The Curious Case of Woodcreepers: Cytogenomic Evidence Based on the Position of NORs
Abstract
1. Introduction
2. Material and Methods
2.1. Specimens Analyzed and Chromosomal Preparations
2.2. Conventional Cytogenetics
2.3. Repetitive DNA Mapping by FISH
2.4. Molecular Phylogeny
3. Results
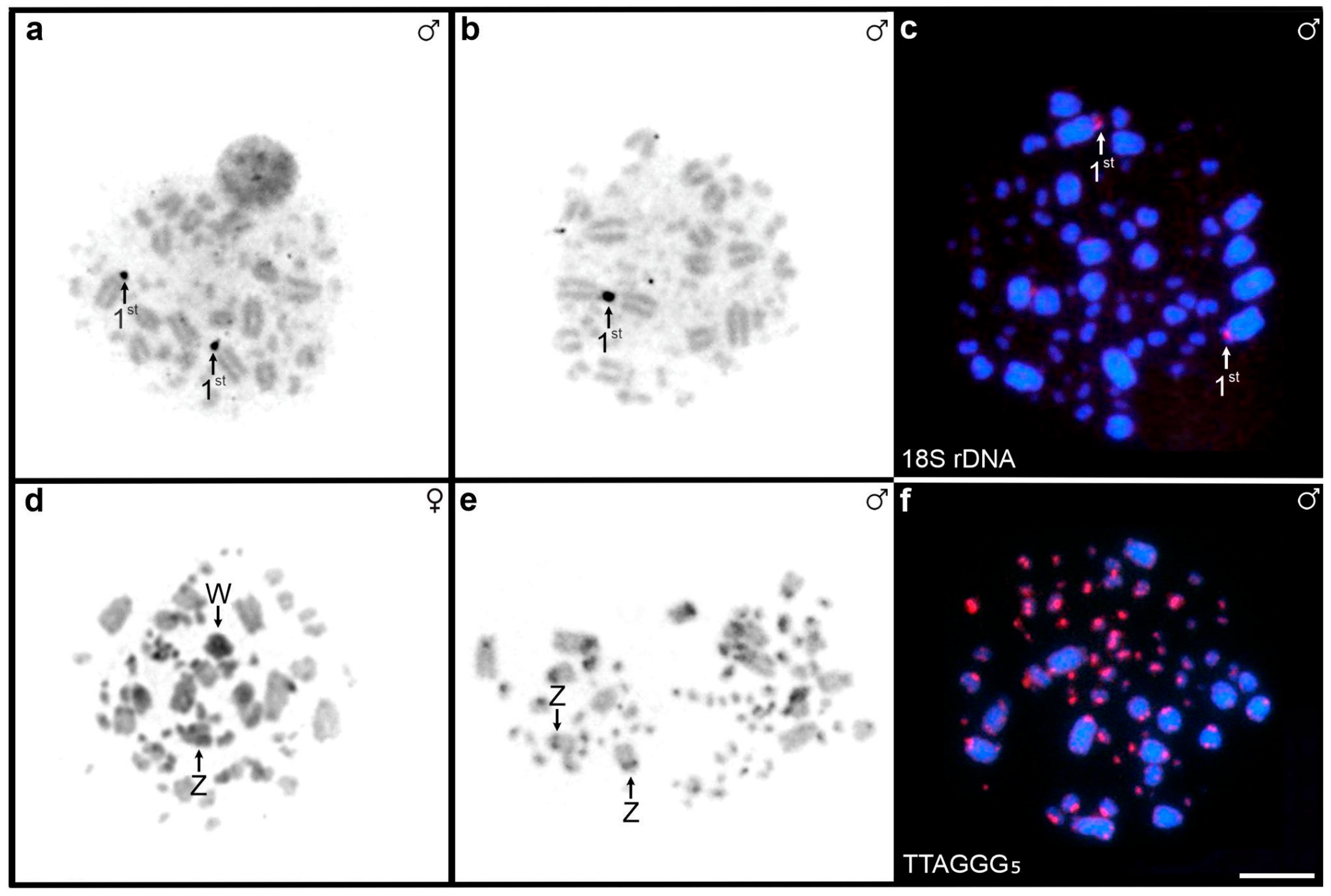
3.1. Cytogenetic Analysis
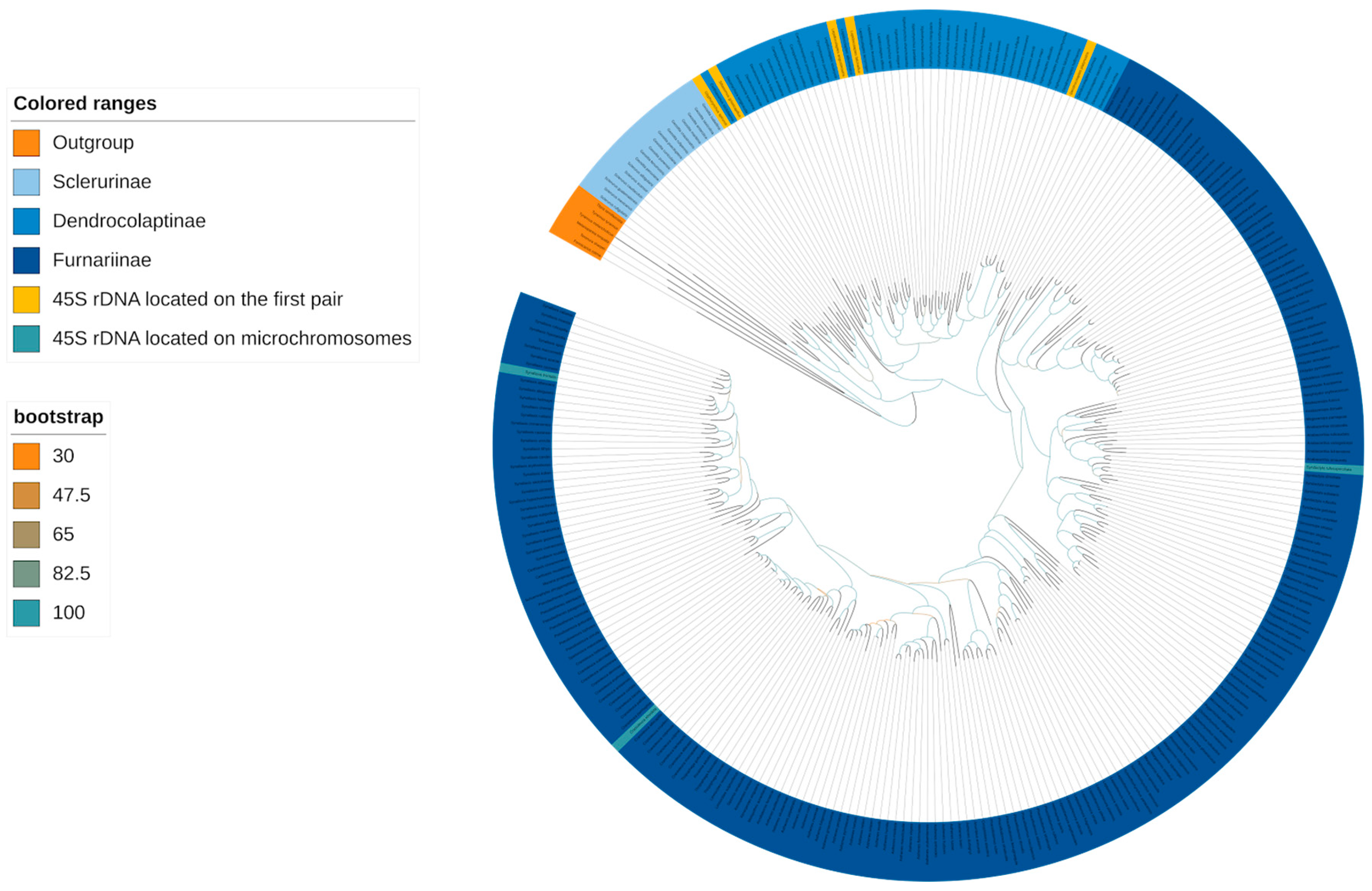
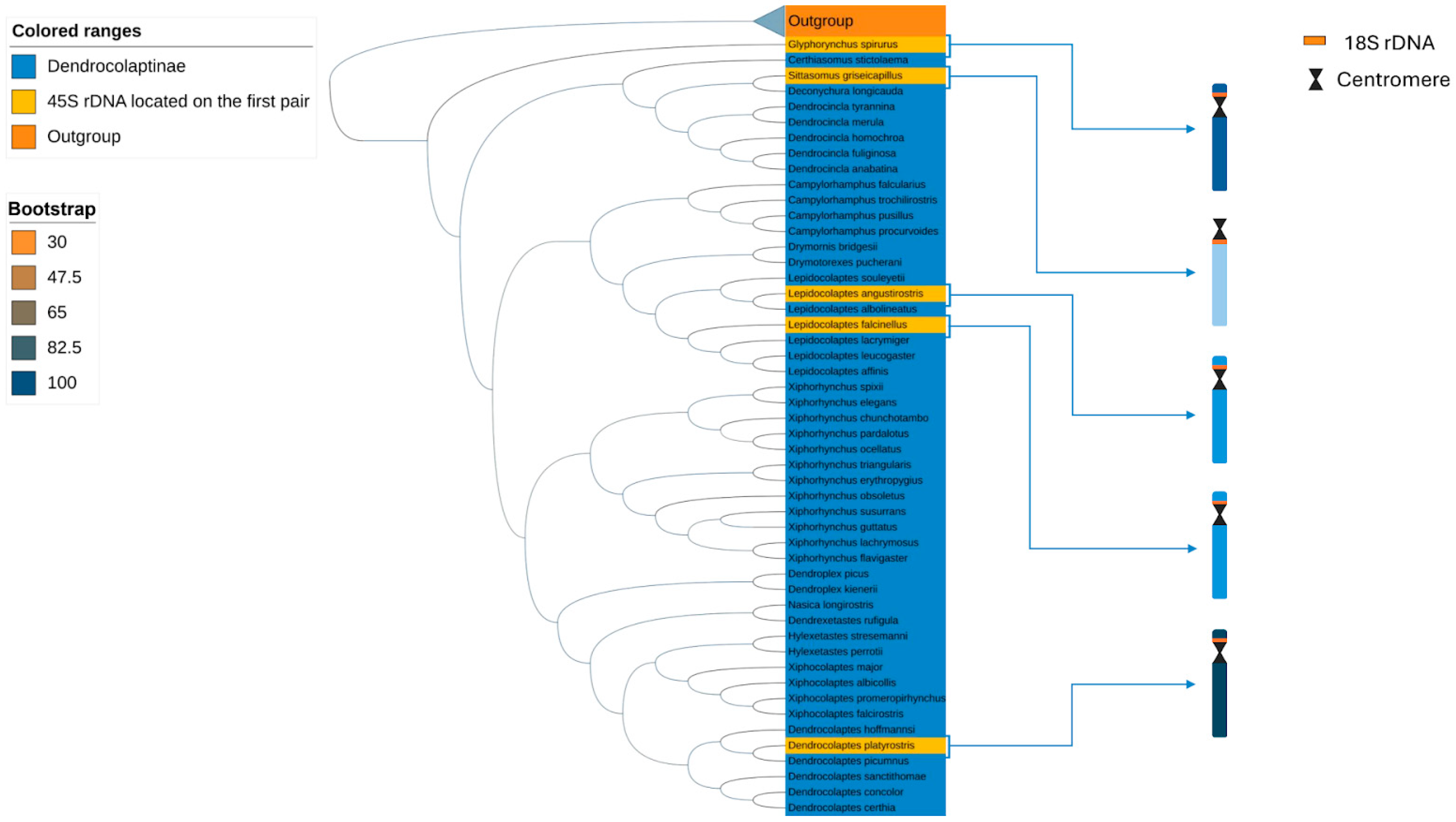
3.2. Phylogeny
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Engel, M.S.; Ceríaco, L.M.P.; Daniel, G.M.; Dellapé, P.M.; Löbl, I.; Marinov, M.; E Reis, R.; Young, M.T.; Dubois, A.; Agarwal, I.; et al. The taxonomic impediment: A shortage of taxonomists, not the lack of technical approaches. Zool. J. Linn. Soc. 2021, 193, 381–387. [Google Scholar] [CrossRef]
- Baker, P.A.; Fritz, S.C.; Dick, C.W.; Eckert, A.J.; Horton, B.K.; Stefano, M.; Camila, C.R.; Carmala, N.G.; David, S.B. The emerging field of geogenomics: Constraining geological problems with genetic data. Earth-Sci. Rev. 2014, 135, 38–47. [Google Scholar] [CrossRef]
- Moyle, R.G.; Chesser, R.T.; Brumfield, R.T.; Tello, J.G.; Marchese, D.J.; Cracraft, J. Phylogeny and phylogenetic classification of the antbirds, ovenbirds, woodcreepers, and allies (Aves: Passeriformes: Infraorder Furnariides). Cladistics 2009, 25, 386–405. [Google Scholar] [CrossRef] [PubMed]
- Irestedt, M.; Fjeldså, J.; Ericson, P.G.P. Phylogenetic relationships of woodcreepers (Aves: Dendrocolaptinae)—Incongruence between molecular and morphological data. J. Avian Biol. 2004, 35, 280–288. [Google Scholar] [CrossRef]
