Abstract
The COVID-19 pandemic, instigated by the emergence of the novel coronavirus, SARS-CoV-2, created an incomparable global health crisis. Due to its highly virulent nature, identifying potential therapeutic agents against this lethal virus is crucial. PLpro is a key protein involved in viral polyprotein processing and immune system evasion, making it a prime target for the development of antiviral drugs to combat COVID-19. To expedite the search for potential therapeutic candidates, this review delved into computational studies. Recent investigations have harnessed computational methods to identify promising inhibitors targeting PLpro, aiming to suppress the viral activity. Molecular docking techniques were employed by researchers to explore the binding sites for antiviral drugs within the catalytic region of PLpro. The review elucidates the functional and structural properties of SARS-CoV-2 PLpro, underscoring its significance in viral pathogenicity and replication. Through comprehensive all-atom molecular dynamics (MD) simulations, the stability of drug–PLpro complexes was assessed, providing dynamic insights into their interactions. By evaluating binding energy estimates from MD simulations, stable drug–PLpro complexes with potential antiviral properties were identified. This review offers a comprehensive overview of the potential drug/lead candidates discovered thus far against PLpro using diverse in silico methodologies, encompassing drug repurposing, structure-based, and ligand-based virtual screenings. Additionally, the identified drugs are listed based on their chemical structures and meticulously examined according to various structural parameters, such as the estimated binding free energy (ΔG), types of intermolecular interactions, and structural stability of PLpro–ligand complexes, as determined from the outcomes of the MD simulations. Underscoring the pivotal role of targeting SARS-CoV-2 PLpro in the battle against COVID-19, this review establishes a robust foundation for identifying promising antiviral drug candidates by integrating molecular dynamics simulations, structural modeling, and computational insights. The continual imperative for the improvement of existing drugs and exploring novel compounds remains paramount in the global efforts to combat COVID-19. The evolution and management of COVID-19 hinge on the symbiotic relationship between computational insights and experimental validation, underscoring the interdisciplinary synergy crucial to this endeavor.
1. Introduction
A new coronavirus known as SARS-CoV-2, which exhibits structural similarities with the virus causing severe acute respiratory syndrome (SARS), emerged during the COVID-19 epidemic in December 2019 [1,2]. SARS-CoV-2 has resulted in a significant global burden of disease and mortality, with the Worldometer coronavirus monitoring tool providing daily updated status information on more than 707.75 million confirmed cases and 7.01 million fatalities (https://www.worldometers.info/coronavirus/, accessed on 25 May 2024) [3]. Following previous coronavirus illness outbreaks, such as SARS in 2002–2003 and Middle East respiratory syndrome (MERS) in 2012, this event presented serious obstacles to the fields of public health, research, and medicine. SARS-CoV-2, the virus that causes COVID-19, started at a seafood market in Wuhan, China, and quickly spread to other parts of the world, sparking a pandemic with significant global implications [4,5,6].
Similar to other coronaviruses, the SARS-CoV-2 virus possesses a distinctive round or multishaped virion particle with a diameter of 120–160 nm that contains the triple spike (S) protein, which oversees virus attachment and membrane fusion during infection [7]. In addition to the S protein, the viral genome codes for three other structural proteins that play distinct roles in the structure and replication of the virus: the membrane (M) protein, the envelope (E) protein, and the nucleocapsid (N) protein [8]. SARS-CoV-2 is a member of the Betacoronavirus genus and possesses a large single-stranded RNA genome that encodes for several different proteins, including accessory and structural proteins [9]. With a ribosomal frame-shifting mechanism, nonstructural proteins (Nsps) are translated from the large polyproteins Pp1a and Pp1ab during the replication process of SARS-CoV-2 [10]. The release and maturation of 15 Nsps, which make up the replicase– transcriptase complex in charge of transcription and replication of the viral genome, depend on appropriate polyprotein processing [10,11].
The spike (S) protein is crucial to the SARS-CoV-2 infection process because of its interaction with the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of host cells, facilitating viral entry [11]. The two primary domains of the S protein are S1 and S2, where S1 mediates ACE2 binding, and S2 facilitates viral membrane fusion with the host cell membrane [12]. The receptor-binding domain (RBD) of the S1 domain interacts with ACE2, and the entry of the virus into host cells depends on the cleavage of the S protein at specific areas [13]. The lipid membrane that envelops coronaviruses is produced by the host cell, and the spike (S) protein on their surface gives them a distinctive halo-like appearance [14]. Among all RNA viruses, the coronavirus genome is one of the largest known [15]. It is a single-stranded RNA with positive polarity [16].
With multiple candidates in development and several vaccines in use globally by early 2022, the development of COVID-19 vaccines moved quickly after the SARS-CoV-2 genetic sequence was made public in January 2020 [17]. The virus has infected millions of people and significantly increased the mortality rates despite vaccine efforts, particularly in low-income nations [18].
One of the nonstructural proteins of SARS-CoV-2, papain-like protease (PLpro), has various functions in the growth and replication of the virus [19]. It contributes to the cleavage of viral polyproteins and, through downregulating type I interferon production, impedes the host’s immune response to the virus [20]. PLpro is a desirable target for therapeutic research because of its capacity to degrade ubiquitin-like interferon-stimulated gene 15 protein (ISG15) from interferon-responsive factor 3 (IRF3) [21].
Recent years have seen an increase in the use of the computational method known as molecular dynamics (MD) simulation to investigate peptides and peptide-like compounds that may be able to suppress SARS-CoV-2 [3,22]. SARS-CoV-2 PLpro has drawn attention as a potential therapeutic target because of its vital function in both the host’s immune response regulation and viral replication [22]. To identify potential drugs to treat SARS-CoV-2, computational techniques such as homology modeling, molecular docking, and MD simulations have been used [23].
Our research study, “Unveiling the Inhibitory Potentials of Peptidomimetic Azanitriles and Pyridyl Esters toward SARS-CoV-2 Main Protease: A Molecular Modeling Investigation,” describes how we evaluated new peptidomimetic nitriles, azatripeptide and azatetrapeptide, as possible inhibitors of the SARS-CoV-2 Mpro [24]. This study used molecular docking, MD simulations, and several post-MD analyses, such as percentage hydrogen occupancy. These methods were used to clarify the binding free energy potentials that the chosen compounds displayed in response to SARS-CoV-2. In addition, we identified the precise residues responsible for the drug-binding characteristics of the selected inhibitors in relation to SARS-CoV-2 MPro [24]. Furthermore, the focus of our current studies, “In Silico Analysis of Repurposed Antiviral Drugs as Prospective Therapeutic Agents for COVID-19: Molecular Docking and Dynamics Simulations Targeting the 3-Chymotryspine-Like Protease (3CLpro) and the Papain-Like Protease (PLpro),” is on how Ritonavir, Lopinavir, Ombitasvir, Paritaprevir, and Ginkgolic Acid might reduce the activity of the 3CLpro and PLpro enzymes. Targeting 3CLpro and PLpro is appropriate because of their critical functions in viral replication. To investigate the potential of these compounds as COVID-19 therapeutic agents, this study sheds light on their binding interactions. This investigation entails examining their capacities to bind to and inhibit the activities of 3CLpro and PLpro enzymes, which are essential targets in the development of coronavirus-targeting antiviral medications.
The COVID-19 pandemic has increased the demand for efficient antiviral drugs, and there has been much interest in the potential therapeutic role of PLpro [25]. In-depth analyses of the virus’s structural and functional features, vaccine developments, and the importance of PLpro targeting in the ongoing fight against COVID-19 are all covered in this review, which also emphasizes the use of computational methods such as MD modeling in drug discovery.
2. Implications for Viral Evolution (Target Enzymes and Receptors)
The molecular interactions between the virus and host cells, particularly those involving viral enzymes and host receptors, have a complex connection to both the evolution of viruses and the control of COVID-19 [26]. To understand this relationship, studies must investigate the various phases of the SARS-CoV-2 life cycle and the potential targets for therapeutic treatments [9]. A crucial step in the life cycle of a virus is the attachment to host cells that occurs at the beginning of the infection [9]. Initially, the human angiotensin-converting enzyme 2 (hACE2) receptor binds to SARS-CoV-2 cells [27]. One possible treatment approach to stop viral infection in its tracks is to interfere with the first virus–host contact [28]. Once within the host cell, the replication of SARS-CoV-2 is dependent on the action of viral enzymes [9]. In this setting, the two crucial proteases are 3CLpro and PLpro [29]. The cleavage of viral polyproteins by these proteases results in the production of nonstructural proteins that are necessary for viral replication [30]. Thus, these proteases are excellent targets for the creation of anti-SARS-CoV-2 drugs that will successfully prevent viral replication, thereby lessening the severity and transmission of COVID-19 (see Figure 1) [31]. When comparing SARS-CoV-2 with its predecessor, SARS-CoV, which produced the SARS pandemic in 2002–2003, the importance of these proteases, particularly PLpro, in the context of the SARS cycle becomes even more evident [32].
