The Crucial Role of Plant Taxonomy in Ensuring the Biodiversity Sustainability: Insights from the Pharmaceutically Significant Genus Paris (Melanthiaceae)
Abstract
:1. Introduction
2. Pharmaceutical Significance and Scientific Importance of the Genus Paris
3. The Scarcity of Taxonomic Expertise Exacerbates the Risk of Extinct for Extant Paris Species
4. Taxonomic Knowledge Gap Poses Obstacles to the Standardized Cultivation of Medicinal Paris
5. Conclusions and Suggestions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Costanza, R.; d’Arge, R.; de Groot, R.; Farber, S.; Grasso, M.; Hannon, B.; Limburg, K.; Naeem, S.; O’Neill, R.V.; Paruelo, J.; et al. The value of the world’s ecosystem services and natural capital. Nature 1997, 387, 253–260. [Google Scholar] [CrossRef]
- Costanza, R.; de Groot, R.; Sutton, P.; van der Ploeg, S.; Anderson, S.J.; Kubiszewski, I.; Farber, S.; Turner, R.K. Changes in the global value of ecosystem services. Glob. Environ. Change 2014, 26, 152–158. [Google Scholar] [CrossRef]
- Daily, G.C. Nature’s Services: Societal Dependence on Natural Ecosystems; Island Press: Washington, DC, USA, 1997. [Google Scholar]
- De Groot, R.S.; Fisher, B.; Christie, M.; Aronson, J.; Braat, L.; Gowdy, J.; Haines-Young, R.; Maltby, E.; Neuville, A.; Polasky, S.; et al. Integrating the ecological and economic dimensions in biodiversity and ecosystem service valuation. In The Economics of Ecosystems and Biodiversity: Ecological and Economic Foundations; Kumar, P., Ed.; Earthscan: London, UK, 2010. [Google Scholar]
- De Groot, R.; Costanza, R.; Broeck, D.V.D.; Aronson, J. A global partnership for ecosystem services. Solutions 2011, 2, 42–43. [Google Scholar]
- Henle, K.; Alard, D.; Clitherow, J.; Cobb, P.; Firbank, L.; Kull, T.; McCracken, D.; Moritz, R.F.; Niemelä, J.; Rebane, M.; et al. Identifying and managing the conflicts between agriculture and biodiversity conservation in Europe—A review. Agric. Ecosyst. Environ. 2008, 124, 60–71. [Google Scholar] [CrossRef]
- Gavin, M.C.; McCarter, J.; Mead, A.; Berkes, F.; Stepp, J.R.; Peterson, D.; Tang, R. Defining biocultural approaches to conservation. Trends Ecol. Evol. 2015, 30, 140–145. [Google Scholar] [CrossRef]
- Kim, H.; Rosa, I.M.D.; Alkemade, R.; Leadley, P.; Hurtt, G.; Popp, A.; van Vuuren, D.P.; Anthoni, P.; Arneth, A.; Baisero, D.; et al. A protocol for an intercomparison of biodiversity and ecosystem services models using harmonized land-use and climate scenarios. Geosci. Model Dev. 2018, 11, 4537–4562. [Google Scholar] [CrossRef]
- De Vos, J.M.; Joppa, L.N.; Gittleman, J.L.; Stephens, P.R.; Pimm, S.L. Estimating the normal background rate of species extinction. Conserv. Biol. 2015, 29, 452–462. [Google Scholar] [CrossRef]
- Petraitis, P.S.; Latham, R.E.; Niesenbaum, R.A. The Maintenance of Species Diversity by Disturbance. Q. Rev. Biol. 1989, 64, 393–418. [Google Scholar] [CrossRef]
- Sarkar, S. Wilderness preservation and biodiversity conservation—Keeping divergent goals distinct. BioScience 1999, 49, 405–412. [Google Scholar] [CrossRef]
- Lindenmayer, D.B.; Margules, C.R.; Botkin, D.B. Indicators of Biodiversity for Ecologically Sustainable Forest Management. Conserv. Biol. 2000, 14, 941–950. [Google Scholar] [CrossRef]
- Perrings, C.; Duraiappah, A.; Larigauderie, A.; Mooney, H. The Biodiversity and Ecosystem Services Science-Policy Interface. Science 2011, 331, 1139–1140. [Google Scholar] [CrossRef]
- Niesenbaum, R.A. The Integration of Conservation, Biodiversity, and Sustainability. Sustainability 2019, 11, 4676. [Google Scholar] [CrossRef]
- Ebach, M.C.; Holdrege, C. DNA barcoding is no substitute for taxonomy. Nature 2005, 434, 697. [Google Scholar] [CrossRef]
- Khuroo, A.A.; Dar, G.H.; Khan, Z.S.; Malik, A.H. Exploring an inherent interface between taxonomy and biodiversity: Current problems and future challenges. J. Nat. Cons. 2007, 15, 256–261. [Google Scholar] [CrossRef]
