Forgotten for Decades: Revalidation and Redescription of Raiamas harmandi (Sauvage, 1880) (Cypriniformes: Danionidae) from the Mekong River Basin
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Morphological Analysis
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Phylogenetic Analysis and Genetic Distance
3. Results
3.1. Molecular Phylogeny and Genetic Distance
3.2. Taxonomy and Morphology
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 16 August 2025).
- Stout, C.C.; Tan, M.; Lemmon, A.R.; Lemmon, E.M.; Armbruster, J.W. Resolving Cypriniformes relationships using an anchored enrichment approach. BMC Evol. Biol. 2016, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Armbruster, J.W. Phylogenetic classification of extant genera of fishes of the order Cypriniformes (Teleostei: Ostariophysi). Zootaxa 2018, 4476, 6–39. [Google Scholar] [CrossRef]
- Jordan, D.S. New genera of fishes. Proc. Acad. Nat. Sci. Phila. 1919, 70, 341–344. [Google Scholar]
- Manda, B.K.; Snoeks, J.; Manda, A.C.; Vreven, E. Hidden species diversity in Raiamas salmolucius (Teleostei: Cyprinidae) from the Congo Basin: Two new species based on morphometric evidence. Ichthyol. Explor. Freshw. 2018, 28, 345–363. [Google Scholar] [CrossRef]
- Hamilton, F. An Account of the Fishes Found in the River Ganges and Its Branches; Archibald Constable: Edinburgh, UK, 1822; pp. 1–405. [Google Scholar]
- Day, F. On the freshwater fishes of Burma—Part I. Proc. Zool. Soc. Lond. 1870, 37, 614–623. [Google Scholar] [CrossRef]
- Liao, T.Y.; Arroyave, J.; Stiassny, M.L.J. Diagnosis of Asian Raiamas (Teleostei: Cyprinidae: Chedrina) with comments on Chedrin relationships and previously proposed diagnostic characters for Opsaridium and Raiamas. Ichthyol. Res. 2012, 59, 328–341. [Google Scholar] [CrossRef]
- Liao, T.Y.; Ünlü, E.; Kullander, S.O. Western boundary of the Subfamily Danioninae in Asia (Teleostei, Cyprinidae): Derived from the systematic position of Barilius mesopotamicus based on molecular and morphological data. Zootaxa 2011, 2880, 31–40. [Google Scholar] [CrossRef]
- Tang, K.L.; Agnew, M.K.; Hirt, M.V.; Sado, T.; Schneider, L.M.; Freyhof, J.; Sulaiman, Z.; Swartz, E.; Vidthayanon, C.; Miya, M.; et al. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol. Phylogenet. Evol. 2010, 57, 189–214. [Google Scholar] [CrossRef]