- Remsen, J.V., Jr.; Cadena, C.D.; Jaramillo, A.; Nores, M.; Pacheco, J.F.; Pérez-Emán, J.; Robbins, M.B.; Stiles, F.G.; Stotz, D.F.; Zimmer, K.J. A Classification of the Bird Species of South America. Am. Ornithol. Union 2018, 15. Available online: https://www.museum.lsu.edu/~Remsen/SACCWordFiles/SACCBaseline01.html (accessed on 12 January 2025).
- Fitzpatrick, J.W. Taxonomy and Geographical Distribution of the Furnariidae (Aves, Passeriformes). Bull. AMNH 1982, 166, 810–813. [Google Scholar]
- Claramunt, S. Discovering exceptional diversifications at continental scales: The case of the endemic families of neotropical suboscine passerines. Evolution 2010, 64, 2004–2019. [Google Scholar] [CrossRef]
- Claramunt, T.; Santiago, J. Testing models of biological diversification: Morphological evolution and cladogenesis in the neotropical furnariidae (Aves: Passeriformes). LSU Dr. Diss. 2010, 29118298, 1–60. [Google Scholar] [CrossRef]
- Winkler, D.W.; Billerman, S.M.; Lovette, I.J.; Keeney, B.K.; Rodewald, P.G.; Schulenberg, T.S. Ovenbirds and Woodcreepers (Furnariidae). In Birds of the World; Billerman, S.M., Keeney, B.K., Rodewald, P.G., Schulenberg, T.S., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Howard, R.; Moore, A. A Complete Checklist of the Birds of the World; Princeton University Press: Princeton, NJ, USA, 1991; Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19910506948 (accessed on 12 January 2025).
- Derryberry, E.P.; Claramunt, S.; Derryberry, G.; Chesser, R.T.; Cracraft, J.; Aleixo, A.; Pérez-Emán, J.; Remsen, J.V., Jr.; Brumfield, R.T. Lineage diversification and morphological evolution in a large-scale continental radiation: The neotropical ovenbirds and woodcreepers (aves: Furnariidae). Evolution 2011, 65, 2973–2986. [Google Scholar] [CrossRef]