- Pendry, C.A.; Dick, J.; Pullan, M.R.; Knees, S.G.; Miller, A.G.; Neale, S.; Watson, M.F. In search of a functional flora—Towards a greater integration of ecology and taxonomy. Plant Ecol. 2007, 192, 161–167. [Google Scholar] [CrossRef]
- Mace, G.M. The role of taxonomy in species conservation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 711–719. [Google Scholar] [CrossRef]
- Wheeler, Q.D.; Raven, P.H.; Wilson, E.O. Taxonomy: Impediment or expedient? Science 2004, 303, 285. [Google Scholar] [CrossRef]
- Soltis, P.S.; Gitzendanner, M.A. Molecular systematics and the conservation of rare species. Conserv. Biol. 1999, 13, 471–483. [Google Scholar] [CrossRef]
- Liu, J.Q. The integrative species concept and species on the speciation way. Biodivers. Sci. 2016, 24, 1004–1008. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, C.; Landis, J.B.; Deng, M.; Chen, J. Plastome phylogenomics of Cephalotaxus (Cephalotaxaceae) and allied genera. Ann. Bot. 2021, 127, 697–708. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, J.; Landis, J.B.; Wang, S.; Yang, Z.; Zhang, Y. Deciphering the taxonomic delimitation of Ottelia acuminata (Hydrocharitaceae) using complete plastomes as super-barcodes. Front. Plant Sci. 2021, 12, 681270. [Google Scholar] [CrossRef]
- Godfray, H.C. Challenges for taxonomy. The discipline will have to reinvent itself if it is to survive and flourish. Nature 2002, 417, 17–19. [Google Scholar] [CrossRef]
- Isaac, N.J.B.; Mallet, J.; Mace, G.M. Taxonomic inflation: Its influence on macroecology and conservation. Trends Ecol. Evol. 2004, 19, 464–469. [Google Scholar] [CrossRef]
- Domínguez, L.F.; Moreno, S.J.C.; Sainz, O.H.; Schwartz, M.W. Effects of dynamic taxonomy on rare species and conservation listing: Insights from the Iberian vascular flora. Biodivers. Conserv. 2007, 16, 4039–4050. [Google Scholar] [CrossRef]
- Dar, G.H.; Khuroo, A.A.; Reddy, C.S.; Malik, A.H. Impediment to Taxonomy and Its Impact on Biodiversity Science: An Indian Perspective. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2012, 82, 235–240. [Google Scholar] [CrossRef]
- Liu, D. Train specialists to fight for biodiversity. Nature 2024, 633, 741. [Google Scholar] [CrossRef] [PubMed]
- Schatz, G.E. Taxonomy and Herbaria in service of plant conservation: Lessons from Madagascar’s endemic families. Ann. Missouri. Bot. Gard. 2002, 89, 145–152. [Google Scholar] [CrossRef]
- Smith, T. The work of taxonomy. Conserv. Biol. 2004, 18, 6–7. [Google Scholar]
- Wilson, E.O. On the future of conservation biology. Conserv. Biol. 2000, 14, 1–3. [Google Scholar] [CrossRef]
- Hara, H. Variations in Paris polyphylla Smith with reference to other Asiatic species. J. Fac. Sci. Univ. Tokyo Sect. 3 1969, 10, 141–180. [Google Scholar]
- Li, H. The Genus Paris (Trilliaceae); Science Press: Beijing, China, 1998. [Google Scholar]
- Liang, S.-Y.; Soukup, V.G. Paris L. In Flora of China; Wu, Z.Y., Raven, P.H., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St Louis, MO, USA, 2000; Volume 24, pp. 88–95. [Google Scholar]
- Ji, Y. A Monograph of Paris (Melanthaceae); Science Press: Beijing, China; Springer: Singapore, 2021. [Google Scholar]
- Ji, Y.; Fritsch, P.W.; Li, H.; Xiao, T.; Zhou, Z. Phylogeny and classification of Paris (Melanthiaceae) inferred from DNA sequence data. Ann. Bot. 2006, 98, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Yang, L.; Chase, M.W.; Liu, C.; Yang, Z.; Yang, J.; Yang, J.; Yi, T. Plastome phylogenomics, biogeography, and clade diversification of Paris (Melanthiaceae). BMC Plant Biol. 2019, 19, 543. [Google Scholar] [CrossRef]
- Ji, Y.; Liu, C.; Yang, J.; Jin, L.; Yang, Z.; Yang, J.-B. Ultra-barcoding discovers a cryptic species in Paris yunnanensis (Melanthiaceae), a medicinally important plant. Front. Plant Sci. 2020, 11, 411. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, J.; Landis, J.B.; Wang, S.; Jin, L.; Xie, P.; Liu, H.; Yang, J.-B.; Yi, T.-S. Genome skimming contributes to clarifying species limits in Paris section Axiparis (Melanthiaceae). Front. Plant Sci. 2022, 13, 832034. [Google Scholar] [CrossRef]