- McClelland, J. Indian Cyprinidae. Asiat. Res. 1839, 19, 217–417. [Google Scholar]
- Blyth, E. Report of Curator, Zoological Department, for May, 1858. J. Asiat. Soc. Bengal 1858, 27, 267–290. [Google Scholar]
- Shrestha, J. Fishes of Nepal; Curriculum Development Centre, Tribhuvan Univ.: Kathmandu, Nepal, 1980. [Google Scholar]
- Tilak, R.; Husain, A. Description of a new species of the Genus Barilius Hamilton (Cyprinidae: Cypriniformes) from Corbett National Park, Uttar Pradesh. Mitt. Zool. Mus. Berl. 1980, 56, 41–44. [Google Scholar]
- Kottelat, M. The fishes of the inland waters of Southeast Asia: A catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles Bull. Zool. 2013, (Suppl. S27), 1–663. [Google Scholar]
- Menon, A.G.K. Check List-Fresh Water Fishes of India; Zoological Survey of India: Calcutta, India, 1999; ISBN 978-81-85874-15-9. [Google Scholar]
- Talwar, P.K.; Jhingran, A.G. Inland Fishes of India and Adjacent Countries; Oxford & IBH Publishing, Co.: New Delhi, India; Bombay, India; Calcutta, India, 1991. [Google Scholar]
- Sauvage, H.E. Notice sur quelques poissons de l’île Campbell et de l’Indo-Chine. Bull. Soc. Philom. Paris 1880, 7, 228–233. [Google Scholar]
- Yang, Y.; Hwang, H. Leuciscinae. In The Cyprinid Fishes of China; Shanghai Science & Technology Press: Shanghai, China, 1964; Volume 1, pp. 7–61. [Google Scholar]
- Kottelat, M. Fishes of Laos; WHT Publications: Colombo, Sri Lanka, 2001. [Google Scholar]
- Aqmal-Naser, M.; Ali, N.; Azmi, N.; Fahmi-Ahmad, M.; Rizal, S.; Ahmad, A. Freshwater fishes (Actinopterygii) of Kenyir Reservoir, Peninsular Malaysia: Updated checklist, taxonomic concerns and alien species. BDJ 2023, 11, e100337. [Google Scholar] [CrossRef]
- Setiawan, A.; Iqbal, M.; Pormansyah, P.; Setiawan, D.; Yustian, I. The burmese trout Raiamas guttatus (Day, 1870) (Cypriniformes: Cyprinidae) in South Sumatra revealed its southernmost record of its distributional range. Ecol. Mont. 2020, 27, 39–44. [Google Scholar] [CrossRef]
- Pasco-Viel, E.; Veran, M.; Viriot, L. Bleeker was right: Revision of the genus Cyclocheilichthys (Bleeker 1859) and resurrection of the genus Anematichthys (Bleeker 1859), based on morphological and molecular data of Southeast Asian Cyprininae (Teleostei, Cypriniformes). Zootaxa 2012, 3586, 41–54. [Google Scholar] [CrossRef]