Despite having a significant sequence identity with SARS-CoV, SARS-CoV-2 and SARS-CoV depend on different proteases, which is a crucial characteristic that distinguishes the two pathogenicity [33]. PLpro is essential for the cleavage of proteins in the host’s innate immune pathways, which permits the replication of SARS-CoV-2 [29]. In addition to viral proteases, RNA-dependent RNA polymerase (RdRp) is another essential enzyme involved in the viral life cycle [34]. This enzyme could be the target of therapeutic intervention, because it is essential for the synthesis of viral RNA [35]. Although this role is well recognized in the setting of SARS-CoV, it is equally significant in the case of SARS-CoV-2 [36]. Furthermore, understanding PLPro’s composition and mechanism of action may aid in the creation of new antiviral medications and improved vaccines [27,30].
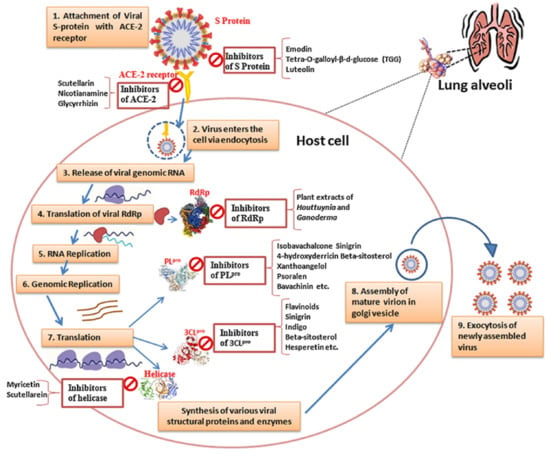
Figure 1.
SARS-CoV-2 life cycle and potential targets for therapeutic treatments, adapted from the source. During viral infection, SARS-CoV-2 first attaches to the host cell surface by binding its spike (S) protein to the human angiotensin-converting enzyme 2 (ACE2) receptor, facilitating viral entry either via endosomes (endocytosis) or direct fusion with the host cell membrane. Once inside the host cell, viral genomic RNA is released into the cytoplasm and translated by host ribosomes, producing polyproteins pp1a and pp1ab, which are then cleaved into nonstructural proteins (Nsp1-16) by viral proteases, including PLpro and Mpro (3CLpro). These non-structural proteins assemble to form the replicase–transcriptase complex (RTC), with Nsp12–16 providing enzymatic functions for viral genome replication and transcription. The viral RNA (gRNA) is first replicated to form a negative-strand RNA and then used for either replication to produce more gRNA or the transcription of subgenomic mRNAs. Subgenomic mRNAs are translated into structural and accessory proteins, including spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins. The structural proteins, along with the nucleoprotein complex formed by the N protein attaching to the gRNA, assemble in the endoplasmic reticulum (ER) and Golgi intermediate compartment (ERGIC) to form mature virions. These virions are then released from the host cell through exocytosis, infecting new cells and perpetuating the viral infection. Throughout these steps, potential treatments include inhibitors targeting the interactions between the viral S protein and ACE2 receptor, viral proteases (PLpro and Mpro), RNA-dependent RNA polymerase (Nsp12), helicase (Nsp13), and other viral enzymes involved in genome replication and transcription [37].
One key factor in the development of COVID-19 is the interaction between the virus and host receptors, particularly the ACE2 receptor [38]. The impact of the virus on distinct organs is largely dependent on ACE2 expression in the lungs, heart, kidneys, and intestines, among other host tissues [39]. It is noteworthy that ACE2 appears to play two roles in infections with SARS-CoV-2 [40]. It protects the lungs from acute respiratory lung injury, including damage caused by the SARS spike protein, but it also increases viral susceptibility [41]. In addition to ACE2, other receptors and proteases mediate the entrance and pathogenesis of SARS-CoV-2, including TMPRSS2, DPP4, and cathepsins B and L [42]. Currently, the exact processes by which these receptors and proteases aid in viral entry remain vague [43]. However, the above literature highlights their potential as therapeutic intervention targets.
The spike protein and activation of viral entry by cellular proteases are other examples of interactions between the virus and host [44,45]. Inhibitors that target TMPRSS2 have demonstrated promise in reducing viral infection. TMPRSS2 is essential for priming the spike protein for viral entry [46,47].
The proteomics and genomics of SARS-CoV-2 are equally intriguing. Proteases such as 3CLpro are essential for breaking down polyproteins into useful proteins [48]. The virus codes for various structural and non-structural proteins. The replicase–transcriptase complex (RTC), which is necessary for viral transcription, replication, and maturation, is formed in part by this process [30,49]. Enzymes such as Nsp3, Nsp4, Nsp5, Nsp12, Nsp13, and Nsp14 are among the many components of the RTC that coordinate various stages of viral replication [50]. These enzymes reveal themselves as excellent possibilities for the creation of antiviral drugs, because they lack near homologs in their hosts [51].
During the crucial step of viral entry, SARS-CoV-2 interacts with various proteases and receptors [52]. Finding inhibitors for these proteases is a promising approach to stop the spread of viruses [53]. Effective antiviral drugs require a thorough understanding of the host receptors and viral enzymes involved in SARS-CoV-2 infection [12,54]. Novel approaches may be discovered to stop viral replication, lessen the severity of COVID-19, and possibly contain this worldwide epidemic and similar outbreaks by focusing on proteins and receptors [55].
In light of the above, numerous enzymes and receptors involved in the viral life cycle and pathogenesis have been clarified by recent studies on SARS-CoV-2 (see Table 1) [1,56]. Key enzymes such as RdRp, PLpro, and Mpro are desirable targets for antiviral medication development, because they are essential for both viral transcription and replication [6]. Furthermore, the significance of this contact in viral infectivity is demonstrated by the fact that the spike (S) protein of SARS-CoV-2 interacts with the hACE2 receptor to enter host cells [4,57]. The virus modifies several cellular signaling pathways, including those linked to immune evasion and inflammation, to control the machinery of the host cell [7]. In the development of efficient therapeutic options against SARS-CoV-2 infection, it is essential to comprehend the roles played by these enzymes, receptors, and their interactions with viral components [8,56]. Repurposing FDA-approved drugs has helped in identifying new targets for medication development and vaccine creation by clarifying the molecular pathways underlying viral pathogenesis [9]. This perspective will ultimately aid the global effort to combat the COVID-19 pandemic and appears to be helpful in future similar outbreaks.

Table 1.
Summary of various enzymes, receptors, and their functions in SARS-CoV-2.
3. Structure and Function of the SARS-CoV-2 PLpro
Proteases found in coronaviruses, such as SARS-CoV and MERS-CoV, were reported to be structurally and functionally similar to the PLpro of SARS-CoV-2 [23]. These proteases are interesting targets for antiviral therapy, because they are essential for viral reproduction and immune system evasion [59]. Although coronaviral proteases have some similarities, the protease domains of each virus display unique structural and biochemical characteristics [25]. In contrast to its equivalents in other coronaviruses, SARS-CoV-2 PLpro has distinct substrate specificity and enzymatic activity profiles [26]. These discrepancies result from changes in the structural configurations and amino acid sequences of the protease active sites, which affect how the enzymes interact with inhibitors and substrates [26]. Through an analysis of the structural and functional properties of PLpro in several coronaviruses, scientists can discern the conserved areas or residues crucial for protease activity, in addition to distinct attributes exclusive to SARS-CoV-2 [27]. This comparative analysis facilitates the discovery and enhancement of antiviral medications with broad-spectrum efficacy against multiple coronaviruses [28]. One way to create pan-coronavirus inhibitors that can stop viral replication in a variety of coronaviruses is to target conserved catalytic residues or substrate-binding sites that are shared by coronaviral proteases [29]. On the other hand, by comprehending the distinct structural components or allosteric sites particular to SARS-CoV-2 PLpro, selective inhibitors are specifically designed to tackle the current pandemic without compromising other similar outbreaks [30].
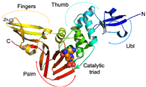
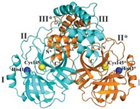
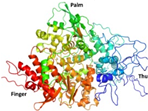
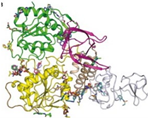
Nevertheless, PLpro remains a key player in the genomic RNA replication of SARS-CoV and is important for the closely related SARS-CoV-2 [23]. Because this sequence is essential for cleaving the replicase substrate, PLpro is an excellent candidate for the creation of antiviral drugs that specifically target SARS-CoV-2 [31]. Three domains have been identified in the crystal structure of SAR-CoV-2 PLpro, including a catalytic cysteine cleavage domain, a potential labile Zn-binding domain, and a ubiquitin domain (see Figure 2) [32]. The labile ZnII ion is essential for preserving the structural integrity of SAR CoV-2 PLpro, whereas the catalytic site is defined by the amino acid triad Cys111–His272–Asp286 [33]. All of these domains are viable targets for therapy, because the ubiquitin domain is connected to the host’s innate immune responses [34].
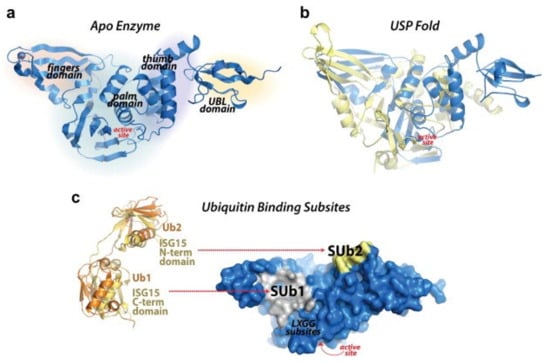
Figure 2.
