for the development of diagnostic methods for SAC sarcocystosis. To this end, sarcocyst immunoreactive protein bands were analyzed via mass spectrometry, and proteins in each band were identified in silico by searching in the parasite transcriptome. Five highly antigenic, hydrophilic B-cell epitopes, predicted not to cross-react with antibodies against other coccidia, were selected for future development of peptide-based serological tests. In addition, conserved domains present in the identified proteins allowed us to unravel metabolic pathways and mechanisms active in the parasitic stages present in sarcocysts, including aerobic respiration, antioxidant activity, signal transduction, protein synthesis and processing, and host–pathogen interactions. This study provides novel information on the biology of S. aucheniae, as well as new protein sequences that can be used for the development of diagnostic tests and chemotherapeutic approaches for SAC sarcocystosis.
1. Introduction
The genus Sarcocystis (Phylum: Apicomplexa, Family Sarcocystidae) comprises more than 200 species of coccidian protozoans infecting mammals, birds, and reptiles [1,2]. They display dixenous life cycles, which encompass a predator and prey, the definitive and the intermediate host, respectively. Typical to these parasitic infections is the formation of sarcocysts in the tissues of the intermediate hosts, which essentially consist of wall-encapsulated vesicles containing live parasites. After a predator ingests sarcocyst-infected tissues of the prey, the parasites undergo sexual reproduction in the host intestine, leading to the release of infective forms in the feces. Upon ingestion of contaminated water or food by the intermediary host, the parasites gain access to its intestinal mucosa and then the general circulation, finally reaching their destination tissues, where they encyst [1,3].
Depending on the Sarcocystis species, sarcocysts formed in the tissues of the intermediate host range in size from microscopic (as in S. capracanis, S. cruzi, and S. tenella, among others, which infect goat, cattle, and sheep, respectively) to macroscopic (as in S. buffalonis, S. cafferi, S. cameli, and S. gigantea, among others, which infect water buffalo, African buffalo, dromedary, and sheep, respectively). Also, the cell wall varies among species, with over 80 different structures, as well as the tissue where sarcocysts are formed, which can be striated muscle, cardiac muscle, or the nervous system [1,3].
Sarcocystis sp. infections are generally asymptomatic but can be occasionally accompanied by eosinophilic myositis (EM), a striated muscle inflammatory disease caused mostly by leukocyte accumulations, particularly eosinophils. EM has been sometimes recorded along with other symptoms such as anorexia, fever, anemia, weakness, weight loss, neurological symptoms, and diarrhea [4,5]. It has mostly been detected in cattle [1]. Clinically, the afflicted animals may look normal. Most observations occur at the abattoir level, leading to carcass condemnation with important economic losses [1].
Sarcocystis neurona is an unusual case among Sarcocystis spp. because it shows tropism towards the gray and white matter of the nervous system of horses, producing a severe neurological syndrome known as equine protozoal myeloencephalitis (EPM). EPM clinical cases have different degrees of severity and can provoke incoordination, lameness, muscular atrophy, paralysis of certain muscles, and, on rare occasions, seizures and collapse [6,7]. Sarcocystis neurona has attracted considerable research efforts, and its genome is the only one sequenced for this genus to date [8].
For three Sarcocystis spp., S. hominis, S. heydorni, and S. suihominis, humans can act as definitive hosts, with intestinal infections that can be accompanied by nausea, abdominal discomfort, and diarrhea. More infrequently, humans can act as intermediate hosts by the ingestion of sporocysts of certain species, such as S. nesbitti. Sarcocyst formation in human muscles is typically asymptomatic but can be associated with fever, myalgia, weakness, eosinophilia, and bronchospasm [9].
Llamas (Lama glama) as well as alpacas (Vicugna pacos) are domestic South American camelids (SACs) raised in the Andean regions of Argentina, Bolivia, Peru, Chile, and Ecuador [10]. These animals portray an ancestral Andean bio-cultural heritage, and their breeding constitutes an important part of the life strategy of rural communities [11]. SACs are well adapted to extreme environmental conditions, such as high altitude, lack of water, steep slopes, high radiation, and frost. They can also thrive under varied climatic and topographic conditions and have been introduced as livestock to other regions of the world, such as Australia [12]. Importantly, due to their unique anatomical and physiological characteristics, their impact on ecosystems is much less than that of European livestock, such as cattle and sheep [13].
Besides their provision of fiber, transport, and manure for fuel, llamas and alpacas have long constituted a major source of animal protein for Andean rural communities [11,14,15]. In addition, given its low cholesterol/protein ratio and good taste, combined with a low SAC ecological impact, this type of meat constitutes an attractive food for local and foreign gourmet cuisine [16]. However, an important limitation of the production and commercialization of SAC meat is the frequent infestation of skeletal muscles with oval-shaped macroscopic cysts that resemble rice grains. The etiological agent of this parasitosis has been molecularly confirmed to be Sarcocystis aucheniae, and the disease is known as SAC sarcocystosis [1,17,18]. Finding of cysts upon postmortem examination leads to confiscation of carcasses by sanitary authorities or devaluation of their commercial value. In addition, ingestion of insufficiently cooked cyst-infested meat provokes gastroenteritis [3,12,14]. This is due to the presence of a peptidic enterotoxin in the cysts, like what has been observed with S. fayeri [19,20]. There is a paucity of information on the pathogenic effects of S. aucheniae in SACs. Infections are considered essentially subclinical, and only two cases of EM have been reported in alpacas [4,5].
Given the constraints imposed by S. aucheniae infections on SAC meat commercialization, the development of diagnostic and control strategies for SAC sarcocystosis can bring important economic benefits to the Andean region [21]. It is possible to detect S. aucheniae DNA in SAC blood using specific and sensitive semi-nested PCR protocols, albeit positive results probably correspond only to the early stages of the infection and are not predictive of the presence or absence of cysts in muscles [22,23]. Serological diagnosis might better reflect previous exposure of the animal to the parasite. The detection of anti-Sarcocystis sp. antibodies in llamas has been carried out using immunofluorescence and an indirect ELISA based on an immunogenic protein fraction of S. aucheniae cysts [24,25]. However, these methods have not yet been validated regarding their capacity to predict cyst infestation, and as they are based on the use of whole parasites or parasite fractions as antigens, they are prone to low reproducibility and are not easily scaled up. Moreover, investigations on proteins relevant to host–pathogen interactions that could be useful for the development of vaccines, diagnostic methods, or chemotherapeutic approaches are very limited [21].
In the present study, an immunoproteomics approach was used to identify B-cell epitopes suitable to be applied for the development of peptide-based serodiagnostic tests for S. aucheniae. At the same time, it was possible to unravel the sequences of several proteins predicted to participate in various metabolic pathways and cellular mechanisms, contributing the first information for the genus Sarcocystis on gene expression in the parasitic stages that thrive inside sarcocysts, as well as new potential drug targets for SAC sarcocystosis.