- Saitoh, K.; Sado, T.; Doosey, M.H.; Bart, H.L.; Inoue, J.G.; Nishida, M.; Mayden, R.L.; Miya, M. Evidence from mitochondrial genomics supports the lower Mesozoic of South Asia as the time and place of basal divergence of cypriniform fishes (Actinopterygii: Ostariophysi): Cypriniform mitogenomics and biogeography. Zool. J. Linn. Soc.-lond. 2011, 161, 633–662. [Google Scholar] [CrossRef]
- Rüber, L.; Kottelat, M.; Tan, H.; Ng, P.K.L.; Britz, R. Evolution of miniaturization and the phylogenetic position of Paedocypris, comprising the world’s smallest vertebrate. BMC Evol. Biol. 2007, 7, 38. [Google Scholar] [CrossRef]
- Yu, P.; Zhou, L.; Yang, X.-L.; Li, L.-L.; Jin, H.; Li, Z.; Wang, Z.-W.; Li, X.-Y.; Zhang, X.-J.; Wang, Y.; et al. Comparative mitogenomic analyses unveil conserved and variable mitogenomic features and phylogeny of Chedrinae fish. Zool. Res. 2022, 43, 30–32. [Google Scholar] [CrossRef]
- Chen, X.J.; Song, L.; Cao, Y.W.; Wang, Q. Phylogenetic relationship and characterization of the complete mitochondrial genome sequence of Opsarius caudiocellatus (Cypriniformes: Danionidae: Chedrinae). Mitochondrial DNA Part B 2022, 7, 2051–2055. [Google Scholar] [CrossRef] [PubMed]
- Flynn, T.; Signal, B.; Johnson, S.L.; Gemmell, N.J. Mitochondrial genome diversity among six laboratory zebrafish (Danio rerio) strains. Mitochondrial DNA Part A 2016, 27, 4364–4371. [Google Scholar] [CrossRef] [PubMed]
- Stiassny, M.L.J.; Schelly, R.C.; Schliewen, U.K. A new species of Raiamas (Teleostei: Cyprinidae) from the lower Congo River, with a phylogenetic assessment of the generic limits of the predatory cyprinid genera Opsaridium, Raiamas, and Leptocypris. Copeia 2006, 3, 370–377. [Google Scholar] [CrossRef]
- Mayden, R.L.; Tang, K.L.; Conway, K.W.; Freyhof, J.; Chamberlain, S.; Haskins, M.; Schneider, L.; Sudkamp, M.; Wood, R.M.; Agnew, M.; et al. Phylogenetic relationships of Danio within the order Cypriniformes: A framework for comparative and evolutionary studies of a model species. J. Exp. Zool. Part B 2007, 308B, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Phil. Trans. R. Soc. B 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. In Bioinformatics Methods and Protocols; Humana Press: Totowa, NJ, USA, 2000; Volume 132, pp. 71–91. ISBN 978-1-59259-192-3. [Google Scholar]