- Snethlage, E. Über die verbreitung der vogelarten in unteramazonien. J. Ornithol. 1913, 61, 469–539. [Google Scholar] [CrossRef]
- Bock, W.J. Check-List of Birds of the World: A Continuation of the Work of James L. Peters 1990, 11, 629–648. Peters 1990, 11, 629–648. [Google Scholar]
- Sibley, C.G.; Ahlquist, J.E.; Monroe, B.L. A Classification of the Living Birds of the World Based on Dna-Dna Hybridization Studies. Auk 1988, 105, 409–423. [Google Scholar] [CrossRef]
- Aleixo, A. Molecular systematics and the role of the “várzea”-“terra-firme” ecotone in the diversification of Xiphorhynchus woodcreepers (Aves: Dendrocolaptidae). Auk 2002, 119, 621–640. [Google Scholar]
- Gill, F.; Donsker, D. IOC World Bird List; IOC: Lausanne, Switzerland, 2021; Volume 11. [Google Scholar]
- Barbosa, M.d.O.; Silva, R.R.D.; Correia, V.C.D.S.; Santos, L.P.D.; Garnero, A.D.V.; Gunski, R.J. Nucleolar organizer regions in Sittasomus griseicapillus and Lepidocolaptes angustirostris (Aves, Dendrocolaptidae): Evidence of a chromosome inversion. Genet. Mol. Biol. 2013, 36, 70–73. [Google Scholar] [CrossRef]
- Kretschmer, R.; de Lima, V.L.C.; de Souza, M.S.; Costa, A.L.; O’brien, P.C.M.; Ferguson-Smith, M.A.; de Oliveira, E.H.C.; Gunski, R.J.; Garnero, A.D.V. Multidirectional chromosome painting in Synallaxis frontalis (Passeriformes, Furnariidae) reveals high chromosomal reorganization, involving fissions and inversions. Comp. Cytogenet. 2018, 12, 97–110. [Google Scholar] [CrossRef]
- Ribas, T.F.A.; Nagamachi, C.Y.; Aleixo, A.; Pinheiro, M.L.S.; O´bRien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Suarez, P.; Pieczarka, J.C.; Rutherford, S. Chromosome painting in Glyphorynchus spirurus (Vieillot, 1819) detects a new fission in Passeriformes. PLoS ONE 2018, 13, e0202040. [Google Scholar] [CrossRef]