2. Results
2.1. Llama Anti-S. aucheniae IgG Antibodies Recognize Six Immunoreactive Protein Bands from Sarcocysts
The reactivity of 43 sera from S. aucheniae-positive llamas against proteins of S. aucheniae cysts was determined in immunoblots (Figure 1). All positive sera reacted with one or more protein bands of 23, 26, 33, 38, 45, and 73 kDa, which were recognized by 21, 15, 63, 53, 14, and 23% of the tested S. aucheniae-positive sera, respectively. No cyst protein bands were recognized by sera of S. aucheniae-negative llamas (data not shown). Triton X-114 phase separation showed that the detected immunoreactive proteins preferentially partitioned into the soluble fraction (Figure 1).
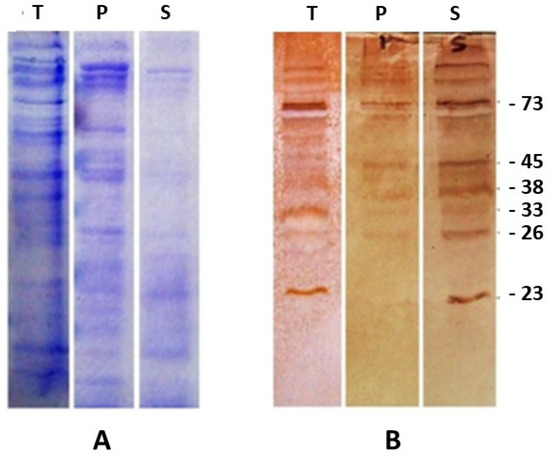
Figure 1.
Detection of immunoreactive protein bands in S. aucheniae sarcocysts. Proteins present in an S. aucheniae sarcocyst lysate (T) were separated by SDS-PAGE and either stained (A) or transferred to a nitrocellulose membrane (B) or first partitioned into membrane (P) and soluble (S) proteins with Triton X-114 and then stained or transferred. The membrane was cut in strips and incubated with sera of S. aucheniae-positive llamas and revealed with peroxidase-labeled anti-llama IgG and a colorimetric substrate (brown bands). A representative experiment is shown.
Incubation with isotype-specific anti-llama immunoglobulin monoclonal antibodies showed that the reaction was, in all cases, due to G-type immunoglobulins (IgG) with no involvement of IgM antibodies (data not shown).
S. aucheniae sarcocyst protein bands of 23, 26, 33, 38, 45, and 73 kDa were subjected to mass spectrometry. Data obtained from the mass spectrogram were analyzed by BLAST against an S. aucheniae transcriptome database available from previous work [21]. Between 3 and 15 peptides could be identified in each immunoreactive protein band. A total of 44 transcripts were identified in the transcriptome database, which, after subtracting those that were found more than once, resulted in a total of 35 newly identified proteins (Table S1). Of these, most were predicted to be either cytoplasmic or secreted, based on the absence of transmembrane domains and GPI-anchor signals and the absence or presence of a signal peptide, respectively, which agrees well with the preferential partitioning of immunoreactive bands into the Triton X-114 soluble fraction (Table S1, Figure 1). However, in some cases, only incomplete peptide sequences could be predicted from the transcriptome database, likely due to partial degradation of the corresponding transcripts, which prevented the analysis of their subcellular localization and other features (Table S1).
2.2. Sarcocystis aucheniae Immunoreactive Bands Contain Soluble B-Cell Epitopes Not Conserved in Other Coccidia
The protein in each immunoreactive band displaying the highest antigenic index was selected for further analysis. The most likely B-cell epitopes recognized by S. aucheniae-infected llama sera were identified in each band using two different algorithms (Table 1).

Table 1.
Sarcocystis aucheniae highly antigenic proteins identified by immunoproteomics and their selected B-cell epitopes. For each identified epitope, the algorithm used is indicated (S: SVMTrip; B: Bepipred), as well as their solubility in water and their propensity of cross-reactivity with proteins of other coccidia. Predicted B-cell epitopes that could be applied to the development of a peptide-based serological test are shown in bold.
To analyze the suitability of the predicted B-cell epitopes for the development of peptide-based serological diagnostic tests, their water solubility, and the degree of conservation with proteins of the closely related coccidia Toxoplasma gondii, Neospora caninum and Eimeria spp. were further investigated. Conservation of 80% or more contiguous amino acids in a B-cell epitope predicted for these coccidia was considered to indicate a propensity for cross-reactivity, while a lack of conservation or conservation with a non-epitope region was considered to indicate no propensity for cross-reactivity. Twelve B-cell epitopes were predicted to have good solubility in water, and five of them have no potential cross-reactions and could therefore be used in peptide-based serologic tests. (Table 1).
2.3. Conserved Domains Reveal Metabolic Pathways and Cellular Mechanisms Active in Sarcocyst Parasitic Stages
Putative roles could be assigned to 33 of the newly identified proteins via the identification of conserved domains and studies carried out on their homologues in T. gondii. This allowed us to infer active metabolic pathways and molecular mechanisms that operate in the parasite stages inside S. aucheniae sarcocysts. Proteins involved in gliding motility and host cell invasion, cyst formation, aerobic respiration, antioxidant activity, energy storage via gluconeogenesis, protein synthesis, processing and regulation, lipid metabolism, and signal transduction, among other cellular mechanisms, could be distinguished. Two identified proteins lacked conserved domains. One of them has homologous proteins in other coccidia, while for the second, no homologous sequence could be found in other coccidian genera (Table 2 and Table S1). In the latter case, the S. neurona genome was also searched to evaluate if this protein is conserved across the Sarcocystis genus, yet no significant hits were retrieved.

Table 2.
S. aucheniae bradyzoite cellular mechanisms and proteins unraveled by immunoproteomics.
3. Discussion
In the present study, we took advantage of a previously obtained transcriptome database for S. aucheniae sarcocyst stages to unravel specific B-cell epitopes that could be used in the development of serological diagnostic tests [21]. Peptides of immunoreactive bands identified by mass spectrometry were used to search in the predicted protein pool derived from this transcriptome, and the most antigenic protein in each band was used for B-cell epitope prediction. This led to the identification of 12 B-cell epitopes of high antigenicity and water solubility. Importantly, contrary to the more common approach of in silico search for B-cell epitopes in whole genomes, the present study identified peptides that are indeed expressed by the parasite in the intermediate host. Five of these epitopes showed no propensity for cross-reactivity with antibodies raised against other coccidia that can also infect SACs, T. gondii, N. caninum, and Eimeria sp. [24,26,27]. This type of study could not, however, be carried out for S. masoni, another SAC parasite that forms microscopic cysts in striated and cardiac muscles, due to the unavailability of genomic sequence data [18]. Although the antigenicity of the identified B-cell epitopes and their lack of cross-reactivity need experimental confirmation, the information provided in this study can be highly useful for the future development of peptide-based serological diagnostic methods, as has been described for T. gondii [28]. In addition, those B-cell epitopes with high antigenicity and water solubility but that showed significant conservation with proteins of other coccidia could be used for the development of peptide-based vaccination approaches. An additional pool of predicted B-cell epitopes was obtained from other proteins present in each immunoreactive band with somewhat lower antigenicity indexes (data not shown). However, they have not been included in the present study, which focuses on the most likely proteins to be recognized by sera of S. aucheniae-positive llamas.
None of the studied immunoreactive protein bands was recognized by all positive sera in immunoblots, which suggests that a combination of B-cell epitopes might be needed to develop a highly sensitive serological test.