- Xiang, C.; Gao, F.; Jakovlić, I.; Lei, H.; Hu, Y.; Zhang, H.; Zou, H.; Wang, G.; Zhang, D. Using PhyloSuite for molecular phylogeny and tree-based analyses. iMeta 2023, 2, e87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.M. The fresh-water fishes of Siam, or Thailand. Bull. U. S. Natl. Mus. 1945, 188, 1–622. [Google Scholar] [CrossRef]
- Li, S. New records of Chinese fishes from the Lancang River, Yunnan Province. Acta Zool. Sin. 1976, 22, 117–118. [Google Scholar]
- Kottelat, M. A review of the species of Indochinese fresh-water fishes described by H.-E. Sauvage. Bull. Mus. Natl. Hist. Nat. Sect. A 1984, 6, 791–822. [Google Scholar] [CrossRef]
- Kuang, F. Danioninae. In Fishes of Yunnan Province I; Science Press: Beijing, China, 1989; pp. 11–35. [Google Scholar]
- Rainboth, W.J. Fishes of the Cambodian Mekong; Food & Agriculture Org.: Rome, Italy, 1996. [Google Scholar]
- Zeng, Y.; Liu, C.; Pu, X.; Lyu, J.; Chen, X. A new record of Danio from the Red River and Salween River basins in Yunnan Province and a taxonomic revision of D. albolineatus in China. Chin. J. Zool. 2025, in press. [Google Scholar]

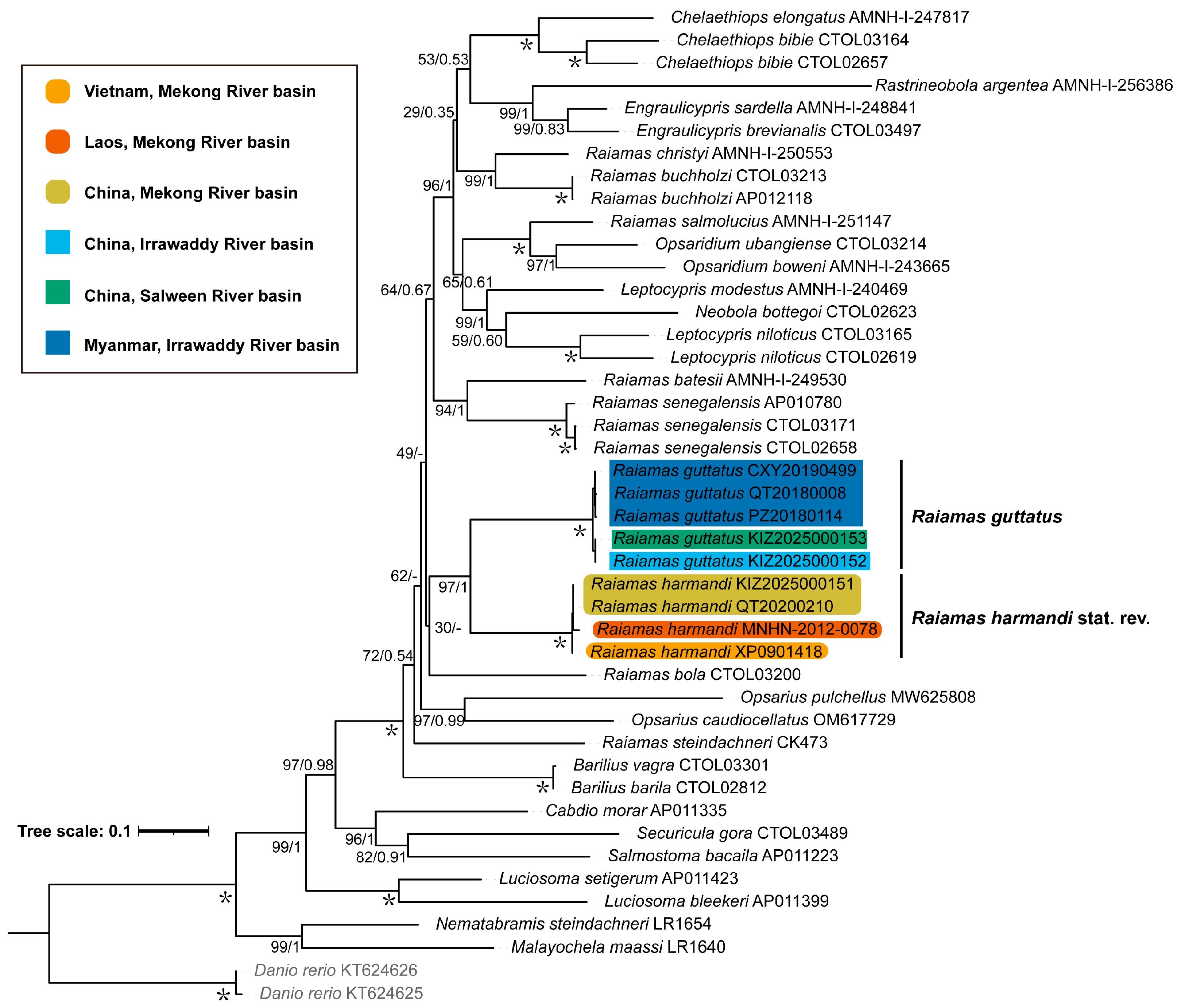


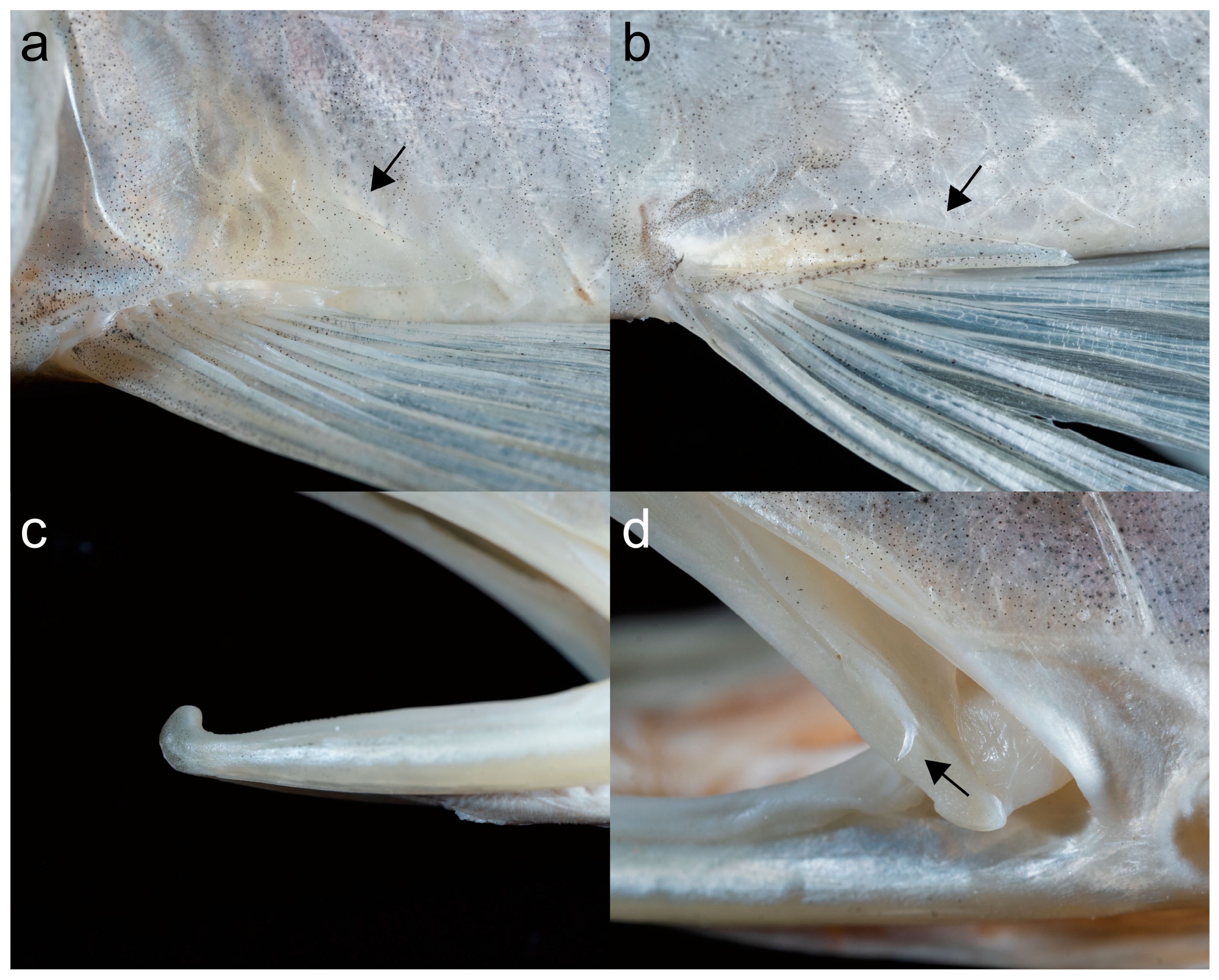

| Species | Voucher No. | COI | Cyt b | Source |
|---|---|---|---|---|
| Ingroup | ||||
| Barilius cf. barila 1 | CTOL02812 | HM224138 | HM224257 | [10] |
| Barilius vagra 1 | CTOL03301 | HM224140 | HM224259 | [10] |
| Chelaethiops bibie 2 | CTOL02657 | HM224141 | HM224260 | [10] |
| Chelaethiops bibie 2 | CTOL03164 | HM224142 | HM224261 | [10] |
| Chelaethiops elongatus 2 | AMNH-I-247817 | JX196996 | JX197006 | [8] |
| Engraulicypris Sardella 2 | AMNH-I-248841 | JX196997 | JX197007 | [8] |
| Engraulicypris brevianalis 2 | CTOL03497 | HM224176 | HM224295 | [10] |
| Leptocypris modestus 2 | AMNH-I-240469 | JX196998 | JX197008 | [8] |
| Leptocypris niloticus 2 | CTOL02619 | HM224174 | HM224293 | [10] |
| Leptocypris niloticus 2 | CTOL03165 | HM224175 | HM224294 | [10] |
| Opsaridium boweni 2 | AMNH-I-243665 | JX197000 | JX197009 | [8] |