Redrawn SARS-CoV-2 papain-like protease structure, adapted from the source. The multidomain architecture of PLpro and its two ubiquitin binding subsites are depicted as follows: (a) The SARS-CoV PLpro monomer (PDB: 2FE8) exhibits four distinct domains, arranged from the N-terminus to the C-terminus: the extended UBL, thumb, palm, and finger domains. (b) An overlay of SARS-CoV PLpro (blue) with USP7 (yellow, PDB: 4M5W) illustrates the USP fold. (c) A solvent-accessible surface representation of SARS-CoV PLpro is shown in blue, with a K48-linked di-Ub molecule (orange) and an ISG15 molecule (yellow) superimposed and depicted as ribbons. The ISG15 structure comprises two tandem UBL folds (Ub1 and Ub2). The two Ub and UBL binding subsides of SARS-CoV PLpro are highlighted in the solvent accessible surface representation, with SUb1 shaded in white and SUb2 shaded in yellow [39].
Dual purpose is one of the primary roles of the conserved cysteine residue in viral PLpro [31]. First, it binds to the ZnII ion, which is necessary to preserve correct protein folding and the stability of the local geometry [35]. Second, it attacks the substrate in the capacity of a nucleophile, changing the structure and functionality of PLpro [36]. Potential antiviral methods to interfere with PLpro activity and hinder viral replication include inhibiting catalytic cysteine at the active site or focusing on the elimination of the labile ZnII ion [37,38].
Furthermore, interest in SARS-CoV-2 drug development focuses on PLpro and other subdomains like Nsp3, Nsp10, and Nsp13 containing potential labile Zn sites as well, expanding potential targets. Zn-ejector drugs pose a challenge due to their weaker selectivity, potentially affecting various Zn-containing proteins in the human body [60]. Substance-based inhibitors, on the other hand, show promise in stopping viral replication, and hence, reducing infection, because they specifically bind to the catalytic cysteine at the active site of SARS-CoV-2 PLpro. Peptide-based inhibitors offer the potential for increased antiviral efficacy and allow for a more rational design, making thiol-targeting inhibitors potentially effective treatments for SARS-CoV-2 [60,61].
Nsp3, which contains the PLpro domain, is a crucial element in the extensive genome of SARS-CoV-2. Between a nucleic acid-binding domain (NAB) and the SARS unique domain (SUD/HVR), in Nsp3, lies a large multidomain protein that includes PLpro. It is often found in two copies, known as PL1pro and PL2pro, and is highly conserved across coronaviruses [26]. The proteolytic cleavage of peptide bonds by these cysteine proteases releases Nsp1, Nsp2, and Nsp3, which are essential for viral replication [62]. The identification and cleavage of PLpro depend on the LXGG motif present in Pp1a/Pp1ab. Beyond its involvement in viral replication, PLpro performs a variety of other tasks that affect host immunological responses, such as deubiquitination and deISG15ylating [63]. This makes it a prime candidate for treatment, because it affects many host cellular pathways, indicating that it plays a crucial role in the growth and replication of viruses [26,63].
Previous research on SARS-CoV-2 antiviral drug development has mostly concentrated on Nsp proteins, such as Nsp3 PLpro, Nsp5 Mpro, and Nsp12 RNA-dependent RNA polymerase, which were found in investigations on other previous coronaviruses [64,65]. The argument for focusing on the SARS-CoV-2 Nsp3 domain, PLpro, is discussed in this context. Many coronaviruses have available structural characterizations of PLpro, which are a great resource for structure-based drug development [36]. Nevertheless, obstacles have arisen in the development of antivirals against SARS-CoV and SARS-CoV-2, requiring new approaches [66].
Despite having less research conducted on it than Mpro, PLpro remains a multifunctional protein that functions as a deubiquitinase and a cysteine protease, which opens the door for the development of powerful PLpro inhibitors [67]. This special combination of roles affects host immune responses in addition to viral protein processing [67]. Thus, PLpro is an appealing target for antivirals, because blocking its functions may prevent viral replication and interfere with the host immune system [68]. The expertise obtained from investigating SARS-CoV PLpro can be quickly used to investigate SARS-CoV-2 PLpro, thereby accelerating the development of new antivirals and targeting drug methods for SARS-CoV-2 [68,69].
Furthermore, SARS-CoV-2 is a positive-sense genomic RNA that encodes 10 open reading frames (ORFs) as well [69,70]. Of these, most of the genome is made up of ORF1ab, which translates into two large polypeptide products, pp1a and pp1ab, in the host cell [71]. As mentioned above, proteolytic cleavage of these polypeptides is necessary to produce the crucial nonstructural proteins (Nsps) required for viral replication [72]. Notably, PLpro (Nsp3) and Mpro (Nsp5) catalyze the autocatalytic cleavage process [73,74]. PLpro greatly aids in viral protein processing, distinguished by its Cy-sHis–Asp catalytic triad, and is essential for immune evasion [75]. The inhibition of PLpro prevents viral replication and interferes with different host immunological responses, underscoring its potential as a crucial target for drug development [68].
Because PLpro is a difficult pharmacological target, there has not been much research on its inhibitors [76]. In contrast to Mpro, its active site pocket is flatter [77]. Finding initial hits with potential PLpro inhibitors for additional optimization can be facilitated by screening for cysteine protease and deubiquitinase inhibitors, given the dual activity of PLpro as a cysteine protease and deubiquitinase [78]. A thorough understanding of the substrate specificity, structure, and mechanism of SARS-CoV-2 PLpro will be essential for designing potent PLpro inhibitors and opening the door for the creation of antiviral drugs to treat COVID-19 and future outbreaks.
Additionally, the response to the SARS-CoV-2 outbreak has prompted a re-evaluation of metal complexes in drug discovery efforts. Although traditionally under-represented in compound libraries, metal complexes exhibit growing promise in medicinal chemistry [79,80]. To this end, over 100 structurally diverse metal complexes were selected for profiling as inhibitors, targeting two critical SARS-CoV-2 replication mechanisms: the spike (S) protein interaction with the ACE2 receptor and the PLpro [79,80,81]. Notably, silver- and gold-containing complexes emerged as potent inhibitors of PLpro activity, with polyoxometalates (POMs) showing a promising activity against both targets [79,80,82]. These findings underscore the potential of metal-based compounds as future SARS-CoV-2 antiviral agents, making them promising avenues for further exploration and drug development. Recent research highlights the diverse therapeutic applications of metal complexes, including gold compounds, against various viruses and parasites, underscoring their broad-spectrum antiviral potential [83]. Furthermore, innovative approaches using metal complexes as cysteine-targeting warheads offer promising avenues for inhibiting SARS-CoV-2-associated cysteine proteases, paving the way for novel therapeutic interventions against viral infections [67].
The knowledge gathered from coronaviral protease comparison studies can help in the rational development of inhibitors with improved pharmacokinetics, specificities, and potencies [40]. Medicinal chemists can tailor inhibitor candidates to enhance their efficacy against SARS-CoV-2 PLpro while avoiding off-target effects and potential toxicity using structural similarities and differences [41]. Furthermore, understanding how inhibitors and proteases of various coronaviruses cross-react helps anticipate and prevent the formation of drug-resistant viral variations [42]. Thus, thorough comparative studies of coronaviral proteases will aid in the creation of novel antiviral tactics that may be used to stop coronavirus epidemics both now and in the future [43].
4. Multifaceted Approach with MD Simulations Targeting SARS-CoV-2 Papain-like Protease (PLpro)
Molecular dynamics (MD) simulation studies have been used more often recently to explore possible inhibitors of the PLpro of SARS-CoV-2. Researchers can investigate the interactions between PLpro and potential inhibitors at the atomic level using MD simulations, which provide insightful information about the dynamic behaviors and structural features of biomolecular systems [27]. Using these simulations, inhibitor–PLpro complex binding patterns, affinities, and stability may be predicted, offering vital information for logical drug design (see Figure 3) [45]. Through the simulation of PLpro activity in several solvent environments and physiological situations, molecular docking studies (MD) provide insights into the flexibility and conformational dynamics of the enzyme, which are critical for comprehending its role and designing potent inhibitors [46]. Furthermore, the characterization of allosteric regions that may regulate PLpro activity and the discovery of crucial amino acid residues involved in ligand recognition are two additional ways in which MD simulations can help clarify the processes of inhibitor binding [47]. MD simulation studies are an effective computational tool for investigating the dynamic and structural characteristics of PLpro and directing the development of new inhibitors that may be therapeutically effective against SARS-CoV-2 [48].
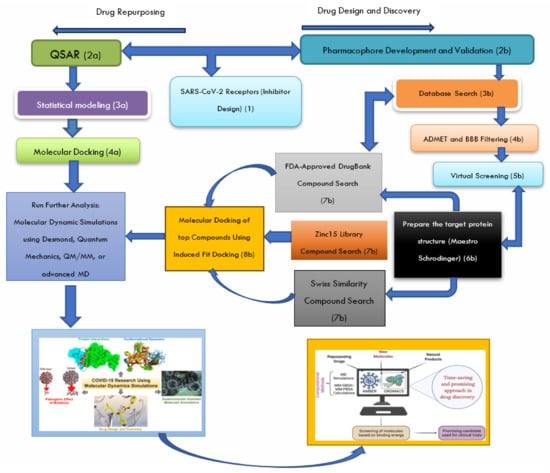
Figure 3.