We have previously identified the array of predicted glycosylphosphatidylinositol (GPI)-anchored proteins using a bioinformatic pipeline that detected transcripts encoding hydrophilic proteins with a signal peptide and a GPI-anchor signal in their N and C-termini, respectively [21]. Some coccidian GPI-anchored proteins bear a SAG/SRS domain (PF04092), initially described for Surface Antigen 1 of T. gondii, with a function in receptor–ligand interactions that regulate host–cell attachment [29]. In the case of S. aucheniae, 13 of the identified GPI-anchored proteins bear such a domain [21]. In the present study, two additional proteins with an SAG/SRS domain were identified (MS21, MS22), but they lack a signal peptide and a GPI anchor signal and thus passed unnoticed in the mentioned previous study. Since SAG/SRS domains are only associated with GPI-anchored-bound proteins, these newly identified ones likely correspond to truncated transcripts. GPI-anchored proteins of other pathogenic protozoa are abundantly present on the parasites’ surfaces and elicit strong humoral responses, thus constituting attractive diagnostic antigens [30,31]. It might seem surprising not to have detected membrane-bound S. aucheniae GPI-anchored proteins, expected to be immunodominant, in the present study. However, it needs to be considered that the experimental approach here applied to obtain an S. aucheniae sarcocyst lysate essentially excludes membrane-bound proteins that remain attached to the glass beads used in the homogenization process. This is likely why only very few of the identified proteins had transmembrane domains and, also, immunoreactive bands mostly partitioned into the Triton X-114 soluble fraction (Figure 1, Table S1). Importantly, the B-cell epitopes in soluble S. aucheniae proteins recognized in the present study can be added to the diagnostic toolbox for this parasite, together with those present in GPI-anchored proteins, which are currently under investigation in our laboratory.
The set of proteins identified by mass spectrometry and searches in the transcriptome database of S. aucheniae sarcocysts were also analyzed regarding molecular weight, homology to proteins of related coccidia, and possible functions. As shown in Table S1, several of the identified proteins show different molecular weights when predicted by either their SDS-PAGE mobility or translation of their corresponding transcripts. This variation could be due to post-translational processing, including protease cleavage or addition of functional groups, aberrant electrophoretic mobility, or, in some cases, truncated transcripts. We recognize that further work is required to establish the detailed basis of this variation.
Sarcocysts contain two parasite stages: the round to oval metrocytes (“mother cells”), specialized in multiplication, and the “banana-shaped” bradyzoites (“slow cells”), which do not divide and are specialized in the invasion of the intestinal cells in the definitive host. Accordingly, metrocytes, the dominant stage in immature cysts, do not display most apical complex organelles associated with invasion, such as micronemes and conoid and polar rings, but have a large nucleus and abundant mitochondria and ribosomes. Bradyzoites, the dominant stage in mature cysts, bear a complete apical complex and show gliding motility [3]. When a medium-sized sarcocyst, such as those used in the present study, is cut open and the semi-liquid content is observed under the microscope, large numbers of banana-shaped parasites with oscillating movement—presumed to be bradyzoites—are observed [17]. However, a number of metrocytes are expected to be present in the sarcocyst to ensure the continuation of parasite proliferation until the cyst reaches its final size. Mass spectrometry data and in silico searches of the sarcocyst transcriptome allowed us to identify several proteins that are expressed in S. aucheniae metrocytes and/or bradyzoites. Thus, identified proteins associated with chromosome separation during mitosis and protein synthesis and folding are likely expressed in the actively dividing metrocytes. On the other hand, proteins associated with special secretory organelles of Apicomplexans (rhoptries, micronemes, and dense granules) are expected to be present in bradyzoites. One such protein (OR538336, Table 2) is the S. aucheniae homologue of T. gondii microneme MIC12 and Eimeria tenella MIC4. Micronemes are rod-like structures clustered at the apical complex of Apicomplexan parasites and appear to be particularly abundant in Sarcocystis spp. [3]. Micronemes undergo exocytosis by fusing with the plasma membrane at the apical end to discharge their contents of adhesin complexes and perforins that participate in critical events for parasitic life, including host cell invasion and egress and gliding motility (see below, [32]). Interestingly, both T. gondii MIC12 and E. tenella MIC4 were found to be strongly immunogenic when screening a protein microarray or a phage display library, respectively, with immune sera against the respective parasites [33,34]. Consistently, in the present study, S. aucheniae MIC12 displayed the highest antigenicity among the proteins of its corresponding immunoreactive band and was one of the proteins selected for epitope B identification (Table 1, Supplementary Table S1).
Apicomplexan parasites use an ATP-powered actin and myosin-based machine for gliding motility, known as a glideosome. It is located at the pellicle, between the plasma membrane and the inner membrane complex, and includes three glideosome-associated proteins: GAP50, GAP45, and GAP40 [35]. The assemblage of the glideosome is strictly regulated by post-translational modifications, which are controlled by a calcium-depended signaling cascade [36]. GAP45, which was identified in S. aucheniae in this study, is responsible for recruiting Myosin A, and it also mediates the association of the inner membrane complex and the plasma membrane, maintaining the structural integrity of the pellicle architecture. Transient adhesions to the substrate or to the host cell surface, through a set of parasitic actin-bound adhesins, generate sufficient traction to allow the parasite to propel itself forward. Gliding motility is used by apicomplexans for migration through host tissues and cells, as well as host cell invasion and egress, which are essential activities of these obligatory intracellular parasites [37].
Dense granules or dense bodies are also considered part of the apical complex, although they are not necessarily located at the apical end and release their contents elsewhere on the parasite surface. In SAC Sarcocystis spp., they have been observed as electron-dense vesicles in the central and caudal regions of bradyzoites [38]. Their secretions are thought to participate in post-invasion processes that allow the parasites to take advantage of their new intracellular environment [39]. An S. aucheniae homologue of Eimeria sp. dense granule protein GRA9 was identified in the present study (Table 2). Notably, homologues of this S. aucheniae protein were not found in other organisms outside the Eimeria genus, indicating polymorphisms among GRA proteins of coccidians. GRA proteins are characterized as being relatively small and have an N-terminal signal peptide [39]. In the present study, no signal peptide was found in S. aucheniae GRA9, likely because the sequence identified is only partial (Supplementary Table S1). Although their exact function is unknown, it has been hypothesized for T. gondii that they are involved in building the parasitophorous vacuole upon parasite invasion of the host cell, contributing to transforming this new space into a metabolically active compartment that can acquire host cell nutrients. They have also been involved in building the tissue cyst wall [40].
This study also evidenced the expression of several proteases in sarcocysts. Proteases fulfill a large variety of essential roles for cell functioning of all organisms, including regulation of fate, localization, and activity of many proteins; modulation of protein–protein interactions; and generation of new bioactive molecules. In addition, in parasites, they can also participate in immune evasion, invasion, and egress from host cells; tissue penetration; degradation of host proteins destined for nutrition; and other processes. Given their essential roles, several proteases have been proposed as chemotherapeutic targets or vaccine candidates against different pathogenic microorganisms [41,42]. Proteases identified in this study include a secreted metalloprotease belonging to the family M16, which is a homologue of T. gondii toxolysin 1. This protease has been shown to be localized in T. gondii rhoptries and to be secreted during invasion, with a proposed role in host protein cleavage [43]. In addition, two components of the parasite proteasome system were identified, mainly ubiquitin and a threonine protease (Table 2). Proteasomes are large, multi-component protein complexes that degrade poly-ubiquitinated proteins in the cytosol and nucleus of all eukaryotic and archaebacterial cells. Interestingly, the use of drugs that target the proteasome has been identified as an effective chemotherapeutic strategy against several protozoan pathogens [44].