- Tura, V.; Kretschmer, R.; Sassi, F.d.M.C.; de Moraes, R.L.R.; Barcellos, S.A.; de Rosso, V.O.; de Souza, M.S.; Cioffi, M.d.B.; Gunski, R.J.; Garnero, A.d.V. Chromosomal Evolution of Suboscines: Karyotype Diversity and Evolutionary Trends in Ovenbirds (Passeriformes, Furnariidae). Cytogenet. Genome Res. 2023, 162, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Degrandi, T.M.; Gunski, R.J.; Garnero, A.d.V.; de Oliveira, E.H.C.; Kretschmer, R.; de Souza, M.S.; Barcellos, S.A.; Hass, I. The distribution of 45S rDNA sites in bird chromosomes suggests multiple evolutionary histories. Genet. Mol. Biol. 2020, 43, e0331. [Google Scholar] [CrossRef]
- Hadjiolov, A.A. Ribosomal Genes. In The Nucleolus and Ribosome Biogenesis; Springer: Vienna, Austria, 1985; Volume 12, pp. 5–53. [Google Scholar] [CrossRef]
- Shaw, P.; Brown, J. Nucleoli: Composition, Function, and Dynamics. Plant Physiol. 2012, 158, 44–51. [Google Scholar] [CrossRef]
- Daniels, L.M.; Delany, M.E. Molecular and cytogenetic organization of the 5S ribosomal DNA array in chicken (Gallus gallus). Chromosom. Res. 2003, 11, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Dyomin, A.G.; Koshel, E.I.; Kiselev, A.M.; Saifitdinova, A.F.; Galkina, S.A.; Fukagawa, T.; Kostareva, A.A.; Gaginskaya, E.R.; Buratti, E. Chicken rRNA Gene Cluster Structure. PLoS ONE 2016, 11, e0157464. [Google Scholar] [CrossRef]
- Nishida-Umehara, C.; Tsuda, Y.; Ishijima, J.; Ando, J.; Fujiwara, A.; Matsuda, Y.; Griffin, D.K. The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosom. Res. 2007, 15, 721–734. [Google Scholar] [CrossRef]