- Huang, Y.; Li, X.; Yang, Z.; Yang, C.; Yang, J.; Ji, Y. Analysis of complete chloroplast genome sequences improves phylogenetic resolution of Paris (Melanthiaceae), Front. Plant Sci. 2016, 7, 1797. [Google Scholar]
- Yang, L.; Yang, Z.; Liu, C.; He, Z.; Zhang, Z.; Yang, J.; Liu, H.; Yang, J.; Ji, Y. Chloroplast phylogenomic analysis provides insights into the evolution of the largest eukaryotic genome holder, Paris japonica (Melanthiaceae). BMC Plant Biol. 2019, 19, 293. [Google Scholar] [CrossRef]
- Cunningham, A.B.; Brinckmann, J.A.; Bi, Y.-F.; Pei, S.-J.; Schippmann, U.; Luo, P. Paris in the spring: A review of the trade, conservation and opportunities in the shift from wild harvest to cultivation of Paris polyphylla (Trilliaceae). J. Ethnopharm. 2018, 222, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, W.; Li, X.; Wei, J.; Jing, S.; Xiao, P. Chemotaxonomic study of the genus Paris based on steroidal saponins. Biochem. Syst. Ecol. 2013, 48, 163–175. [Google Scholar] [CrossRef]
- Yang, Z.; Yang, L.; Liu, C.; Qin, X.; Liu, H.; Chen, J.; Ji, Y. Transcriptome analyses of Paris polyphylla var. chinensis, Ypsilandra thibetica, and Polygonatum kingianum characterize their steroidal saponin biosynthesis pathway. Fitoterapia 2019, 135, 52–63. [Google Scholar] [CrossRef]
- Cong, Y.; Liu, X.; Kang, L.; Yu, Z.; Zhao, Z.; Li, J.; Ma, B.; Cong, Y. Pennogenin tetraglycoside stimulates secretion-dependent activation of rat platelets: Evidence for critical roles of adenosine diphosphate receptor signal pathways. Thromb. Res. 2012, 129, e209–e216. [Google Scholar] [CrossRef]
- Qin, X.J.; Yu, M.Y.; Ni, W.; Yan, H.; Chen, C.-X.; Cheng, Y.-C.; He, L.; Liu, H.-Y. Steroidal saponins from stems and leaves of Paris polyphylla var. yunnanensis. Phytochemistry 2016, 121, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.J.; Ni, W.; Chen, C.X.; Liu, H.-Y. Seeing the light: Shifting from wild rhizomes to extraction of active ingredients from above-ground parts of Paris polyphylla var. yunnanensis. J. Ethnopharm. 2018, 224, 134–139. [Google Scholar] [CrossRef]
- Deng, D.; Lauren, D.R.; Cooney, J.M.; Jensen, D.J.; Wurms, K.V.; Upritchard, J.E.; Cannon, R.D.; Wang, M.Z.; Li, M.Z. Antifungal saponins from Paris polyphylla Smith. Plant. Med. 2008, 74, 1397–1402. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.J.; Sun, D.J.; Ni, W.; Chen, C.-X.; Hua, Y.; He, L.; Liu, H.-Y. Steroidal saponins with antimicrobial activity from stems and leaves of Paris polyphylla var. yunnanensis. Steroids 2012, 77, 1242–1248. [Google Scholar] [CrossRef]
- Chen, M.; Ye, K.; Zhang, B.; Xin, Q.; Li, P.; Kong, A.-N.; Wen, X.; Yang, J. Paris saponin II inhibits colorectal carcinogenesis by regulating mitochondrial fission and NF-kappaB pathway. Pharmacol. Res. 2019, 139, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Liu, B.R.; Hu, W.J.; Yu, L.X.; Qian, X.P. In vitro anticancer activity of aqueous extracts and ethanol extracts of fifteen traditional Chinese medicines on human digestive tumor cell lines. Phytother. Res. 2007, 21, 1102–1104. [Google Scholar] [CrossRef]
- Xie, Z.Z.; Li, M.M.; Deng, P.F.; Wang, S.; Wang, L.; Lu, X.-P.; Hu, L.-B.; Chen, Z.; Jie, H.-Y.; Wang, Y.-F.; et al. Paris saponin-induced autophagy promotes breast cancer cell apoptosis via the Akt/mTOR signaling pathway. Chem. Biol. Interact. 2017, 264, 1–9. [Google Scholar] [CrossRef]
- Zhang, D.; Li, K.; Sun, C.; Cao, G.; Qi, Y.; Lin, Z.; Guo, Y.; Liu, Z.; Chen, Y.; Liu, J.; et al. Anti-cancer effects of Paris polyphylla ethanol extract by inducing cancer cell apoptosis and cycle arrest in prostate cancer cells. Curr. Urol. 2018, 11, 144–150. [Google Scholar] [CrossRef]