Schematic pathway describing the steps involved in setting up molecular dynamics (MD) simulations for SARS-CoV-2 PLpro receptors, as adapted from the source: The process for setting up MD simulations for SARS-CoV-2 PLpro receptors (1) involves several sequential steps. It begins with pharmacophore screening (development and validation) (2b) and database exploration (3b) to identify compounds with structural similarities to the natural ligand of the PLpro receptor (7a, b, c). Subsequently, compounds were subjected to Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) and Blood-Brain Barrier (BBB) filtering during in silico studies to prioritize drug candidates with favorable pharmacokinetic properties (4b). Virtual screening is then employed to rapidly evaluate large compound libraries for their likelihood of binding to the PLpro receptor/enzyme (5b), followed by molecular docking to predict the binding mode and affinity of small molecules to the receptor (8b). For drug repurposing, Quantitative Structure–Activity Relationship (QSAR) modeling is used to establish mathematical relationships between the chemical structures and biological activities (2a–3a), aiding in the identification of potential drug candidates. Molecular docking predicts ligand binding modes and affinities to target proteins by exploring ligand conformations and scoring for optimal binding configurations (4a). The selected candidates undergo MD simulations to evaluate the binding affinity and stability, followed by the prediction of binding energy using the Generalized Born Surface Area (MM-GBSA) solvation calculation. This comprehensive approach offers an efficient means of screening large compound databases or repurposed drugs, thus providing valuable insights into potential drug effectiveness before experimental testing [56].
In the midst of the SARS-CoV-2 pandemic, researchers have taken a variety of approaches to find efficient ways to combat the virus [49]. Targeting PLpro, the virus’s essential protease enzyme, is important for this mission using computational approaches [50]. Several important factors unique to SARS-CoV-2 PLpro determine whether one modeling technique, such as template-free versus template-based modeling, is better suited for finding SARS-CoV-2 PLpro inhibitors than another [51]. Template-based modeling, sometimes referred to as homology modeling, creates a model of the target protein, the SARS-CoV-2 PLpro enzyme, using experimentally determined homologous protein structures as templates [52]. Template-based modeling can yield accurate models of SARS-CoV-2 PLpro by leveraging proteases closely related to other coronaviruses with known structures [53]. When the target and template proteins have a high degree of sequence identity, this approach is particularly helpful [51,54]. However, template-based modeling may not be useful or feasible if there are no close homologs or templates available for SARS-CoV-2 PLpro [55].
Furthermore, from-the-beginning modeling, commonly referred to as template-free modeling, builds protein structures exclusively from the amino acid sequence without the use of structural templates [84]. In the absence of appropriate templates, this method may be useful for estimating the structure of SARS-CoV-2 PLpro [84]. Template-free modeling explores the conformational space of a protein sequence and finds the most energetically favorable fold using physics-based energy functions and conformational sampling techniques [85]. However, compared with template-based approaches, template-free modeling is typically less precise and dependable, especially for larger and more complicated proteins such as PLpro or Mpro [30,51]. Even with these drawbacks, template-free modeling might nevertheless offer insightful information about the composition and operation of PLpro, particularly when combined with experimental data and other computational techniques [86]. Ultimately, the requirements of the study objectives and the available computational tools and skills should serve as a guide for choosing a modeling technique [87].
To tackle the problems caused by this multifunctional protein (PLpro), scientists are using thiol-reacting inhibitors, substrate-based peptides, FDA-approved drug repurposing, and MD simulations [88]. Finding thiol-reacting inhibitors is a potential path, especially if they can covalently bind to the conserved cysteine residues in SARS-CoV-2-PLpro [89]. By forming a Cys–inhibitor complex through this covalent connection, PLpro is rendered inactive, and viral replication is impeded [39]. The capacity of Zn-ejector compounds to remove zinc ions (ZnII) from the PLpro active site has drawn significant interest [60]. Disulfiram (DSF) and other disulfide-based thiol-reacting inhibitors are notable among them [60]. The potential of DSF, an FDA-approved drug for treating persistent alcohol abuse, as a Zn ejector has been thoroughly investigated [61]. DSF has potential as a Zn-ejector drug against CoV-2-PLpro because of the close genetic and structural similarities between SARS-CoV-2-PLpro and SARS-CoV-PLpro [60]. Recent investigations have revealed the multifunctionality of DSF. In addition to acting as a Zn-ejector, it modifies cysteines, causing damage to the structural and functional integrity of SARS-CoV-2 PLpro [90,91]. Because of their combined effects, which limit viral replication, additional FDA-approved sulfur-based drugs are being investigated for their potential to be effective against COVID-19 [92]. Tests have been conducted on compounds such as diethyldithiocarbamate (DDC), captopril, 2,2′-dithiodipyridine, 2,2′-dithiobis (benzothiazole), and 6-thioguanine (6-TG) [60]. Notably, in cell cultures, 6-TG demonstrated a significant inhibition of SARS-CoV-2 PLpro [93]. Moreover, even at low micromolar concentrations, the FDA-approved drug auranofin used to treat rheumatoid arthritis has shown a strong ability to hinder SARS-CoV-2 replication in human cells [94].
Moreover, SARS-CoV-2 PLpro, which recognizes and cleaves peptide bonds at LXGG patterns between Nsps, is a crucial component of the virus replication mechanism [26]. These motifs are the targets of small peptides that have been developed to function as substrate-based inhibitors [60]. The creation of two highly selective tetrapeptides that effectively inhibit SARS-CoV-2 PLpro, VIR250 and VIR251, is the result of recent breakthroughs [50]. The purpose of these peptides is to bind to the catalytic Cys111 residue via thioether linkage, thus blocking the enzyme’s ability to cleave polyprotein and stopping the spread of the virus [95]. Despite its greater potency, VIR251 displays less selectivity for binding SARS-CoV-2 PLpro because of variations in the catalytic site’s amino acid orientation [96].
Repurposing FDA-approved drugs is an alternative therapy approach being studied for COVID-19 [97]. The FDA has approved Remdesivir, an RdRp inhibitor, as the only antiviral drug for treating COVID-19 [98]. However, its efficacy in patients is rather moderate [99]. Ritonavir has been used to block SARS-CoV-2 PLpro, which is well known to have anti-HIV-1 properties [100]. This drug affects the coordination of signals within infected cells by targeting polyprotein produced by viral proteases [101].
Understanding the stability and interactions between small molecules and proteins is made possible through MD simulations [102]. The computational methods used to elucidate the kinetics of binding and interactions between drugs and proteins are shown in Table 2.

Table 2.
MD simulation studies of SARS-CoV-2 PLpro inhibitors.
Because they are commercially available and have a track record of safety, FDA-approved drugs are perfect repurposing candidates [113]. One important goal is to identify the most effective options among these drugs based on their robust binding ability to SARS-CoV-2 PLpro [50]. To assess the stability, binding affinity, and interactions of drug– PLpro binding, molecular mechanics/generalized Born surface area (MM/GBSA) binding energy estimates were obtained after MD simulations [114].
Considering the above, MD simulations have been used to study the stability and interactions of small molecules bonded to proteins in more detail [115]. Because they are readily available on the market, have a track record of safety, and may be used directly without requiring further preclinical or clinical testing, FDA-approved drugs are the main alternatives for treatment in emergencies and outbreaks of diseases [116]. This review found that MD simulations over 50 ns show promising drugs based on the anticipated docking scores for COVID-19 PLpro [117]. Throughout the MD simulated time, drug– PLpro binding energies (ΔGbinding) were assessed based on the molecular mechanics/generalized Born surface area (MM/GBSA) technique [118]. Furthermore, estimates were made of the drug–PLpro binding stabilities, affinities, and interactions [119].
In recent studies, three antioxidants and cell protectants (NAD+, quercitrin, and oxiglutatione), three antivirals (ritonavir, moroxydine, and zanamivir), two antimicrobials (doripenem and sulphaguanidine), two anticancer drugs, three benzimidazole anthelmintics, one antacid (famotidine), three antihypertensive ACE receptor blockers (candesartan, losartan, and valsartan), and other various systemically or topically acting drugs were among the top results expected to bind with SARS-CoV-2 PLpro strongly [120]. These drug binding patterns were superior to those of 6-mercaptopurine (6-MP), a previously discovered SARS CoV PLpro inhibitor, indicating the possibility of using these drugs as an alternative way to treat COVID-19 [120,121]. Moreover, studies have shown that the repurposing of ritonavir, an anti-human immunodeficiency virus type 1 (HIV-1) provides a multifaceted approach to stop the growth of viruses (blocking the transcription and replication of viral RNA) [122,123].
Another study using the Supernatural Database conducted a high-throughput virtual screening (HTVS) program with the aim of finding inhibitors that target PLpro. The study revealed that two substances, SN00334175 and SN00162745, showed docking scores of 10.58 and 9.93 kcal/mol, respectively, according to the XP docking results [124]. Furthermore, Van der Waals energy and hydrophobic energy components were identified as significant contributors to the overall binding free energy in the PRIME MMGB-SA investigations [125]. SN00334175/7JN2 and SN00162745/7JN2 were stabilized by ligand binding, which formed interactions with Gly266, Asn267, Tyr268, Tyr273, Thr301, Asp302, Lys157, Leu162, Asp164, Arg166, Glu167, Pro248, and Tyr264. This result was obtained from a 100-ns MD simulation of these complexes [125].