The activity of cellular proteases is tightly controlled by an array of protease inhibitors. One of them, serpin, an inhibitor of serine proteases such as trypsin, is also expressed in sarcocysts. In T. gondii, serpin has been shown to have anti-inflammatory effects, and a recombinant form of this protease has been proposed as an asthma therapy [45].
Fructose-1,6-bisphosphatase, another identified protein expressed in sarcocysts, catalyzes the reaction from fructose 1,6-bisphosphate into fructose-6-phosphate as part of gluconeogenesis. This process has been associated in T. gondii with the synthesis in bradyzoites of amylopectin, a complex polysaccharide that forms part of tissue cysts. Interestingly, fructose-1,6-bisphosphatase appears to be present only in the coccidian branch of the Apicomplexa and absent in other Apicomplexan protozoa such as Plasmodium, Theileria, or Cryptospordium [46]. So far, the composition of sarcocyst walls has not been unraveled. However, amylopectin granules have been described in Sarcocystis sp. bradyzoites [47,48]. These might serve as carbon source storage and be secreted to form the sarcocyst wall as well, although these assumptions need experimental confirmation.
Heat shock proteins (HSPs) are phylogenetically conserved molecular chaperones participating in the correct folding of nascent proteins. Under stress conditions, they play an essential role in cell survival, participating in either repairing or denaturing damaged proteins, thus maintaining protein homeostasis. For this reason, HSPs of T. gondii and other intracellular protozoan parasites have been proposed as targets for new drug discovery [49,50]. The four S. aucheniae HSPs identified in this study, HSP60, two HSP70 variants, and HSP90 (Table 2), might provide amenable foci for drug development against SAC sarcocystosis.
Our study also unraveled the expression in sarcocysts of two enzymes that take part in lipid metabolism. One of them, phosphatidylserine (PS) decarboxylase, catalyzes the synthesis of phosphatidylethanolamine (PE) from PS—one way, in addition to de novo synthesis and scavenging from host cells, that parasites use for the acquisition of PE, the second most abundant phospholipid in cell membranes [51,52]. The other is a homologue of T. gondii phosphatidylcholine-sterol O-acyltransferase (TgLCAT). When there is an excess of cholesterol in the parasite environment, TgLCAT produces cholesteryl esters, transferring acyl groups from phospholipids to free cholesterol, which can be stored in lipid bodies. In addition, TgLCAT has been shown to possess phospholipase A2 activity, with a role in parasite egress from the host cell by producing lysolecithin, which has membrane-destabilizing effects [53]. It remains to be investigated whether TgLCAT homologues in other coccidias, such as Sarcocystis sp., display a similar role.
Several of the cellular mechanisms active in metrocytes and/or bradyzoites unraveled in this study, such as parasite multiplication, protein synthesis, apical complex formation, gliding motility, and gluconeogenesis, require the provision of energy. No previous studies have investigated the type of cellular respiration carried out by S. aucheniae parasites inside sarcocysts. We here show that several enzymes that participate in glycolysis (glyceraldehyde 3-phosphate dehydrogenase, fructose-bisphosphate aldolase, enolase), the tricarboxylic acid cycle (lactate/malate dehydrogenase), and the phosphorylation chain (membrane-bound ATPase) are expressed in sarcocysts, indicating the occurrence of aerobic respiration. In agreement, the genes encoding the different enzymes of the aerobic respiration pathway have been identified in the S. neurona genome [8]. Additionally, superoxide dismutase, an enzyme that protects cells from oxidative damage, was also identified. These observations indicate an aerobic milieu for parasites inside the sarcocyst and suggest the existence of a tight connection between the host circulatory system and a porous sarcocyst wall that assures oxygen diffusion inside the cyst.
Two identified proteins lacked conserved domains, and thus their putative function could not be inferred. One has homologues in other coccidia, and its function might be related to coccidia-specific cellular pathways. The second has no homologues in T. gondii, N. caninum, Eimeria sp., or S. neurona, indicating that this protein could be specific for S. aucheniae. However, given that the analyzed transcript is truncated, a better analysis of the conservation of this protein, as well as its predicted location and function, will be possible once the S. aucheniae genome has been sequenced and the complete gene sequence is available.
Previous research from our group identified the S. aucheniae GPI biosynthetic pathway by in silico searches in a transcriptome database. Since both free and protein-bound GPIs are of high importance for host cell recognition and invasion by the parasite, this pathway constitutes a potential chemotherapeutic target for SAC sarcocystosis. Also, as mentioned above, GPI-anchored proteins of this parasite constitute attractive diagnostic and vaccination targets for this and other protozoan infections [21]. The present study adds novel information to other cellular mechanisms displayed by S. aucheniae, providing a glimpse of the parasite life inside a sarcocyst, which is expected to serve in the design of new control tools. Importantly, this is the first dataset of proteins of a species belonging to the genus Sarcocystis verified to be expressed in sarcocysts since the only available gene expression studies were carried out in S. neurona in vitro cultured cells [54,55]. Moreover, our study has also unraveled hydrophilic and specific B-cell epitopes that can be applied to peptide-based serological methods for SAC sarcocystosis.
4. Materials and Methods
4.1. Serum Samples
Serum samples were obtained from llamas (Lama glama) destined for meat commercialization at an Argentine slaughterhouse in the province of Jujuy. An aliquot of 5 mL of blood was aseptically collected by a veterinarian from the jugular vein of each living animal following good laboratory practices, and serum was separated by centrifugation and stored at −20 °C after addition of 0.02% (w/v) sodium azide (final concentration). Before use in immunoblots, llama sera were pre-adsorbed with an Escherichia coli homogenate to decrease unspecific reactions. To this aim, an overnight E. coli culture in Luria–Bertani (LB) broth was centrifuged at 1000× g for 15 min, and the supernatant was discarded. The pellet was suspended in 5 mL lysis buffer (50 mM K2HPO4, 400 mM NaCl, 100 mM KCL, 10% glycerol, 0.5% Triton X-100) with the addition of 1 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mg/mL lysozyme, and sonicated in ice (2 × 15 s, maximal potency). After centrifugation at 2000× g for 20 min, the supernatants were collected, and the protein concentration was measured by the Micro BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Each serum sample was incubated for 1 h at 37 °C with 0.1 mg/mL of this supernatant immediately before use in immunoblots. After the sacrifice of llamas, visual inspection of the skeletal muscles of the neck was carried out in each sampled animal to search for S. aucheniae macroscopic sarcocysts. A total of 43 and 7 sera corresponding to sarcocyst-positive and sarcocyst-negative llamas, respectively, were selected as positive and negative sera for further analysis via immunoblotting.