- Yan, H.; Ni, W.; Yu, L.L.; Xiao, L.-G.; Ji, Y.-H.; Liu, H.-Y. Parisvaniosides A–E, five new steroidal saponins from Paris vaniotii. Steroids 2022, 177, 108949. [Google Scholar] [CrossRef]
- Yu, L.L.; Li, Y.X.; Gao, W.T.; Ling, S.-S.; Ni, W.; Ji, Y.-H.; Liu, H.-Y. Steroidal saponins with cytotoxic activity from the stems and leaves of Paris fargesii. New J. Chem. 2022, 46, 19136–19146. [Google Scholar] [CrossRef]
- Yu, L.L.; Ling, S.S.; Gao, W.T.; Li, Y.-X.; Xiao, L.-G.; Ni, W.; Ji, Y.-H.; Liu, H.-Y. Parisfargosides A–E, five new cholestane glycosides from the rhizomes of Paris fargesii. Fitoterapia 2022, 158, 105174. [Google Scholar] [CrossRef]
- Miao, K.; Wang, T.; Tang, L.; Hou, L.; Ji, Y. Establishing the first reference library for utilizing high-throughput sequencing technologies in identifying medicinal and endangered Paris species (Melanthiaceae). Ind. Crops Prod. 2024, 218, 118871. [Google Scholar] [CrossRef]
- Pellicer, J.; Fay, M.F.; Leitch, I.J. The largest eukaryotic genome of them all? Bot. J. Linn. Soc. 2010, 164, 10–15. [Google Scholar] [CrossRef]
- Dodsworth, S.; Leitch, A.R.; Leitch, I.J. Genome size diversity in angiosperms and its influence on gene space. Curr. Opin. Genet. Devel. 2015, 35, 73–78. [Google Scholar] [CrossRef]
- Leitch, I.J.; Beaulieu, J.M.; Chase, M.W.; Leitch, A.R.; Fay, M.F. Genome size dynamics and evolution in monocots. J. Bot. 2010, 2010, 862516. [Google Scholar] [CrossRef]
- Pellicer, J.; Kelly, L.J.; Leitch, I.J.; Zomlefer, W.B.; Fay, M.F. A universe of dwarfs and giants: Genome size and chromosome evolution in the monocot family Melanthiaceae. New Phytol. 2014, 201, 1484–1497. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2020; Volume I. [Google Scholar]
- He, J.; Zhang, S.; Wang, H.; Chen, C.; Chen, S. Advances in studies on and uses of Paris polyphylla var. yunnanensis (Trilliaceae). Acta Bot. Yunnanica 2006, 28, 271–276. [Google Scholar]
- Takhtajan, A. A revision of Daiswa (Trilliaceae). Brittonia 1983, 35, 255–270. [Google Scholar] [CrossRef]
- Yang, Y.; Zhai, Y.H.; Liu, T.; Zhang, F.; Ji, Y. Detection of Valeriana jatamansi as an adulterant of medicinal Paris by length variation of chloroplast psbA-trnH region. Plant. Med. 2011, 77, 87–91. [Google Scholar] [CrossRef]
- Liu, T.; Ji, Y. Molecular authentication of the medicinal plant Paris polyphylla Smith var. yunnanensis (Melanthiaceae) and its related species by polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP). J. Med. Plants Res. 2012, 6, 1181–1186. [Google Scholar]
- Li, H.; Su, B.; Zhang, Z.; Yang, M. An assessment on the rarely medical Paris plants in China with exploring the future development of its plantation. J. West China For. Sci. 2015, 44, 3. [Google Scholar]
- Turner, I.M. Species loss in fragments of tropical rain forest: A review of the evidence. J. Appl. Ecol. 1996, 33, 200–209. [Google Scholar] [CrossRef]
- Stork, N.E.; Coddington, J.A.; Colwell, R.K.; Chazdon, R.L.; Dick, C.W.; Peres, C.A.; Sloan, S.; Willis, K. Vulnerability and resilience of tropical forest species to land-use change. Conserv. Biol 2009, 23, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- Kunwar, R.M.; Adhikari, Y.P.; Sharma, H.P.; Rimal, B.; Devkota, H.P.; Charmakar, S.; Acharya, R.P.; Baral, K.; Ansari, A.S.; Bhattarai, R.; et al. Distribution, use, trade and conservation of Paris polyphylla Sm. in Nepal. Glob. Ecol. Conserv. 2020, 23, e01081. [Google Scholar] [CrossRef]
- IUCN (International Union for Conservation of Nature). The IUCN Red List Categories and Criteria Version 3.1., 2nd ed.; IUCN: Gland, Switzerland, 2012. [Google Scholar]
- Qin, H. Seed Plant of China: Cheklist, Uses and Conservation Status; Hebei Science & Technology Press: Shijiazhuang, China, 2021; Volume 3, pp. 1364–1366. [Google Scholar]
- IUCN (International Union for Conservation of Nature). The IUCN Red List of Threatened Species Version 2019-1; IUCN: Gland, Switzerland, 2019; Available online: www.iucnredlist.org (accessed on 1 October 2016).