From the same perspective, a study screened 21 antiviral, antifungal, and anticancer compounds using an in silico molecular docking technique to identify potential inhibitors of SARS-CoV-2 PLpro [126]. This study showed that neobavaisoflavone has the highest binding energy to SARS-CoV-2 PLpro [122]. These compounds may bind close to the ISGylation, ubiquitination, and PLpro critical catalytic triad of SARS-CoV-2: Trp106, Asn109, Cys111, Met208, Lys232, Pro247, Tyr268, Gln269, His272, Asp286, and Thr301 [127]. These compounds may be useful options for therapeutic intervention against COVID-19, because inhibiting PLpro is a crucial tactic in the battle against viruses [127].
The multifunctional technique using MD simulations to target SARS-CoV-2-PLpro is a comprehensive approach to suppress viral replication and control COVID-19 [120]. In the fight against the current pandemic, thiol-reacting inhibitors, substrate-based peptides, FDA-approved drug repurposing, and MD simulations have been combined to provide information on potential candidates and modes of action.
6. Clinical and Preclinical Studies: Integrating MD Simulation Insights
The translation of computational results into clinically relevant outcomes is a vital link between theoretical understanding and useful healthcare decisions [141]. Because they clarify molecular interactions, forecast medication efficacy, and optimize drug candidates, computational techniques, including molecular docking, MD simulations, and machine learning algorithms, are essential to the drug development process [142]. Due to the widespread effects of COVID-19, a significant amount of research has been conducted using computational and experimental methods to better understand the illness and create treatments, vaccinations, and diagnostic tools [143]. In the first year of the pandemic, drug repurposing was not able to quickly address global problems, despite a great deal of computational attention being directed toward this area [144]. However, a few well-known drugs have been used in clinics to treat COVID-19 patients, and several drugs with new uses are still being clinically tested [145]. In this setting, the need to deliver experimentally testable ideas to expedite the identification of novel therapies is highlighted, along with the requirement for powerful computational tools [146]. As the world struggled to deal with the substantial death toll and disruptions brought on by COVID-19 in 2020, further research and clinical trials resulted in the creation of vaccinations [147]. Although the evolving coronavirus has raised questions about vaccination efficacy, it has refocused the attention to drug repositioning [143]. Using matrix completion approaches, computational models have been developed to predict drug–virus relationships for drug repositioning. This process provides a useful tool for doctors to choose possible antiviral treatments [148]. Using chemical and genetic structures, this study contains a curated database of the known correlations between viruses and antivirals [149]. After confirmation using regular experimental protocols, ongoing clinical studies agree with the computational techniques’ accurate anticipation of potential antiviral drugs for COVID-19.
Targeting SARS-CoV-2 PLpro has been the focus of extensive studies because of the urgent need to discover new therapeutic techniques to attack COVID-19 [129]. This review also looked at preclinical and clinical research on PLpro inhibition and the knowledge obtained from MD simulations. In addressing SARS-CoV-2 PLpro, GRL-0617, a non-covalent inhibitor, demonstrated potential for SARS-CoV PLpro [150]. Because of the structural similarities between the two viral proteases and the compound’s efficacy against SARS-CoV-2 PLpro, there is potential to repurpose existing PLpro inhibitors for SARS-CoV-2 [27,129,151]. The conserved Tyr268 in SARS-CoV-2 PLpro and GRL-0617 share the same binding mechanism, which was further supported using computational simulations [89].
Although vaccines such as those from Moderna, Johnson & Johnson, and Pfizer/BioNTech provide promise in controlling the pandemic, their efficacy against novel virus strains remains uncertain, because they mostly target the spike protein [152]. Moreover, vaccinations act as prevention measures, which makes the creation of antiviral drugs for treating COVID-19 necessary [153].
Recent research has shown that there are irreversible inhibitors with great selectivity for SARS-CoV-2 PLpro, such as VIR250 and VIR251 [50]. These structural discoveries provide the basis for logical drug design approaches that efficiently target SARS-CoV-2 PLpro [154]. In a multicenter prospective study, a study registered with ClinicalTrials.gov, NCT04276688, showed that early triple-antiviral therapy was safe and superior to lopinavir–ritonavir alone in alleviating symptoms and shortening the duration of viral shedding and hospital stay in patients with mild to moderate COVID-19 [155,156]. In another study, the potential of Cyanovirin-N to suppress SARS-CoV-2 was recommended because of its strong binding energies with the spike protein, Mpro, and PLpro [157,158]. In a similar perspective, other putative PLpro inhibitors were identified through a virtual screening of FDA-approved medications. Among these, Mefloquine showed promise as a treatment agent because of its efficacy against SARS-CoV-2 in cell-based experiments [136]. Furthermore, structural similarities between the PLpro of SARS-CoV-2 and Human Coronavirus 229E (HCoV-229E) were found through molecular modeling studies, indicating that the latter could be a good surrogate for drug discovery investigations [159]. This strategy resulted in the discovery of Mefloquine, which demonstrated encouraging outcomes against SARS-CoV-2 PLpro. However, additional research is required to fully explore its therapeutic potential.
Moreover, certain pharmaceuticals, including Dronedarone, Dasatinib, and Clofazimine, were identified as possible PLpro inhibitors through the screening of clinical and preclinical medications with Clofazimine, which has recently been identified as a broad-spectrum inhibitor of coronaviruses and may be a promising candidate for coronaviruses that have emerged and may emerge in the future [160,161]. The above-mentioned substances demonstrated antiviral efficacy against SARS-CoV-2 both in vivo and in vitro, implying that they may be useful in treatment. Furthermore, the lead compound Jun12682 was found using structure-based drug design; it effectively inhibited the protease, deubiquitinase, and deISGylase activities of PLpro. This substance consistently showed effectiveness against nirmatrelvir-resistant mutants and SARS-CoV-2 variants, indicating its potential to address present and future viral challenges [78].
In other studies, five compounds were identified after a detailed analysis of the screening results: MFCD00832476, MFCD02180753, Ergotamine, Pacritinib, and Bemcentinib inhibit PLpro [162,163]. At a 20 ns MD simulation, these compounds were stable within the active sites of the protease proteins [162]. In contrast, 147 substances were shown to be possible SARS-CoV-2 PLpro inhibitors in a different investigation. Dasatinib was one of these agents; in clinical situations, it demonstrated antiviral effectiveness against SARS-CoV-2, but it was unclear if Dasatinib interacted with PLpro directly [136,164]. Nonetheless, the research recommends that one of the most crucial approaches for future COVID-19 treatment and development, preventive research, is computer-aided small-molecule drug discovery.
However, there are several difficulties in applying computational results to clinical practice, especially in the early stages of clinical trials [165]. The safety and effectiveness of potential medications in humans must be confirmed through experimental validation in clinical trials, even though computational models offer useful predictions [166]. Clinical trials evaluate the safety, dose, effectiveness, and side effects of drugs in various patient populations through extensive testing conducted in several phases [167]. Issues such as patient recruitment, the study design, regulatory approval, and financial limitations may hamper the clinical translation of computational insights [168]. Moreover, disparities between computational forecasts and experimental results highlight the necessity of iteratively improving and validating computational models to improve their predictive precision and reliability in clinical settings [143]. Despite these challenges, the integration of computational methods into clinical research holds immense promise for accelerating the development of novel therapeutics and improving patient outcomes in diverse disease contexts.
7. Challenges and Future Directions: Guided by MD Simulations
There have been numerous challenges in finding efficient coronavirus protease inhibitors, especially those that target the PLpro and Mpro of SARS-CoV-2 [129]. The main causes of these obstacles are the inhibitory efficacy and pharmacokinetic properties of potential drugs [169]. To be deemed a promising drug or drug candidate, a molecule needs to accomplish two things: it needs to successfully reach a target in the body and trigger the desired biological reactions [170,171]. These are important factors to consider while developing antiviral drugs, particularly considering the critical need to eradicate COVID-19 and similar viruses [172,173].
The kinetics of enzyme–ligand interactions determine the suitability of the small timescale used in the MD simulation for finding PLpro inhibitors [174]. Although they are less effective at simulating longer-term phenomena such as complex binding kinetics or protein folding, short timescale simulations can nonetheless provide important insights into the early phases of a protein–ligand interaction or the dynamics of certain protein domains [175]. Short timescale simulations could be useful, for example, if the goal is to comprehend the early phases of PLpro inhibition or investigate the flexibility of binding pockets [23]. However, longer timescale simulations or improved sampling approaches may be required to adequately capture uncommon events and transitions when investigating the stability of PLpro-inhibitor complexes over lengthy periods of time or revealing allosteric processes [176].