- Garnero, A.D.V.; Gunski, R.J. A case of chromosomal polymorphism. Int. J. Cytol. 2000, 43, 64–70. [Google Scholar]
- Guerra, M.; Souza, M.D. Como observar cromossomos: Um guia de técnicas em citogenética vegetal, animal e humana. Ribeirão Preto FUNPEC 2002, 201, 20–39. [Google Scholar]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Howell, W.M.; e Black, D.A. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: A 1-step method. Experientia 1972, 36, 1014–1015. [Google Scholar] [CrossRef] [PubMed]
- Oliveira de Rosso, V.; Tura, V.; Severo Salau, H.; de Oliveira Machado, L.; Pimentel Torres, F.; Gunski, R.J.; Del Valle Garnero, A. Atypical Presence of Interstitial Telomeric Sequences in Thamnophilus Species (Passeriformes: Thamnophilidae). Cytogenet. Genome Res. 2025, 4, 1–11. [Google Scholar] [CrossRef]
- Kretschmer, R.; Ferguson-Smith, M.A.; De Oliveira, E.H.C. FISH in Birds. In Cytogenetics and Molecular Cytogenetics; CRC Press: Boca Raton, FL, USA, 2022; pp. 263–279. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Chernomor, O.; von Haeseler, A.; Minh, B.Q. Terrace aware data structure for phylogenomic inference from supermatrices. Syst. Biol. 2016, 65, 997–1008. [Google Scholar] [CrossRef]
- Kück, P.; Longo, G.C. FASconCAT-G: Extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front. Zool. 2014, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Degrandi, T.M.; Barcellos, S.A.; Costa, A.L.; Garnero, A.D.; Hass, I.; Gunski, R.J. Introducing the Bird Chromosome Database: An Overview of Cytogenetic Studies in Birds. Cytogenet. Genome Res. 2021, 160, 199–205. [Google Scholar] [CrossRef]