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Kress, W.J.; García-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2015, 30, 25–35. [Google Scholar] [CrossRef]
- Xu, S.-Z.; Li, Z.-Y.; Jin, X.-H. DNA barcoding of invasive plants in China: A resource for identifying invasive plants. Mol. Ecol. Resour. 2018, 18, 128–136. [Google Scholar] [CrossRef]
- Mosal, K.A.; Gairola, S.J.; Jamdade, R.; El-Keblawy, A.; Al Shaer, K.I.; Al Harthi, E.K.; Shabana, H.A.; Mahmoud, T. The promise of molecular and genomic techniques for biodiversity research and DNA barcoding of the Arabian Peninsula Flora. Front. Plant Sci. 2019, 9, 1929. [Google Scholar]
- Weigand, H.; Beermann, A.J.; Čiampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geigerg, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 2019, 78, 499–524. [Google Scholar] [CrossRef]
- Nijman, V. An overview of international wildlife trade from Southeast Asia. Biodiv. Conserv. 2010, 19, 1101–1114. [Google Scholar] [CrossRef]
- Damania, R.; Bulte, E.H. The economics of wildlife farming and endangered species conservation. Ecol. Econ. 2007, 62, 461–472. [Google Scholar] [CrossRef]
- Tensen, L. Under what circumstances can wildlife farming benefit species conservation? Glob Ecol. Conserv. 2016, 6, 286–298. [Google Scholar] [CrossRef]
- Hinsley, A.; De-Boer, H.J.; Fay, M.F.; Gale, S.W.; Gardiner, L.M.; Gunasekara, R.S.; Kumar, P.; Masters, S.; Metusala, D.; Roberts, D.L.; et al. A review of the trade in orchids and its implications for conservation. Bot. J. Linn. Soc. 2017, 186, 435–455. [Google Scholar] [CrossRef]
- Liu, H.; Gale, S.W.; Cheuk, K.L.; Fischer, G.A. Conservation impacts of commercial cultivation of endangered and overharvested plants. Conserv. Biol. 2019, 33, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Shi, N.; Xie, P.; Zhang, G.; Liu, H.; Ji, Y. Plastome sequencing for accurate and effective authentication of Polygonatum kingianum (Asparagaceae). Ind. Crops Prod. 2022, 184, 1150056. [Google Scholar] [CrossRef]
- Wang, S.; Zhou, N.; Shi, N.; Zhang, G.; Liu, H.; Guo, X.; Ji, Y. Testing and using complete plastomes for authentication of medicinal Polygonatum species (Asparagaceae). Ind. Crops Prod. 2023, 197, 116557. [Google Scholar] [CrossRef]