Furthermore, the parameterization of force fields and other simulation parameters significantly affects the interpretation of MD simulation results for PLpro inhibitors [14,16]. The choice of ion parameters, water models, force field parameters, and how electrostatic interactions are handled can all have a significant impact on the accuracy and reliability of the simulation results. Therefore, to ensure reproducibility and transparency, it is essential to provide comprehensive information on the parameterization technique used in MD simulations. This includes specifying the force field employed (e.g., CHARMM, AMBER, or OPLS), the version and parameter sets utilized, any customizations made to the force field, and the rationale behind these choices [177]. In addition, understanding the simulation process and correctly interpreting the outcomes depend on the thorough documentation of the simulation setup, including the solvent treatment, boundary conditions, integration time steps, and equilibration protocols [178]. Researchers can improve the robustness and credibility of their MD simulation studies on PLpro inhibitors by providing comprehensive details on parameterization and simulation techniques [179]. This will enable the rigorous interpretation and confirmation of the findings.
Baicalein, disulfiram, carbofur, ebselen, tideglusib, shikonin, and PX-12 are only a few of the compounds that have been proven to exhibit drug-like qualities and fit the requirements to be considered for drug development [180]. Nevertheless, these substances still struggle to attain the appropriate level of potency and selectivity against coronavirus proteases [181]. This paradox is also shown in the case of SARS-CoV-2 PLpro inhibitors, where it is still difficult to strike a precise balance between drug-linked qualities and the capacity to elicit strong and targeted protease inhibition [182]. It is critical to address these problems, because they have an immediate influence on the viability of creating antiviral drugs [182].
Studies on the structure–activity relationship (SAR) have provided important new information on the properties of protease inhibitors [183]. Covalent and peptidomimetic inhibitors have demonstrated the ability to selectively and potently inhibit the active sites of these enzymes [24]. However, regarding pharmacokinetic factors, these inhibitors often fail, even when they show promise [184]. They are not acceptable as drug candidates because of either their poor metabolism, excretion, absorption, distribution, or toxicological properties [24,184]. On the other hand, low-molecular-weight or non-peptidomimetic compounds show potential in meeting drug-like criteria [173]; however, they encounter difficulties in obtaining the required potency and selectivity against coronavirus proteases [40]. To close this gap, it is imperative to concentrate on the lead optimization of low-molecular-weight, non-peptidomimetic drugs [185].
To address these issues in the creation of protease inhibitors, fragment-based drug design (FBDD) appears to be a potential strategy [186]. This involves identifying low-molecular-weight compounds as potential protease inhibitors, using a combination of computational and experimental methods to select advantageous fragments from peptidomimetic compounds, and incorporating these fragments into the lead optimization process of low-molecular-weight compounds [187]. The last phase involves carefully optimizing hybrid compounds to achieve desirable pharmacokinetic and pharmacodynamic characteristics [188]. This approach may offer a viable way to create protease inhibitors that are more potent in the future. The choice of regimen of applicability should be based on their advantages and limitations, as mentioned in Table 3.

Table 3.
Summary comparison and contrast of various regimens of applicability of different computational techniques.
In recent research efforts, drug development has been greatly aided by in silico studies; however, it is crucial to validate these results with in vivo research [201]. These in vivo investigations demonstrate the therapeutic potential of these drug candidates and confirm their method of action [202]. Therefore, a multidisciplinary strategy that incorporates in vivo research, pharmacokinetics, pharmacodynamics, and computational drug design is crucial [203]. When combined, these tactics offer the best chance of surmounting current obstacles and advancing the development of potent antiviral drugs for COVID-19 treatment [204]. Without a doubt, this effort will support the ongoing fight against the worldwide epidemic and aid in the creation of vital drug compounds.
8. Conclusions and Author Insights into Targeting SARS-CoV-2 Papain-like Protease
To find efficient cures and solutions, the global response to the COVID-19 epidemic has sparked uncommon experimental and computational research endeavors. Of the several strategies used, machine learning (ML) has drawn interest because of its potential for drug repurposing, that is, using drugs already approved by the FDA to fight SARS-CoV-2 [205]. Machine learning is used to forecast possible virus inhibitors, highlighting the critical function of trustworthy databases [48]. The effectiveness of ML in virtual drug screening also clarifies the shortcomings of databases and ML models, offering helpful guidance to researchers negotiating this challenging terrain [206].
This review examined the crucial role of SARS-CoV-2 PLpro in the development and control of COVID-19. The discovery of prospective antiviral drugs and our understanding of this important enzyme have both benefited greatly from the application of innovative computational techniques [142,207,208]. In previous research, a tertiary structure model for SARS-CoV-2 PLpro was created using homology modeling, and the catalytic region of the PLpro protein was found to have binding sites for antiviral drugs using molecular docking techniques [209]. In the process of creating new drugs, this computational method, which combines modeling and docking, is a useful first step that lays the groundwork for further experimental studies [210].
Moreover, all-atom MD simulations were performed to guarantee the stability of the drug–PLpro complexes in a water solvent environment [211]. The MD simulations provided important insights into the dynamics and interactions between drug candidates and the PLpro protein. The most stable complexes were found using the binding energy estimations from MD simulations, which are crucial in the search for effective antiviral drugs that inhibit SARS-CoV-2 by inhibiting PLpro [212]. This versatile computational method is effective in the search for new antiviral drugs [213].
Although the promise of certain therapeutic candidates targeting SARS-CoV-2 PLpro has been revealed using computational approaches, it is crucial to stress that these results need to undergo thorough in vitro and in vivo examinations [214]. To assess the therapeutic value of these recommended drugs, real-world experimental settings must validate their safety and effectiveness [215]. These crucial next steps need to be completed in accordance with acceptable scientific methods to guarantee that encouraging computational results will materialize, with real progress against COVID-19 [216,217].
This review contributes to the idea that the existing range of drugs intended to suppress PLpro is insufficient. Their weak actions against this enzyme necessitate careful definition and refinement before they may be considered clinically useful. Therefore, to fully utilize these drugs in the therapeutic context, researchers must optimize and improve their performance. Unbinding events of PLpro inhibitors, such as GRL0617 and its derivatives, have been recorded through a thorough understanding of the unbinding pathways attained via the SuMD simulation method, providing important insights into drug behavior sand opportunities for improvement [218].
Apart from optimizing existing drugs, investigating sub-structurally similar molecules is a potentially fruitful path [219]. Various computational techniques have been used to evaluate the possibility of compounds similar to ritonavir that were obtained from the PubChem database as antiviral drugs for SARS-CoV-2 [220]. Once again, the use of homology modeling, molecular docking, and MD studies together proved to be quite helpful in identifying new drug candidates that may be effective in treating SARS-CoV-2 by specifically targeting PLpro.
Nevertheless, there are several inherent limitations to employing computational models to explore possible inhibitions targeting SARS-CoV-2 PLpro. First, the overall quality and comprehensiveness of input data have a major impact on the accuracy and dependability of computational models [144]. There may be omissions or inconsistencies in the data used, which may introduce bias and uncertainty into the model predictions given the complexity of biological systems and our limited understanding of the interactions involved [145]. Furthermore, because of computing limitations, computer models sometimes oversimplify or overlook complicated biological processes, which may result in simplified representations that fail to fully capture the complexity of PLpro inhibition mechanisms [146]. These simplified assumptions could lead to incomplete or inaccurate forecasts of putative inhibitory substances [147].
Furthermore, there are restrictions on the validity and generalizability of computer models used to predict PLpro inhibitors [148]. Usually, models are evaluated and trained on datasets or scenarios that cannot adequately capture the variety and unpredictability of real PLpro interactions [132,149]. Consequently, these models may perform poorly when used with fresh or untested data, which would lower their projected accuracy and restrict their usefulness [221]. Therefore, the absence of standardization and reproducibility in computer modeling techniques complicates the evaluation of the precision and dependability of model predictions [222]. The identification of PLpro inhibitors may be questioned in the absence of strong validation protocols, which could impede the development of these compounds into useful therapeutics [223]. While computational modeling offers valuable insights into potential PLpro inhibitions, addressing these limitations is crucial to ensure the validity and utility of the findings in advancing therapeutic strategies against SARS-CoV-2.
It is crucial to understand that these results are preliminary and need to be rigorously validated by experiments. Moreover, future outbreaks and the continuous battle against this worldwide pandemic would require a concentrated effort to improve the currently available drugs and investigate new chemical compounds. This multidisciplinary strategy, which combines experimental validation with computational insights, is critical for the development and control of COVID-19.
Author Contributions
Conceptualization, A.G.-A.M., N.N.M. and H.M.K.; writing—original draft preparation, A.G.-A.M., S.C.U. and N.N.M.; writing—review and editing, A.G.-A.M., N.A.M. and S.C.U.; supervision, R.B.K. and H.M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study received no external funding.