| Opsaridium ubangiense 2 | CTOL03214 | HM224193 | HM224312 | [10] |
| Raiamas batesii 2 | AMNH-I-249530 | JX197002 | JX197010 | [8] |
| Raiamas bola 1 | CTOL03200 | HM224212 | HM224329 | [10] |
| Raiamas buchholzi 2 | CTOL03213 | HM224213 | HM224330 | [10] |
| Raiamas buchholzi 2 | no6265 | AP012118 | AP012118 | Direct Submission |
| Raiamas christyi 2 | AMNH-I-250553 | JX197003 | JX197011 | [8] |
| Raiamas guttatus 1* | KIZ2025000152 | PV960713 | PV976866 | This study |
| Raiamas guttatus 1* | KIZ2025000153 | PV960714 | PV976867 | This study |
| Raiamas guttatus 1 | CXY20190499 | PV960710 | PV976868 | This study |
| Raiamas guttatus 1 | PZ20180114 | PV960711 | PV976869 | This study |
| Raiamas guttatus 1 | QT20180008 | PV960712 | PV976870 | This study |
| Raiamas harmandi 1 | QT20200210 | PV960707 | PV976864 | This study |
| Raiamas harmandi 1* | KIZ2025000151 | PV960709 | PV976863 | This study |
| Raiamas harmandi 1 | XP0901418 | PV960708 | PV976865 | This study |
| Raiamas harmandi 1 | MNHN-2012-0078 | JQ346159 | JQ346143 | [23] |
| Raiamas salmolucius 2 | AMNH-I-251147 | JX197004 | JX197012 | [8] |
| Raiamas senegalensis 2 | CTOL02658 | HM224215 | HM224332 | [10] |
| Raiamas senegalensis 2 | CTOL03171 | HM224216 | HM224333 | [10] |
| Raiamas senegalensis 2 | N/A | AP010780 | AP010780 | [24] |
| Raiamas steindachneri 2 | CK473 | AP012113 | AP012113 | Direct Submission |
| Rastrineobola argentea 2 | AMNH-I-256386 | JX197005 | JX197013 | [8] |
| Luciosoma bleekeri 1 | CBM-ZF-11202 | AP011399 | AP011399 | [10] |
| Luciosoma setigerum 1 | CBM-ZF-11273 | AP011423 | AP011423 | [10] |
| Salmostoma bacaila 1 | CBM-ZF-11516 | AP011223 | AP011223 | [10] |
| Cabdio morar 1 | CBM-ZF-11391 | AP011335 | AP011335 | [10] |
| Malayochela maassi 1 | LR1640 | FJ753486 | EF151098 | [25] |
| Nematabramis steindachneri 1 | LR1654 | FJ753496 | EF151106 | [25] |
| Neobola bottegoi 2 | CTOL02623 | HM224178 | HM224296 | [10] |
| Opsarius pulchellus 1 | N/A | MW625808 | MW625808 | [26] |
| Opsarius caudiocellatus 1 | N/A | OM617729 | OM617729 | [27] |
| Securicula gora 1 | CTOL03489 | HM224250 | HM224381 | [10] |
| Outgroup | ||||
| Danio rerio | N/A | KT624625 | KT624625 | [28] |
| Danio rerio | N/A | KT624626 | KT624626 | [28] |
| COI | Cyt b | |||||||