- Wolf, D.E.; Takebayashi, N.; Rieseberg, L.H. Predicting the risk of extinction through hybridization. Conserv. Biol. 2000, 15, 1039–1053. [Google Scholar] [CrossRef]
- Hedrick, P.W. Recent developments in conservation genetics. For. Ecol. Manag. 2004, 197, 3–19. [Google Scholar] [CrossRef]
- Canter, P.H.; Thomas, H.; Ernst, E. Bringing medicinal plants into cultivation: Opportunities and challenges for biotechnology. Trends Biotechnol. 2005, 23, 180–185. [Google Scholar] [CrossRef]
- Zhang, B.G.; Peng, Y.; Zhang, Z.; Liu, H.; Qi, Y.; Liu, S.; Xiao, P. GAP production of TCM herbs in China. Plant. Med. 2010, 76, 1948–1955. [Google Scholar] [CrossRef]
- Franchet, A. Monographie du Genere Paris; Memoire de la Societe Philomathique Centaire: Paris, France, 1888. [Google Scholar]
- Handel-Mazzetti, H.R.E. Symbolae Sinicae: Botanische Ergebnisse der Expedition der Akademie der Wissenschaften in Wein nach Südwest-China; Springer: Wien, Austria, 1936. [Google Scholar]
- Zhou, N.; Tang, L.; Xie, P. Genome skimming as an efficient tool for authenticating commercial products of the pharmaceutically important Paris yunnanensis (Melanthiaceae). BMC Plant Biol. 2023, 23, 344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Miao, K.; Hou, L.; Liu, H.; Chen, J.; Ji, Y. Phylotranscriptomic analyses reveal the evolutionary complexity of Paris L. (Melanthiaceae), a morphologically distinctive genus with significant pharmaceutical importance. Ann. Bot. 2024, 134, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, G.W.; Freckleton, R.P. Declines in the numbers of amateur and professional taxonomists: Implications for conservation. Anim. Conserv. 2002, 5, 245–249. [Google Scholar] [CrossRef]
- Sites, J.W.; Marshall, J.C. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol. Evol. 2003, 18, 462–470. [Google Scholar] [CrossRef]
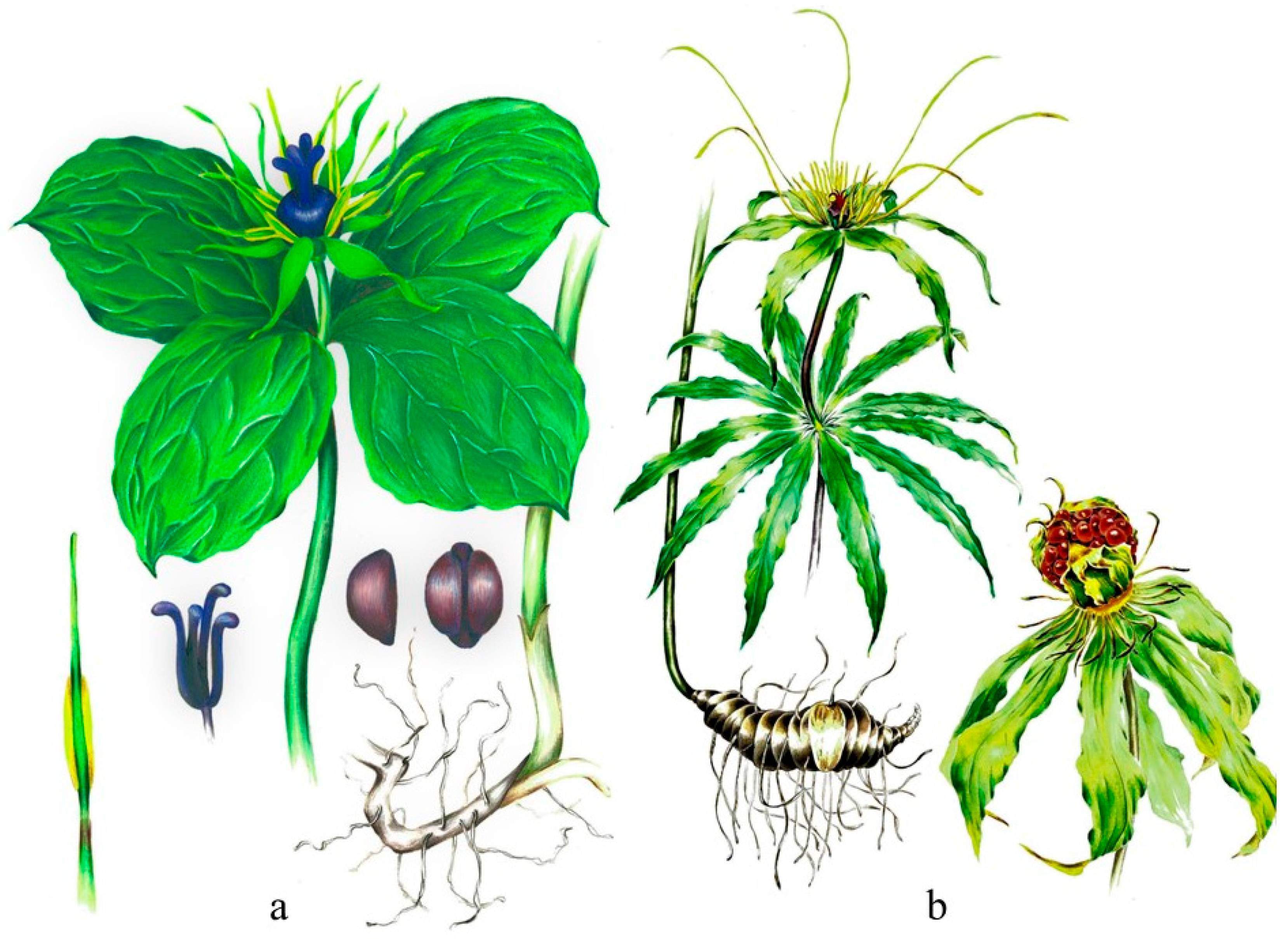
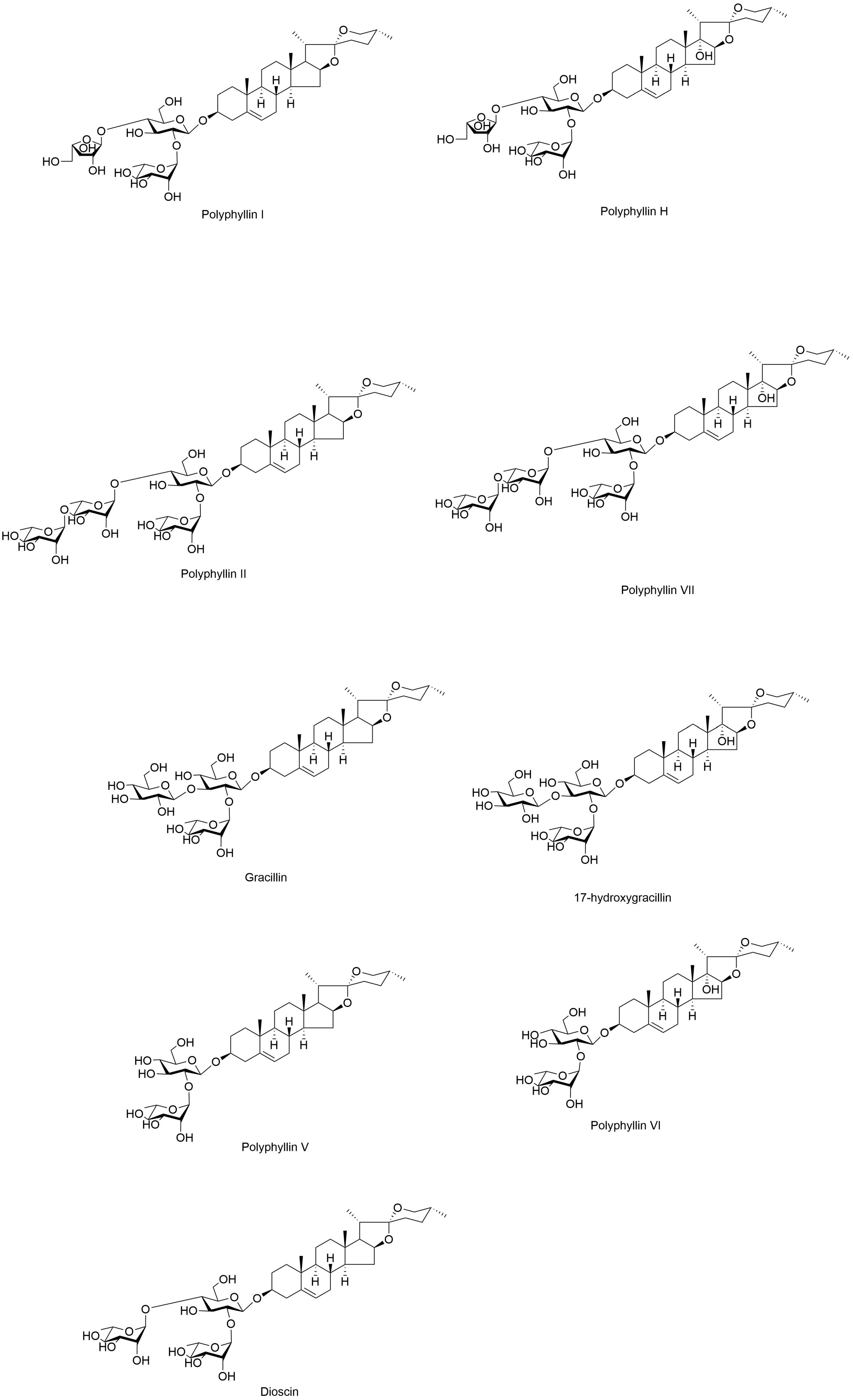
| Species | Longitude Range (°E) | Latitude Range (°N) | Elevation Range (m) |
|---|---|---|---|
| P. bashanensis | 101.762–110.812 | 29.138–32.880 | 1400–2750 |