Acknowledgments
The authors acknowledge the College of Health Sciences of the University of Kwazulu-Natal for their support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pal, M.; Berhanu, G.; Desalegn, C.; Kandi, V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): An update. Cureus 2020, 12, e7423. [Google Scholar] [CrossRef]
- Gusev, E.; Sarapultsev, A.; Solomatina, L.; Chereshnev, V. SARS-CoV-2-specific immune response and the pathogenesis of COVID-19. Int. J. Mol. Sci. 2022, 23, 1716. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 virus use multiple receptors to enter host cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiang, R.; Huo, S.; Zhou, Y.; Jiang, S.; Wang, Q.; Yu, F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduct. Target. Ther. 2021, 6, 233. [Google Scholar] [CrossRef]
- Mushebenge, A.G.-A.; Ugbaja, S.C.; Mbatha, N.A.; Khan, R.B.; Kumalo, H.M. Unravelling Insights into the Evolution and Management of SARS-CoV-2. BioMedInformatics 2024, 4, 385–409. [Google Scholar] [CrossRef]
- Cannalire, R.; Cerchia, C.; Beccari, A.R.; Di Leva, F.S.; Summa, V. Targeting SARS-CoV-2 proteases and polymerase for COVID-19 treatment: State of the art and future opportunities. J. Med. Chem. 2020, 65, 2716–2746. [Google Scholar] [CrossRef]
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Signal Transduct. Target. Ther. 2020, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Rajarshi, K.; Khan, R.; Singh, M.K.; Ranjan, T.; Ray, S.; Ray, S. Essential functional molecules associated with SARS-CoV-2 infection: Potential therapeutic targets for COVID-19. Gene 2021, 768, 145313. [Google Scholar] [CrossRef]
- Anand, U.; Jakhmola, S.; Indari, O.; Jha, H.C.; Chen, Z.S.; Tripathi, V.; Pérez de la Lastra, J.M. Potential Therapeutic Targets and Vaccine Development for SARS-CoV-2/COVID-19 Pandemic Management: A Review on the Recent Update. Front. Immunol. 2021, 12, 658519. [Google Scholar] [CrossRef]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar] [CrossRef]
- Davidson, A.M.; Wysocki, J.; Batlle, D. Interaction of SARS-CoV-2 and other coronavirus with ACE (angiotensin-converting enzyme)-2 as their main receptor: Therapeutic implications. Hypertension 2020, 76, 1339–1349. [Google Scholar] [CrossRef] [PubMed]
- Sarker, J.; Das, P.; Sarker, S.; Roy, A.K.; Momen, A.R. A review on expression, pathological roles, and inhibition of TMPRSS2, the serine protease responsible for SARS-CoV-2 spike protein activation. Scientifica 2021, 2021, 2706789. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.; Rozano, L.; Falasca, M.; Mancera, R.L. Does the SARS-CoV-2 spike protein receptor binding domain interact effectively with the DPP4 (CD26) receptor? A molecular docking study. Int. J. Mol. Sci. 2021, 22, 7001. [Google Scholar] [CrossRef] [PubMed]
- Mouffouk, C.; Mouffouk, S.; Mouffouk, S.; Hambaba, L.; Haba, H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2). Eur. J. Pharmacol. 2021, 891, 173759. [Google Scholar] [CrossRef] [PubMed]
- Chitranshi, N.; Gupta, V.K.; Rajput, R.; Godinez, A.; Pushpitha, K.; Shen, T.; Mirzaei, M.; You, Y.; Basavarajappa, D.; Gupta, V. Evolving geographic diversity in SARS-CoV2 and in silico analysis of replicating enzyme 3CL pro targeting repurposed drug candidates. J. Transl. Med. 2020, 18, 278. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Mitra, D.; Paul, M.; Chaudhary, P.; Kamboj, A.; Thatoi, H.; Janmeda, P.; Jain, D.; Panneerselvam, P.; Shrivastav, R. Potential inhibitors of SARS-CoV-2 (COVID 19) proteases PLpro and Mpro/3CLpro: Molecular docking and simulation studies of three pertinent medicinal plant natural components. Curr. Res. Pharmacol. Drug Discov. 2021, 2, 100038. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y. Host cellular RNA helicases regulate SARS-CoV-2 infection. J. Virol. 2022, 96, e00002-22. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, P.; Desikan, R.; Dixit, N.M. Targeting TMPRSS2 and Cathepsin B/L together may be synergistic against SARS-CoV-2 infection. PLoS Comput. Biol. 2020, 16, e1008461. [Google Scholar] [CrossRef]
- Pišlar, A.; Mitrović, A.; Sabotič, J.; Pečar Fonović, U.; Perišić Nanut, M.; Jakoš, T.; Senjor, E.; Kos, J. The role of cysteine peptidases in coronavirus cell entry and replication: The therapeutic potential of cathepsin inhibitors. PLoS Pathog. 2020, 16, e1009013. [Google Scholar] [CrossRef]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andréo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A. Implications of Spike-glycoprotein processing at S1/S2 by Furin, at S2’by Furin and/or TMPRSS2 and shedding of ACE2: Cell-to-cell fusion, cell entry and infectivity of SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Chao, T.-L.; Li, C.-L.; Chiu, M.-F.; Kao, H.-C.; Wang, S.-H.; Pang, Y.-H.; Lin, C.-H.; Tsai, Y.-M.; Lee, W.-H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020, 33, 108254. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Mushebenge, A.G.; Ugbaja, S.C.; Mtambo, S.E.; Ntombela, T.; Metu, J.I.; Babayemi, O.; Chima, J.I.; Appiah-Kubi, P.; Odugbemi, A.I.; Ntuli, M.L.; et al. Unveiling the Inhibitory Potentials of Peptidomimetic Azanitriles and Pyridyl Esters towards SARS-CoV-2 Main Protease: A Molecular Modelling Investigation. Molecules 2023, 28, 2641. [Google Scholar] [CrossRef] [PubMed]
- Behnam, M.A. Protein structural heterogeneity: A hypothesis for the basis of proteolytic recognition by the main protease of SARS-CoV and SARS-CoV-2. Biochimie 2021, 182, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Osipiuk, J.; Azizi, S.-A.; Dvorkin, S.; Endres, M.; Jedrzejczak, R.; Jones, K.A.; Kang, S.; Kathayat, R.S.; Kim, Y.; Lisnyak, V.G. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021, 12, 743. [Google Scholar] [CrossRef]
- Prajapati, J.; Patel, R.; Rao, P.; Saraf, M.; Rawal, R.; Goswami, D. Perceiving SARS-CoV-2 Mpro and PLpro dual inhibitors from pool of recognized antiviral compounds of endophytic microbes: An in silico simulation study. Struct. Chem. 2022, 33, 1619–1643. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L. Broad-spectrum prodrugs with anti-SARS-CoV-2 activities: Strategies, benefits, and challenges. J. Med. Virol. 2022, 94, 1373–1390. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Chen, S.; Ouyang, H.; Ren, L. Possible targets of pan-coronavirus antiviral strategies for emerging or re-emerging coronaviruses. Microorganisms 2021, 9, 1479. [Google Scholar] [CrossRef]
- Calleja, D.J.; Lessene, G.; Komander, D. Inhibitors of SARS-CoV-2 PLpro. Front. Chem. 2022, 10, 876212. [Google Scholar] [CrossRef]
- Gao, X.; Qin, B.; Chen, P.; Zhu, K.; Hou, P.; Wojdyla, J.A.; Wang, M.; Cui, S. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B 2021, 11, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Pradhan, A.; Maurya, V.K.; Kumar, S.; Theengh, A.; Puri, B.; Saxena, S.K. Therapeutic approaches for SARS-CoV-2 infection. Methods 2021, 195, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Dubey, R.; Dubey, K. SARS-CoV-2: Potential drug targets and its virtual screening. Model. Control Drug Dev. COVID-19 Outbreak Prev. 2022, 366, 203–244. [Google Scholar]
- Liu, J.; Cheng, Y.; Zheng, M.; Yuan, B.; Wang, Z.; Li, X.; Yin, J.; Ye, M.; Song, Y. Targeting the ubiquitination/deubiquitination process to regulate immune checkpoint pathways. Signal Transduct. Target. Ther. 2021, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.J.; Mio, M.J.; Prince, R.B.; Hughes, T.S.; Moore, J.S. A field guide to foldamers. Chem. Rev. 2001, 101, 3893–4012. [Google Scholar] [CrossRef] [PubMed]
- Sanders, B.C.; Pokhrel, S.; Labbe, A.D.; Mathews, I.I.; Cooper, C.J.; Davidson, R.B.; Phillips, G.; Weiss, K.L.; Zhang, Q.; O’Neill, H. Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2. Nat. Commun. 2023, 14, 1733. [Google Scholar] [CrossRef] [PubMed]
- De Paiva, R.E.; Neto, A.M.; Santos, I.A.; Jardim, A.C.; Corbi, P.P.; Bergamini, F.R. What is holding back the development of antiviral metallodrugs? A literature overview and implications for SARS-CoV-2 therapeutics and future viral outbreaks. Dalton Trans. 2020, 49, 16004–16033. [Google Scholar] [CrossRef]