4.2. Electrophoretic Separation of S. aucheniae Cyst Proteins
Ten S. aucheniae cysts of about 2–3 mm in diameter were separated from the muscle fibers of a llama, suspended in 500 µL PBS, and homogenized by FastPrep-24 (MP Biomedicals, Santa Ana, CA, USA) using 3 pulses of 25 s each in the presence of glass powder (Sigma-Aldrich, St. Louis, MO, USA). Protease inhibitor cocktail (1 mM, Sigma-Aldrich) and 1 mM PMSF were added to prevent protein degradation. Protein concentration was determined using the Micro BCA Protein Assay Kit and a curve of different concentrations of bovine serum albumin (BSA). Cyst proteins were separated by preparative electrophoresis in five 12% polyacrylamide minigels (0.48 mg per gel) under denaturing conditions (SDS-PAGE) using a Mini PROTEAN 3 electrophoresis system (Bio-Rad, Hercules, CA, USA), at 120 V. An aliquot of Page Prestained Protein Ladder (Thermo Fisher Scientific) was run in the single individual well as a molecular weight standard. One minigel was stained overnight with Colloidal Coomassie blue (Bio-Rad) and de-stained following standard procedures, while the rest were used for immunoblots, as described below.
4.3. Determination of Immunoreactivity of Llama Sera against S. aucheniae Cyst Proteins via Western Blot
Four minigels with electrophoresed S. aucheniae cyst proteins were subjected to immunoblotting. To this end, proteins were transferred to Hybond nitrocellulose membranes (Thermo Fisher Scientific) using a Bio-Rad wet miniblotter. Membranes were blocked with PBS-T-milk (PBS/0.1% Tween-20/3% skim milk) for 1 h at 37 °C, with stirring, and then cut vertically into 4 mm wide strips. Each strip was incubated with a 1:50 dilution in PBS-T-milk of a different serum sample, previously pre-adsorbed with an E. coli lysate. After three washes with PBS-T, strips were exposed to a 1:5000 dilution of peroxidase labeled-anti-llama IgG antibodies (Bethyl Laboratories, Montgomery, AL, USA) in PBS-T-milk, for 1 h at room temperature, with stirring. Membranes were then washed with PBS containing decreasing concentrations of Tween-20 (0.2, 0.1, 0.05, and 0%) and then incubated with a colorimetric substrate (0.62 mg/mL 3,3′-diaminobenzidine tetrahydrochloride (DAB, Sigma-Aldrich) in 20 mM Tris-HCl/150 mM NaCl, pH 7.4 (TBS), containing 0.03% (v/v) hydrogen peroxide). After the appearance of brown spots, reactions were stopped with double-distilled water. Protein bands recognized by 10% or more of the S. aucheniae-positive llama sera were considered for further analyses, and their sizes were recorded by comparison with the molecular weight standard.
4.4. Determination of Immunoglobulin Isotype Reacting with S. aucheniae Proteins
Twelve nitrocellulose membrane strips with S. aucheniae proteins were obtained, as described above. Six representative sera were selected and incubated with two strips each in 1:50 dilution. After washings, as before, strips were incubated with in-house prepared mouse ascites (1:100) containing monoclonal antibodies either against llama IgG or IgM in PBS-T-milk for 1 h with stirring [56]. After 3 washes under stirring with PBS-T, strips were incubated with anti-mouse IgG conjugated with horseradish peroxidase (Bethyl Laboratories) in a 1:1500 dilution in PBS-T-milk. Washes and incubation with the colorimetric substrate took place as described above.
4.5. Discrimination between Soluble and Membrane-Bound Proteins by Triton X-114 Partition
To analyze the hydrophobicity/hydrophilicity of the immunoreactive bands, 1 mL of S. aucheniae cyst protein sample (6 mg) was subjected to Triton X-114 partition following standard procedures [57]. The phases containing soluble and membrane-associated proteins were subjected to SDS-PAGE and immunoblot using representative sera, following the above-described procedures.
4.6. Mass Spectrometry Analysis of Immunoreactive Bands
Selected bands were excised from the Colloidal Coomassie blue-stained minigel with a scalpel and sent to the Service of the School of Exact and Natural Sciences, University of Buenos Aires (https://exactas.uba.ar/sic/espectrometria-de-masas-cgl-masass/) to be processed and analyzed by mass spectrometry (MS) in an LC-MS/MS Q Exactive (Orbitrap) equipment (Thermo Fisher Scientific). Peptides were identified by bioinformatic searches in the S. aucheniae transcriptome available from a previous study [21].
4.7. Bioinformatic Analysis
A total of 66 transcripts were identified in the S. aucheniae transcriptome database using the peptides obtained by mass spectrometry as queries. Double transcripts and those corresponding to the host (Lama glama), as assessed by BLASTp, were eliminated. In a few cases where different peptides identified transcripts of different sizes corresponding to the same protein, according to BLASTp and conserved domain analyses, peptides were aligned by Clustal omega multiple sequence alignments (www.ebi.ac.uk/Tools/msa/clustalo/ [55], edited and integrated into a single peptide. This analysis resulted in 35 predicted peptides that were deposited in the Genbank under accession numbers OR538333-OR538367. Antigenicity of each peptide was calculated by Scratch Protein Predictor (scratch.proteomics.ics.uci.edu/). Then, B-cell epitopes were identified in the most antigenic protein of each immunoreactive band by two different algorithms, SVMTrip [58] and Bepipred Linear Epitope Predictor 2.0 [59]. The selection criteria for SVMTrip were a length of 20 residues and an antigenicity threshold of 0.9, and for Bepipred, a minimum length of 7 amino acids with an antigenicity score above 0.63. Water solubility of identified B-cell epitopes was assessed by PepCalc (pepcalc.com). Conservation of B-cell epitopes in Toxoplasma gondii (taxid 5811), Neospora caninum (taxid 29175), and Eimeria spp. (taxid 5800) was analyzed by BLASTp (blast.ncbi.nlm.nih.gov/Blast.cgi [60,61] and by Clustal omega [62], followed by manual localization of epitopes. The propensity of cross-reactivity was estimated as highly likely when 80% or more contiguous identical amino acids were found in a protein of other coccidia corresponding to a B-cell epitope. All identified peptides were in silico characterized to predict molecular weight (web.expasy.org/protparam/), presence of a signal peptide (SignalP_5.1, www.cbs.dtu.dk/services/SignalP/, [63]), transmembrane domains (TMHMM, /www.cbs.dtu.dk/services/TMHMM/ [64]), presence of a GPI anchor (big PI Predictor; mendel.imp.ac.at/gpi/gpi_server.html [65]; GPI SOM; gpi.unibe.ch [66]; PredGPI, busca.biocomp.unibo.it/predgpi/ [67]), and presence of conserved domains (InterPro, www.ebi.ac.uk/interpro/ [68]; Pfam, pfam.xfam.org [69]; and Conserved Domains database of the National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/cdd). Conservation with S. neurona was checked by BLAST at VeuPathDB (https://veupathdb.org/veupathdb/app).