|---|---|---|---|---|---|---|---|---|
| Species | Intraspecific | 1 | 2 | 3 | Intraspecific | 1 | 2 | 3 |
| 1 Raiamas harmandi | 0–0.005 | – | – | – | 0–0.009 | – | – | – |
| 2 Raiamas guttatus | 0–0.003 | 0.140–0.149 | – | – | 0–0.009 | 0.161–0.170 | – | – |
| 3 Raiamas bola | – | 0.163–0.165 | 0.162–0.164 | – | – | 0.200–0.211 | 0.214–0.216 | – |
| Characters | Raiamas harmandi | R. guttatus | |||
|---|---|---|---|---|---|
| Holotype | Mean ± SD | Range | Mean ± SD | Range | |
| Standard length (SL, mm) | 160.0 | 167.4 ± 45.2 | 88.6–240.2 | 158.8 ± 28.0 | 121.7–210.6 |
| In Percent of SL (%) | |||||
| Body depth | 21.1 | 20.9 ± 2.4 | 16.9–26.2 | 22.4 ± 2.1 | 17.4–26.6 |
| Pectoral-fin length | 19.4 | 18.0 ± 1.0 | 16.3–20.0 | 19.0 ± 0.9 | 17.2–20.6 |
| Prepectoral length | 28.4 | 27.3 ± 0.9 | 25.6–29.4 | 28.1 ± 1.2 | 25.8–30.4 |
| Pelvic-fin length | 14.1 | 13.0 ± 0.8 | 11.9–14.3 | 12.6 ± 0.6 | 11.6–14.4 |
| Prepelvic length | 49.8 | 50.4 ± 1.6 | 45.6–53.6 | 51.4 ± 1.3 | 49.6–54.6 |
| Dorsal-fin length | 16.0 | 17.6 ± 0.8 | 16.0–19.0 | 16.6 ± 0.7 | 15.3–17.6 |
| Dorsal-fin base length | 10.8 | 10.9 ± 0.7 | 9.8–12.2 | 10.6 ± 0.6 | 9.6–11.6 |
| Predorsal length | 57.9 | 56.8 ± 1.6 | 51.3–59.4 | 57.7 ± 1.3 | 54.3–60.1 |
| Anal-fin length 1 | 14.8 | 14.6 ± 0.9 | 13.0–16.5 | 14.5 ± 0.6 | 12.8–15.3 |
| Anal-fin length 2 | 4.6 | 5.7 ± 0.6 | 4.3–6.7 | 5.2 ± 0.5 | 4.1–6.2 |
| Anal-fin base length | 13.4 | 13.4 ± 0.6 | 12.1–14.5 | 15.1 ± 1.1 | 13.2–17.2 |
| Preanal length | 70.0 | 70.4 ± 2.2 | 64.6–74.5 | 71.9 ± 1.7 | 69.2–76.2 |
| Caudal peduncle length | 17.9 | 17.2 ± 1.0 | 15.5–19.4 | 16.7 ± 1.1 | 13.9–18.7 |
| Caudal peduncle depth | 9.6 | 8.7 ± 0.5 | 7.9–9.8 | 9.2 ± 0.4 | 8.2–9.8 |
| Head length (HL) | 27.8 | 27.0 ± 0.9 | 25.4–29.2 | 27.5 ± 1.1 | 25.8–29.7 |
| In Percent of HL (%) | |||||
| Head depth | 61.5 | 61.5 ± 2.0 | 57.1–66.6 | 60.6 ± 2.4 | 56.6–65.0 |
| Head width | 41.9 | 41.9 ± 2.3 | 38.8–48.7 | 44.1 ± 3.1 | 38.3–50.0 |
| Snout length | 26.8 | 28.7 ± 1.9 | 24.7–33.4 | 28.7 ± 1.5 | 25.7–31.7 |
| Interorbital width | 26.8 | 25.9 ± 1.3 | 23.6–28.2 | 26.7 ± 1.1 | 23.2–28.2 |
| Upper-jaw length | 67.1 | 64.2 ± 2.5 | 57.9–68.6 | 59.2 ± 1.3 | 56.3–61.9 |
| Eye diameter | 18.2 | 17.4 ± 2.2 | 13.9–22.1 | 17.0 ± 1.7 | 14.1–20.4 |