- Tagliarini, M. Aplicação de Pintura Cromossômica em Espécies da Família Accipitridae (Aves, Falconiformes): Considerações Filogenéticas e Evolutivas; Universidade Federal do Pará: Belém Pará, Brazil, 2013; Available online: http://repositorio.ufpa.br/handle/2011/5839 (accessed on 2 April 2025).
- Nishida, C.; Ishishita, S.; Yamada, K.; Griffin, D.K.; Matsuda, Y. Dynamic Chromosome Reorganization in the Osprey (Pandion haliaetus, Pandionidae, Falconiformes): Relationship Between Chromosome Size and the Chromosomal Distribution of Centromeric Repetitive DNA Sequences. Cytogenet. Genome 2014, 142, 179–189. Available online: https://karger.com/cgr/article/142/3/179/61551 (accessed on 12 January 2025). [CrossRef]
- Nie, W.; O’bRien, P.C.M.; Ng, B.L.; Fu, B.; Volobouev, V.; Carter, N.P.; Ferguson-Smith, M.A.; Yang, F. Avian comparative genomics: Reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes) An atypical species with low diploid number. Chromosome Res. 2009, 17, 99–113. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, T.D.; Kretschmer, R.; Bertocchi, N.A.; Degrandi, T.M.; de Oliveira, E.H.C.; Cioffi, M.D.B.; Garnero, A.D.V.; Gunski, R.J. Genomic organization of repetitive DNA in woodpeckers (Aves, Piciformes): Implications for karyotype and ZW sex chromosome differentiation. PLoS ONE 2017, 12, e0169987. [Google Scholar] [CrossRef] [PubMed]
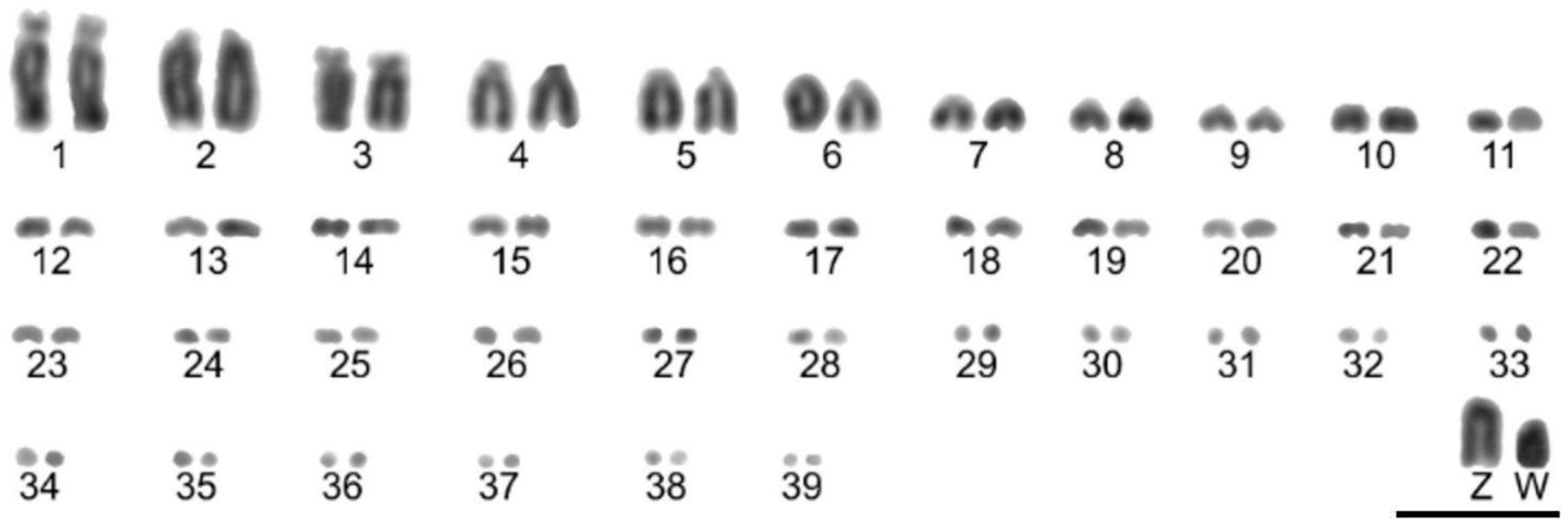
| L. falcinellus | L. angustirostris | S. griseicapillus | G. spirurus | S. frontalis | S. rufosuperciliata | C. obsoleta | |
|---|---|---|---|---|---|---|---|
| 2n = 80 | 2n = 82 | 2n = 82 | 2n = 80 | 2n = 82 | 2n = 82 | 2n = 82 | |
| 1º | SM | A | T | A | T | A | A |
| 2º | A | T | T | A | A | SM | SM |
| 3º | SM | A | A | A | T | SM | SM |