5. Conclusions
This study has focused on S. aucheniae, a parasite well-known to SAC producers but mostly neglected by the scientific community. The development of control and diagnostic methods is essential to strengthen the Andean regional economies that depend on the commercialization of SAC meat. The B-cell epitopes that we have unraveled can be applied in future experimental approaches for the development of new serological methods. On the other hand, the discovered proteins and cellular mechanisms active in sarcocysts throw new light on the biology of these successful parasites and provide possible targets for vaccines and/or chemotherapeutic interventions against SAC sarcocystosis. Importantly, our results can contribute to the research carried out on other Sarcocystis spp. of veterinary and/or medical importance that also urgently require the development of control strategies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/parasitologia3040034/s1, Table S1: S. aucheniae proteins identified by immunoproteomics.
Author Contributions
Conceptualization, M.F.-C.; investigation: C.D.-F., S.N.W., S.R., P.d.A., A.F. and M.F.-C.; writing—original draft preparation, C.D.-F. and S.N.W.; writing—review and editing, M.F.-C. and L.S.; supervision, M.F.-C.; funding acquisition, M.F.-C., L.S. and S.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Science and Technology of Argentina (MINCyT, PICT 2014-3747), the National Research Council of Argentina (CONICET, PIP-2021_11220200101925CO), and the National Institute of Agricultural Technology, Argentina (INTA, PD-i113 and PD-114).
Institutional Review Board Statement
Blood has been taken from llamas as a routine veterinary health check up in an abboiture before animal slaughter. This procedure was carried out by an veterinarian applying good veterinarian practices and does not require approval by an Ethics committee in accordance with Argentine regulations.
Informed Consent Statement
Not applicable.
Data Availability Statement
Identified Sarcocystis aucheniae protein sequences have been deposited in the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers OR538333-OR538367.
Acknowledgments
The authors gratefully acknowledge the assistance of Romina Mamani in obtaining llama sera.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dubey, J.P.; Calero-Bernal, R.; Rosenthal, B.M.; Speer, C.A.; Fayer, R. Sarcocystosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2016; ISBN 9781498710152. [Google Scholar]
- Wassermann, M.; Raisch, L.; Lyons, J.A.; Natusch, D.J.D.; Richter, S.; Wirth, M.; Preeprem, P.; Khoprasert, Y.; Ginting, S.; Mackenstedt, U.; et al. Examination of Sarcocystis spp. of Giant Snakes from Australia and Southeast Asia Confirms Presence of a Known Pathogen—Sarcocystis Nesbitti. PLoS ONE 2017, 12, e0187984. [Google Scholar] [CrossRef] [PubMed]
- Decker Franco, C.; Schnittger, L.; Florin-Christensen, M. Sarcocystis. In Parasitic Protozoa of Farm Animals and Pets; Springer International Publishing: Cham, Switzerland, 2018; pp. 103–124. ISBN 9783319701325. [Google Scholar]
- Gabor, M.; Gabor, L.J.; Srivastava, M.; Booth, M.; Reece, R. Chronic Myositis in an Australian Alpaca (Llama pacos) Associated with Sarcocystis spp. J. Vet. Diagn. Investig. 2010, 22, 966–969. [Google Scholar] [CrossRef] [PubMed]
- La Perle, K.M.D.; Silveria, F.; Anderson, D.E.; Blomme, E.A.G. Dalmeny Disease in an Alpaca (Lama pacos): Sarcocystosis, Eosinophilic Myositis and Abortion. J. Comp. Pathol. 1999, 121, 287–293. [Google Scholar] [CrossRef] [PubMed]
- MacKay, R.J.; Howe, D.K. Equine Protozoal Myeloencephalitis. Vet. Clin. Equine Pract. 2022, 38, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P. Sarcocystis neurona, Neospora spp. and Toxoplasma gondii Infections in Horses and Equine Protozoal Myeloencephalitis (EPM): Five Decades of Personal Experience, Perspectives and Update. Parasitology 2022, 149, 717–728. [Google Scholar] [CrossRef]
- Blazejewski, T.; Nursimulu, N.; Pszenny, V.; Dangoudoubiyam, S.; Namasivayam, S.; Chiasson, M.A.; Chessman, K.; Tonkin, M.; Swapna, L.S.; Hung, S.S.; et al. Systems-Based Analysis of the Sarcocystis neurona Genome Identifies Pathways That Contribute to a Heteroxenous Life Cycle. mBio 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Rosenthal, B.M. Zoonotic Sarcocystis. Res. Vet. Sci. 2021, 136, 151–157. [Google Scholar] [CrossRef]
- Wheeler Jane, C. Evolution and Present Situation of the South American Camelidae. Biol. J. Linn. Soc. 1995, 54, 271–295. [Google Scholar] [CrossRef]
- Vilá, B.; Arzamendia, Y. South American Camelids: Their Values and Contributions to People. Sustain. Sci. 2022, 17, 707–724. [Google Scholar] [CrossRef]
- Saeed, M.A.; Rashid, M.H.; Vaughan, J.; Jabbar, A. Sarcocystosis in South American Camelids: The State of Play Revisited. Parasit. Vectors 2018, 11, 146. [Google Scholar] [CrossRef]
- Fraser, M.D.; Vallin, H.E.; Roberts, B.P. Animal Board Invited Review: Grassland-Based Livestock Farming and Biodiversity. Animal 2022, 16, 100671. [Google Scholar] [CrossRef] [PubMed]
- Leguía, G. The Epidemiology and Economic Impact of Llama Parasites. Parasitol. Today 1991, 7, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Vilca, M. Producción, Tecnología e Higiene de La Carne. In Avances y Perspectivas del Conocimiento de los Camélidos Sudamericanos; FAO: Santiago de Chile, Chile, 1991; pp. 42–49. [Google Scholar]
- Popova, T.; Tejeda, L.; Peñarrieta, J.M.; Smith, M.A.; Bush, R.D.; Hopkins, D.L. Meat of South American Camelids—Sensory Quality and Nutritional Composition. Meat Sci. 2021, 171, 108285. [Google Scholar] [CrossRef] [PubMed]