| Distance between nostril and snout tip | 18.9 | 19.9 ± 1.7 | 16.7–24.6 | 20.0 ± 1.3 | 18.0–23.3 |
| Horizontal eye to operculum distance | 29.7 | 32.5 ± 2.3 | 28.2–37.1 | 33.5 ± 2.3 | 28.4–38.1 |
| Opercular length | 24.8 | 20.8 ± 1.5 | 18.2–24.8 | 22.0 ± 1.4 | 19.4–24.3 |
| Counts | Holotype | Median | Range | Median | Range |
| Total number of vertebrae | 42 | 43 | 41–44 | 43.5 | 42–44 |
| Abdominal vertebrae | 22 | 22 | 21–23 | 22 | 21–23 |
| Caudal vertebrae | 20 | 21 | 20–22 | 21 | 21–22 |
| Dorsal-fin rays | iii, 7 | iii, 7 | iii, 7 | iii, 7 | iii, 7 |
| Anal-fin rays | iii, 10 | iii, 10 | iii, 10–iii, 11 | iii, 11 | iii, 10–iii, 12 |
| Pectoral-fin rays | i, 15 | i, 14 | i, 13–i, 15 | i, 13.5 | i, 13–i, 14 |
| Pelvic-fin rays | i, 8 | i, 8 | i, 8 | i, 8 | i, 8 |
| Lateral line scales | 45 | 47 | 45–50 | 48 | 46–50 |
| Lateral line–dorsal-fin origin scales | 9½ | 9½ | 8½–9½ | 9½ | 8½–9½ |
| Lateral line–pelvic-fin origin scales | 3 | 3 | 3–4 | 3 | 3–4 |
| Predorsal scales | 25 | 26 | 25–28 | 22 | 21–23 |
| Circumpeduncular scales | 19 | 18 | 18–20 | 18 | 18–19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-X.; Xu, Y.-Y.; Zeng, Y.-Y.; Naing Oo, T.; Chen, X.-Y. Forgotten for Decades: Revalidation and Redescription of Raiamas harmandi (Sauvage, 1880) (Cypriniformes: Danionidae) from the Mekong River Basin. Taxonomy 2025, 5, 42. https://doi.org/10.3390/taxonomy5030042
Liu C-X, Xu Y-Y, Zeng Y-Y, Naing Oo T, Chen X-Y. Forgotten for Decades: Revalidation and Redescription of Raiamas harmandi (Sauvage, 1880) (Cypriniformes: Danionidae) from the Mekong River Basin. Taxonomy. 2025; 5(3):42. https://doi.org/10.3390/taxonomy5030042
Chicago/Turabian StyleLiu, Cai-Xin, Yi-Yang Xu, Yu-Yang Zeng, Thaung Naing Oo, and Xiao-Yong Chen. 2025. "Forgotten for Decades: Revalidation and Redescription of Raiamas harmandi (Sauvage, 1880) (Cypriniformes: Danionidae) from the Mekong River Basin" Taxonomy 5, no. 3: 42. https://doi.org/10.3390/taxonomy5030042
APA StyleLiu, C.-X., Xu, Y.-Y., Zeng, Y.-Y., Naing Oo, T., & Chen, X.-Y. (2025). Forgotten for Decades: Revalidation and Redescription of Raiamas harmandi (Sauvage, 1880) (Cypriniformes: Danionidae) from the Mekong River Basin. Taxonomy, 5(3), 42. https://doi.org/10.3390/taxonomy5030042