| 4º | A | A | T | A | SM | SM | SM |
| 5º | A | T | T | A | A | SM | SM |
| 6º | T | T | T | A | A | A | A |
| 7º | T | T | T | A | A | A | A |
| 8º | T | T | T | A | M | - | SM |
| 9º | T | T | T | A | T | - | SM |
| 10º | T | T | T | A | T | - | SM |
| Z | T | A | A | A | M | SM | A |
| W | T | T | T | A | M | - | - |
| NORs/18S | 1st pair p arm | 1st pair p arm | 1st pair q arm | 1st pair p arm | 1 pair of micro | 1 pair of micro | 1 pair of micro |
| References | Present study | [17] | [19] | [18,20] | [20] | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garnero, A.d.V.; de Rosso, V.O.; Salau, H.S.; Rosa de Lara, P.A.; Tura, V.; Pimentel Torres, F.; Gunski, R.J. The Curious Case of Woodcreepers: Cytogenomic Evidence Based on the Position of NORs. Taxonomy 2025, 5, 41. https://doi.org/10.3390/taxonomy5030041
Garnero AdV, de Rosso VO, Salau HS, Rosa de Lara PA, Tura V, Pimentel Torres F, Gunski RJ. The Curious Case of Woodcreepers: Cytogenomic Evidence Based on the Position of NORs. Taxonomy. 2025; 5(3):41. https://doi.org/10.3390/taxonomy5030041
Chicago/Turabian StyleGarnero, Analía del Valle, Vitor Oliveira de Rosso, Hybraim Severo Salau, Paulo Afonso Rosa de Lara, Victoria Tura, Fabiano Pimentel Torres, and Ricardo José Gunski. 2025. "The Curious Case of Woodcreepers: Cytogenomic Evidence Based on the Position of NORs" Taxonomy 5, no. 3: 41. https://doi.org/10.3390/taxonomy5030041
APA StyleGarnero, A. d. V., de Rosso, V. O., Salau, H. S., Rosa de Lara, P. A., Tura, V., Pimentel Torres, F., & Gunski, R. J. (2025). The Curious Case of Woodcreepers: Cytogenomic Evidence Based on the Position of NORs. Taxonomy, 5(3), 41. https://doi.org/10.3390/taxonomy5030041