- Carletti, T.; Martin, M.; Romero, S.; Morrison, D.A.; Marcoppido, G.; Florin-Christensen, M.; Schnittger, L. Molecular Identification of Sarcocystis aucheniae as the Macrocyst-Forming Parasite of Llamas. Vet. Parasitol. 2013, 198, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Moré, G.; Regensburger, C.; Gos, M.L.; Pardini, L.; Verma, S.K.; Ctibor, J.; Serrano-Martínez, M.E.; Dubey, J.P.; Venturini, M.C. Sarcocystis masoni, n. Sp. (Apicomplexa: Sarcocystidae), and Redescription of Sarcocystis Aucheniae from Llama (Lama glama), Guanaco (Lama guanicoe) and Alpaca (Vicugna pacos). Parasitology 2016, 143, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Irikura, D.; Saito, M.; Sugita-Konishi, Y.; Ohnishi, T.; Sugiyama, K.; Watanabe, M.; Yamazaki, A.; Izumiyama, S.; Sato, H.; Kimura, Y.; et al. Characterization of Sarcocystis fayeri’s Actin-Depolymerizing Factor as a Toxin That Causes Diarrhea. Genes Cells 2017, 22, 825–835. [Google Scholar] [CrossRef] [PubMed]
- Kamata, Y.; Saito, M.; Irikura, D.; Yahata, Y.; Ohnishi, T.; Bessho, T.; Inui, T.; Watanabe, M.; Sugita-Konishi, Y. A Toxin Isolated from Sarcocystis fayeri in Raw Horsemeat May Be Responsible for Food Poisoning. J. Food Prot. 2014, 77, 814–819. [Google Scholar] [CrossRef]
- Decker Franco, C.; Wieser, S.N.; Soria, M.; de Alba, P.; Florin-Christensen, M.; Schnittger, L. In Silico Identification of Immunotherapeutic and Diagnostic Targets in the Glycosylphosphatidylinositol Metabolism of the Coccidian Sarcocystis aucheniae. Transbound Emerg. Dis. 2020, 67, 165–174. [Google Scholar] [CrossRef]
- Martin, M.; Decker Franco, C.; Romero, S.; Carletti, T.; Schnittger, L.; Florin-Christensen, M. Molecular Detection of Sarcocystis aucheniae in the Blood of Llamas from Argentina. Rev. Argent Microbiol. 2016, 48, 200–205. [Google Scholar] [CrossRef]
- Decker Franco, C.; Romero, S.; Ferrari, A.; Schnittger, L.; Florin-Christensen, M. Detection of Sarcocystis Aucheniae in Blood of Llama Using a Duplex Semi-Nested PCR Assay and Its Association with Cyst Infestation. Heliyon 2018, 4, e00928. [Google Scholar] [CrossRef]
- Moré, G.; Pardini, L.; Basso, W.; Marín, R.; Bacigalupe, D.; Auad, G.; Venturini, L.; Venturini, M.C. Seroprevalence of Neospora caninum, Toxoplasma gondii and Sarcocystis sp. in Llamas (Lama glama) from Jujuy, Argentina. Vet. Parasitol. 2008, 155, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Romero, S.; Carletti, T.; Decker Franco, C.; Moré, G.; Schnittger, L.; Florin-Christensen, M. Seropositivity to Sarcocystis Infection of Llamas Correlates with Breeding Practices. Vet. Parasitol. Reg. Stud. Rep. 2017, 10, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Dubey, J.P. A Review of Coccidiosis in South American Camelids. Parasitol. Res. 2018, 117, 1999–2013. [Google Scholar] [CrossRef] [PubMed]
- Cañal, V.; Beltrame, M.O. Gastrointestinal Parasite Diversity of South American Camelids (Artiodactyla: Camelidae): First Review throughout the Native Range of Distribution. Int. J. Parasitol. Parasites Wildl. 2022, 19, 222–242. [Google Scholar] [CrossRef] [PubMed]
- Can, H.; Aksoy Gökmen, A.; Döşkaya, M.; Erkunt Alak, S.; Değirmenci Döşkaya, A.; Karakavuk, M.; Köseoğlu, A.E.; Karakavuk, T.; Gül, C.; Güvendi, M.; et al. Development of a New Serotyping ELISA for Toxoplasma gondii Type II, Type III and Africa 1 Lineages Using in Silico Peptide Discovery Methods, Well Categorized Feline and Human Outbreak Serum Samples. BMC Infect Dis. 2022, 22, 110. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.J.; Korcsmaros, T.; Carding, S.R. Mechanisms and Pathways of Toxoplasma gondii Transepithelial Migration. Tissue Barriers 2017, 5, e1273865. [Google Scholar] [CrossRef] [PubMed]
- Dangoudoubiyam, S.; Oliveira, J.B.; Víquezà, C.; Gó Mezgarcíà, A.; González, O.; Romeroà, J.J.; Kwok, O.C.H.; Dubey, J.P.; Howe, D.K. Detection of Antibodies against Sarcocystis neurona, Neospora spp., and Toxoplasma gondii in Horses from Costa Rica. J. Parasitol. 2011, 97, 522–524. [Google Scholar] [CrossRef]
- Dominguez, M.; Echaide, I.; de Echaide, S.T.; Wilkowsky, S.; Zabal, O.; Mosqueda, J.J.; Schnittger, L.; Florin-Christensen, M. Validation and Field Evaluation of a Competitive Enzyme-Linked Immunosorbent Assay for Diagnosis of Babesia bovis Infections in Argentina. Clin. Vaccine Immunol. 2012, 19, 924–928. [Google Scholar] [CrossRef]
- Dubois, D.J.; Soldati-Favre, D. Biogenesis and Secretion of Micronemes in Toxoplasma gondii. Cell Microbiol. 2019, 21, e13018. [Google Scholar] [CrossRef]
- Döşkaya, M.; Liang, L.; Jain, A.; Can, H.; Gülçe Iz, S.; Felgner, P.L.; Deǧirmenci Döşkaya, A.; Davies, D.H.; Gürüz, A.Y. Discovery of New Toxoplasma gondii Antigenic Proteins Using a High Throughput Protein Microarray Approach Screening Sera of Murine Model Infected Orally with Oocysts and Tissue Cysts. Parasit. Vectors 2018, 11, 393. [Google Scholar] [CrossRef]
- Juárez-Estrada, M.A.; Tellez-Isaias, G.; Graham, D.M.; Laverty, L.; Gayosso-Vázquez, A.; Alonso-Morales, R.A. Identification of Eimeria tenella Sporozoite Immunodominant Mimotopes by Random Phage-Display Peptide Libraries—A Proof of Concept Study. Front. Vet. Sci. 2023, 10, 1223436. [Google Scholar] [CrossRef] [PubMed]
- Frénal, K.; Dubremetz, J.F.; Lebrun, M.; Soldati-Favre, D. Gliding Motility Powers Invasion and Egress in Apicomplexa. Nat. Rev. Microbiol. 2017, 15, 645–660. [Google Scholar] [CrossRef]
- Baum, J.; Papenfuss, A.T.; Baum, B.; Speed, T.P.; Cowman, A.F. Regulation of Apicomplexan Actin-Based Motility. Nat. Rev. Microbiol. 2006, 4, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Heintzelman, M.B. Gliding Motility in Apicomplexan Parasites. Semin Cell Dev. Biol. 2015, 46, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.R.; Barrios-Arpi, M.; Rodríguez, J.; Balcázarnakamatsu, S.; Zarria, J.; Namiyama, G.; Taniwaki, N.; Gonzales-Viera, O. Ultrastructural Description of Sarcocystis sp. in Cardiac Muscle of Naturally Infected Alpacas (Vicugna pacos). Iran. J. Parasitol. 2019, 14, 174–179. [Google Scholar]
- Blackman, M.J.; Bannister, L.H. Apical Organelles of Apicomplexa: Biology and Isolation by Subcellular Fractionation. Mol. Biochem. Parasitol. 2001, 117, 11–25. [Google Scholar] [CrossRef]