| P. caobangensis | 98.998–114.603 | 18.795–29.553 | 300–2900 |
| P. chinensis | 98.835–122.402 | 18.579–39.079 | 150–2800 |
| P. cronquistii | 101.632–108.812 | 22.679–29.138 | 200–1950 |
| P. delavayi | 102.111–115.866 | 24.161–32.332 | 700–2900 |
| P. dunniana | 106.699–109.732 | 18.591–25.416 | 400–1100 |
| P. fargesii | 103.018–121.639 | 22.049–31.884 | 500–2100 |
| P. forrestii | 82.183–101.718 | 20.783–30.120 | 600–3200 |
| P. incompleta | 38.080–45.720 | 40.830–45.050 | 600–2000 |
| P. japonica | 136.771–139.966 | 35.616–38.785 | 1000–2100 |
| P. lancifolia | 101.744–121.639 | 22.484–36.921 | 1100–2300 |
| P. liiana | 98.835–108.316 | 21.734–25.137 | 1200–2200 |
| P. luquanensis | 102.585–102.733 | 25.940–26.569 | 2300–2800 |
| P. mairei | 87.904–106.291 | 24.509–33.313 | 1800–3500 |
| P. marmorata | 86.730–101.992 | 25.744–28.958 | 1500–3100 |
| P. polyphylla | 81.760–110.041 | 23.360–36.061 | 1100–2800 |
| P. qiliangiana | 103.628–110.812 | 31.034–34.145 | 720–1140 |
| P. quadrifolia | −0.266–108.000 | 40.066–70.366 | 50–1600 |
| P. tetraphylla | 129.960–143.310 | 32.000–44.820 | 200–1400 |
| P. thibetica | 85.266–104.395 | 25.275–35.717 | 1600–3600 |
| P. vaniotii | 102.111–112.691 | 24.998–31.590 | 700–3000 |
| P. verticillata | 85.364–145.855 | 29.138–60.716 | 600–3600 |
| P. vietnamensis | 98.420–111.547 | 20.667–24.900 | 600–2000 |
| P. xichouensis | 104.679–105.115 | 22.750–23.391 | 1200–1500 |
| P. yanchii | 99.305 | 25.891 | 2300–2800 |
| P. yunnanensis | 94.006–109.997 | 20.783–30.075 | 1000–3200 |
| Species | 1C Value (pg) | Species | 1C Value (pg) |
|---|---|---|---|
| P. bashanensis | 29.38 [35] | P. luquanensis | 64.60 [35] |
| P. caobangensis | 64.13–65.30 [35] | P. mairei | 55.96 [35] |
| P. chinensis | 55.34 [35] | P. marmorata | 70.36 [35] |
| P. cronquistii | 63.44 [35] | P. polyphylla | 54.22 [61] |
| P. delavayi | 61.79 [35] | P. quadrifolia | 50.52 [61] |
| P. dunniana | 59.98 [35] | P. tetraphylla | 40.75 [61] |
| P. fargesii | 60.83 [35] | P. thibetica | 51.30 [35] |
| P. forrestii | 55.37–87.27 [35] | P. vaniotii | 55.38–60.80 [35] |
| P. incompleta | 42.25 [61] | P. verticillata | 31.21 [61] |
| P. japonica | 152.23 [58] | P. vietnamensis | 62.90 [35] |