- Mercier, C.; Adjogble, K.D.Z.; Däubener, W.; Delauw, M.F.C. Dense Granules: Are They Key Organelles to Help Understand the Parasitophorous Vacuole of All Apicomplexa Parasites? Int. J. Parasitol. 2005, 35, 829–849. [Google Scholar] [CrossRef]
- Sajid, M.; Blackman, M.J.; Doyle, P.; He, C.; Land, K.M.; Lobo, C.; Mackey, Z.; Ndao, M.; Reed, S.L.; Shiels, B.; et al. Proteases of Parasitic Protozoa—Current Status and Validation. In Antiparasitic and Antibacterial Drug Discovery: From Molecular Targets to Drug Candidates; Wiley-VCH Verlag GmbH & Co.: Weinheim, Germany, 2009; pp. 175–209. [Google Scholar] [CrossRef]
- Florin-Christensen, M.; Sojka, D.; Ganzinelli, S.; Šnebergerová, P.; Suarez, C.E.; Schnittger, L. Degrade to Survive: The Intricate World of Piroplasmid Proteases. Trends Parasitol. 2023, 39, 532–546. [Google Scholar] [CrossRef]
- Hajagos, B.E.; Turetzky, J.M.; Peng, E.D.; Cheng, S.J.; Ryan, C.M.; Souda, P.; Whitelegge, J.P.; Lebrun, M.; Dubremetz, J.F.; Bradley, P.J. Molecular Dissection of Novel Trafficking and Processing of the Toxoplasma gondii Rhoptry Metalloprotease Toxolysin-1. Traffic 2012, 13, 292–304. [Google Scholar] [CrossRef]
- Xie, S.C.; Dick, L.R.; Gould, A.; Brand, S.; Tilley, L. The Proteasome as a Target for Protozoan Parasites. Expert Opin. Ther. Targets 2019, 23, 903–914. [Google Scholar] [CrossRef]
- Soto, A.S.; Fenoy, I.M.; Sanchez, V.R.; March, F.; Perrone Sibilia, M.D.; Aldirico, M.D.L.A.; Picchio, M.S.; Arcon, N.; Acosta, P.L.; Polack, F.P.; et al. Toxoplasma gondii Serine-Protease Inhibitor-1: A New Adjuvant Candidate for Asthma Therapy. PLoS ONE 2017, 12, e0187002. [Google Scholar] [CrossRef]
- Fleige, T.; Pfaff, N.; Gross, U.; Bohne, W. Localisation of Gluconeogenesis and Tricarboxylic Acid (TCA)-Cycle Enzymes and First Functional Analysis of the TCA Cycle in Toxoplasma gondii. Int. J. Parasitol. 2008, 38, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Ghaffar, F.; Bashtar, A.R.; Al-Quraishy, S.; Al Nasr, I.; Mehlhorn, H. Sarcocystis Infecting Reptiles in Saudi Arabia: 11-Light and Electron Microscopic Study on Sarcocysts of Sarcocystis turcicii sp. nov. Infecting the Gecko Hemidactylus turcicus linnaeus. Parasitol. Res. 2009, 104, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Qian, W.; Li, X.; Wang, T.; Ding, K.; Huang, T. Morphological and Molecular Characterization of Sarcocystis miescheriana from Pigs in the Central Region of China. Parasitol. Res. 2013, 112, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Ashwinder, K.; Kho, M.T.; Chee, P.M.; Lim, W.Z.; Yap, I.K.S.; Choi, S.B.; Yam, W.K. Targeting Heat Shock Proteins 60 and 70 of Toxoplasma gondii as a Potential Drug Target: In Silico Approach. Interdiscip. Sci. 2016, 8, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Anas, M.; Kumari, V.; Gupta, N.; Dube, A.; Kumar, N. Protein Quality Control Machinery in Intracellular Protozoan Parasites: Hopes and Challenges for Therapeutic Targeting. Cell Stress Chaperones 2019, 24, 891–904. [Google Scholar] [CrossRef] [PubMed]
- Welti, R.; Mui, E.; Sparks, A.; Wernimont, S.; Isaac, G.; Kirisits, M.; Roth, M.; Roberts, C.W.; Botté, C.; Maréchal, E.; et al. Lipidomic Analysis of Toxoplasma gondii Reveals Unusual Polar Lipids. Biochemistry 2007, 46, 13882–13890. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Liang, X.; Brouwers, J.F.; Miron, R.C.; Shen, B.; Gupta, N. Synthesis vs. Salvage of Ester- and Ether-Linked Phosphatidylethanolamine in the Intracellular Protozoan Pathogen Toxoplasma gondii. Commun. Biol. 2023, 6, 306. [Google Scholar] [CrossRef]
- Pszenny, V.; Ehrenman, K.; Romano, J.D.; Kennard, A.; Schultz, A.; Roos, D.S.; Grigg, M.E.; Carruthers, V.B.; Coppens, I. A Lipolytic Lecithin:Cholesterol Acyltransferase Secreted by Toxoplasma Facilitates Parasite Replication and Egress. J. Biol. Chem. 2016, 291, 3725–3746. [Google Scholar] [CrossRef]
- Li, L.; Brunk, B.P.; Kissinger, J.C.; Pape, D.; Tang, K.; Cole, R.H.; Martin, J.; Wylie, T.; Dante, M.; Fogarty, S.J.; et al. Gene Discovery in the Apicomplexa as Revealed by EST Sequencing and Assembly of a Comparative Gene Database. Genome Res. 2003, 13, 443–454. [Google Scholar] [CrossRef]
- Hunt, A.G.; Howe, D.K.; Brown, A.; Yeargan, M. Transcriptional Dynamics in the Protozoan Parasite Sarcocystis neurona and Mammalian Host Cells after Treatment with a Specific Inhibitor of Apicomplexan MRNA Polyadenylation. PLoS ONE 2021, 16, e0259109. [Google Scholar] [CrossRef]
- Friedrich, A.; Ledesma, M.; Landone, I.; Ferrari, A.; Leoni, J. Production of a Monoclonal Antibody against Serum Immunoglobulin M of South American Camelids and Assessment of Its Suitability in Two Immunoassays. J. Vet. Diagn. Investig. 2014, 26, 646–650. [Google Scholar] [CrossRef]
- Wang, L.; Coppel, R.L. Triton X-114 Phase Partitioning Triton X-114 Phase Partitioning for Antigen Characterization. In Malaria Methods and Protocols: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2002; pp. 581–585. [Google Scholar]
- Yao, B.; Zheng, D.; Liang, S.; Zhang, C. SVMTriP: A Method to Predict B-Cell Linear Antigenic Epitopes. Methods Mol. Biol. 2020, 2131, 299–307. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: Improving Sequence-Based B-Cell Epitope Prediction Using Conformational Epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Madden, T. The BLAST Sequence Analysis Tool; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013. [Google Scholar]
- McGinnis, S.; Madden, T.L. BLAST: At the Core of a Powerful and Diverse Set of Sequence Analysis Tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Eisenhaber, B.; Bork, P.; Eisenhaber, F. Prediction of Potential GPI-Modification Sites in Proprotein Sequences. J. Mol. Biol. 1999, 292, 741–758. [Google Scholar] [CrossRef]
- Fankhauser, N.; Mäser, P. Identification of GPI Anchor Attachment Signals by a Kohonen Self-Organizing Map. Bioinformatics 2005, 21, 1846–1852. [Google Scholar] [CrossRef]
- Pierleoni, A.; Martelli, P.; Casadio, R. PredGPI: A GPI-Anchor Predictor. BMC Bioinform. 2008, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).