| P. lancifolia | 50.13–54.80 [35] | P. yanchii | 53.75 [35] |
| P. liiana | 52.98–59.14 [35] | P. yunnanensis | 50.80–57.72 [35] |
| Species | Conservation Status | Species | Conservation Status |
|---|---|---|---|
| P. bashanensis | Vulnerable | P. mairei | Vulnerable |
| P. caobangensis | Vulnerable | P. marmorata | Vulnerable |
| P. chinensis | Vulnerable | P. polyphylla | Vulnerable |
| P. cronquistii | Vulnerable | P. qiliangiana | Vulnerable |
| P. delavayi | Vulnerable | P. quadrifolia | Least Concern |
| P. dunniana | Endangered | P. tetraphylla | Least Concern |
| P. fargesii | Vulnerable | P. thibetica | Vulnerable |
| P. forrestii | Vulnerable | P. vaniotii | Vulnerable |
| P. incompleta | Least Concern | P. verticillata | Least Concern |
| P. japonica | Least Concern | P. vietnamensis | Vulnerable |
| P. lancifolia | Vulnerable | P. xichouensis | Critically Endangered |
| P. liiana | Vulnerable | P. yanchii | Critically Endangered |
| P. luquanensis | Critically Endangered | P. yunnanensis | Vulnerable |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, Y.; Yang, Z.; Zhang, X.; Pei, S. The Crucial Role of Plant Taxonomy in Ensuring the Biodiversity Sustainability: Insights from the Pharmaceutically Significant Genus Paris (Melanthiaceae). Taxonomy 2025, 5, 32. https://doi.org/10.3390/taxonomy5020032
Ji Y, Yang Z, Zhang X, Pei S. The Crucial Role of Plant Taxonomy in Ensuring the Biodiversity Sustainability: Insights from the Pharmaceutically Significant Genus Paris (Melanthiaceae). Taxonomy. 2025; 5(2):32. https://doi.org/10.3390/taxonomy5020032
Chicago/Turabian StyleJi, Yunheng, Zhiwei Yang, Xinqi Zhang, and Shengji Pei. 2025. "The Crucial Role of Plant Taxonomy in Ensuring the Biodiversity Sustainability: Insights from the Pharmaceutically Significant Genus Paris (Melanthiaceae)" Taxonomy 5, no. 2: 32. https://doi.org/10.3390/taxonomy5020032
APA StyleJi, Y., Yang, Z., Zhang, X., & Pei, S. (2025). The Crucial Role of Plant Taxonomy in Ensuring the Biodiversity Sustainability: Insights from the Pharmaceutically Significant Genus Paris (Melanthiaceae). Taxonomy, 5(2), 32. https://doi.org/10.3390/taxonomy5020032