- Gupta, Y.; Maciorowski, D.; Zak, S.E.; Jones, K.A.; Kathayat, R.S.; Azizi, S.-A.; Mathur, R.; Pearce, C.M.; Ilc, D.J.; Husein, H. Bisindolylmaleimide IX: A novel anti-SARS-CoV2 agent targeting viral main protease 3CLpro demonstrated by virtual screening pipeline and in-vitro validation assays. Methods 2021, 195, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Báez-Santos, Y.M.; St John, S.E.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Amin, S.A.; Banerjee, S.; Ghosh, K.; Gayen, S.; Jha, T. Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Biorg. Med. Chem. 2021, 29, 115860. [Google Scholar] [CrossRef]
- Mengist, H.M.; Dilnessa, T.; Jin, T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021, 9, 622898. [Google Scholar] [CrossRef] [PubMed]
- Stefani, C.; Fantoni, T.; Bissoli, M.; Thomas, J.; Ruggiero, A. HIV and SARS-CoV-2 Co-Infection: From Population Study Evidence to In Vitro Studies. Life 2022, 12, 2089. [Google Scholar] [CrossRef] [PubMed]
- Jamal, Q.M.S. Antiviral Potential of Plants against COVID-19 during Outbreaks-An Update. Int. J. Mol. Sci. 2022, 23, 13564. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.S.; Ho, J.; Wills, S.; Kawall, A.; Sharma, A.; Chavada, K.; Ebert, M.C.; Evoli, S.; Singh, A.; Rayalam, S. Aloin isoforms (A and B) selectively inhibits proteolytic and deubiquitinating activity of papain like protease (PLpro) of SARS-CoV-2 In Vitro. Sci. Rep. 2022, 12, 2145. [Google Scholar] [CrossRef] [PubMed]
- Singh, E.; Jha, R.K.; Khan, R.J.; Kumar, A.; Jain, M.; Muthukumaran, J.; Singh, A.K. A computational essential dynamics approach to investigate structural influences of ligand binding on Papain like protease from SARS-CoV-2. Comput. Biol. Chem. 2022, 99, 107721. [Google Scholar] [CrossRef] [PubMed]
- Hasan, A.H.; Hussen, N.H.; Shakya, S.; Jamalis, J.; Pratama, M.R.F.; Chander, S.; Kharkwal, H.; Murugesan, S. In silico discovery of multi-targeting inhibitors for the COVID-19 treatment by molecular docking, molecular dynamics simulation studies, and ADMET predictions. Struct. Chem. 2022, 33, 1645–1665. [Google Scholar] [CrossRef]
- Narayanan, A.; Narwal, M.; Majowicz, S.A.; Varricchio, C.; Toner, S.A.; Ballatore, C.; Brancale, A.; Murakami, K.S.; Jose, J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022, 5, 169. [Google Scholar] [CrossRef] [PubMed]
- Shaheer, M.; Singh, R.; Sobhia, M.E. Protein degradation: A novel computational approach to design protein degrader probes for main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2022, 40, 10905–10917. [Google Scholar] [CrossRef] [PubMed]
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef]
- Rut, W.; Lv, Z.; Zmudzinski, M.; Patchett, S.; Nayak, D.; Snipas, S.J.; El Oualid, F.; Huang, T.T.; Bekes, M.; Drag, M. Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Sci. Adv. 2020, 6, eabd4596. [Google Scholar] [CrossRef]
- Waman, V.P.; Sen, N.; Varadi, M.; Daina, A.; Wodak, S.J.; Zoete, V.; Velankar, S.; Orengo, C. The impact of structural bioinformatics tools and resources on SARS-CoV-2 research and therapeutic strategies. Brief. Bioinform. 2021, 22, 742–768. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Cui, H.; Gao, Z.; Liu, M.; Lu, S.; Mkandawire, W.; Narykov, O.; Sun, M.; Korkin, D. Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses 2020, 12, 360. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Feig, M. Modeling of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins by machine learning and physics-based refinement. BioRxiv 2020. [Google Scholar] [CrossRef]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef]
- Mondal, S.K.; Mukhoty, S.; Kundu, H.; Ghosh, S.; Sen, M.K.; Das, S.; Brogi, S. In silico analysis of RNA-dependent RNA polymerase of the SARS-CoV-2 and therapeutic potential of existing antiviral drugs. Comput. Biol. Med. 2021, 135, 104591. [Google Scholar] [CrossRef]
- Mushebenge, A.G.-A.; Ugbaja, S.C.; Mbatha, N.A.; B. Khan, R.; Kumalo, H.M. Assessing the Potential Contribution of In Silico Studies in Discovering Drug Candidates That Interact with Various SARS-CoV-2 Receptors. Int. J. Mol. Sci. 2023, 24, 15518. [Google Scholar] [CrossRef]
- Mushebenge, A.G.-A.; Ugbaja, S.C.; Mbatha, N.A.; Khan, R.B.; Kumalo, H.M. A Comprehensive Analysis of Structural and Functional Changes Induced by SARS-CoV-2 Spike Protein Mutations. COVID 2023, 3, 1454–1472. [Google Scholar] [CrossRef]
- Essalmani, R.; Jain, J.; Susan-Resiga, D.; Andréo, U.; Evagelidis, A.; Derbali, R.M.; Huynh, D.N.; Dallaire, F.; Laporte, M.; Delpal, A. Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity. J. Virol. 2022, 96, e00128-22. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudvand, S.; Shokri, S. Interactions between SARS coronavirus 2 papain-like protease and immune system: A potential drug target for the treatment of COVID-19. Scand. J. Immunol. 2021, 94, e13044. [Google Scholar] [CrossRef] [PubMed]
- Maiti, B.K. Can Papain-like Protease Inhibitors Halt SARS-CoV-2 Replication? ACS Pharmacol. Transl. Sci. 2020, 3, 1017–1019. [Google Scholar] [CrossRef]
- Lanz, J.; Biniaz-Harris, N.; Kuvaldina, M.; Jain, S.; Lewis, K.; Fallon, B.A. Disulfiram: Mechanisms, Applications, and Challenges. Antibiotics 2023, 12, 524. [Google Scholar] [CrossRef] [PubMed]
- Kakavandi, S.; Zare, I.; VaezJalali, M.; Dadashi, M.; Azarian, M.; Akbari, A.; Ramezani Farani, M.; Zalpoor, H.; Hajikhani, B. Structural and non-structural proteins in SARS-CoV-2: Potential aspects to COVID-19 treatment or prevention of progression of related diseases. Cell Commun. Signal. 2023, 21, 110. [Google Scholar] [CrossRef]
- Barretto, N.; Jukneliene, D.; Ratia, K.; Chen, Z.; Mesecar, A.D.; Baker, S.C. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 2005, 79, 15189–15198. [Google Scholar] [CrossRef] [PubMed]
- Kandwal, S.; Fayne, D. Genetic conservation across SARS-CoV-2 non-structural proteins–Insights into possible targets for treatment of future viral outbreaks. Virology 2023, 581, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Tam, D.; Lorenzo-Leal, A.C.; Hernández, L.R.; Bach, H. Targeting SARS-coV-2 non-structural proteins. Int. J. Mol. Sci. 2023, 24, 13002. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Wang, M.; Liu, J.; Ma, P.; Pang, S.; Liu, W.; Liu, A. Diagnostics and analysis of SARS-CoV-2: Current status, recent advances, challenges and perspectives. Chem. Sci. 2023, 14, 6149–6206. [Google Scholar] [CrossRef] [PubMed]
- Karges, J.; Cohen, S.M. Rhenium (V) complexes as cysteine-targeting coordinate covalent warheads. J. Med. Chem. 2023, 66, 3088–3105. [Google Scholar] [CrossRef] [PubMed]
- Van Vliet, V.J.; De Silva, A.; Mark, B.L.; Kikkert, M. Viral deubiquitinating proteases and the promising strategies of their inhibition. Virus Res. 2024, 344, 199368. [Google Scholar] [CrossRef]
- Zmudzinski, M.; Rut, W.; Olech, K.; Granda, J.; Giurg, M.; Burda-Grabowska, M.; Kaleta, R.; Zgarbova, M.; Kasprzyk, R.; Zhang, L. Ebselen derivatives inhibit SARS-CoV-2 replication by inhibition of its essential proteins: PLpro and Mpro proteases, and nsp14 guanine N7-methyltransferase. Sci. Rep. 2023, 13, 9161. [Google Scholar] [CrossRef] [PubMed]
- Naidu, A.S.; Wang, C.-K.; Rao, P.; Mancini, F.; Clemens, R.A.; Wirakartakusumah, A.; Chiu, H.-F.; Yen, C.-H.; Porretta, S.; Mathai, I. Precision nutrition to reset virus-induced human metabolic reprogramming and dysregulation (HMRD) in long-COVID. NPJ Sci. Food 2024, 8, 19. [Google Scholar] [CrossRef]
- Darvishi, M.; Hajilou, F. Review on Virology of Coronaviridae. Clinic 2024, 1, 520–2644. [Google Scholar]
- Van Huizen, M.; Bloeme-ter Horst, J.R.; de Gruyter, H.L.; Geurink, P.P.; van der Heden van Noort, G.J.; Knaap, R.C.; Nelemans, T.; Ogando, N.S.; Leijs, A.A.; Urakova, N. Deubiquitinating activity of SARS-CoV-2 papain-like protease does not influence virus replication or innate immune responses in vivo. PLoS Pathog. 2024, 20, e1012100. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wu, G. Potential 3-chymotrypsin-like cysteine protease cleavage sites in the coronavirus polyproteins pp1a and pp1ab and their possible relevance to COVID-19 vaccine and drug development. FASEB J. 2021, 35, e21573. [Google Scholar] [CrossRef] [PubMed]
- Ziebuhr, J.; Schelle, B.; Karl, N.; Minskaia, E.; Bayer, S.; Siddell, S.G.; Gorbalenya, A.E.; Thiel, V. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 2007, 81, 3922–3932. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choudhary, S.; Nehul, S.; Singh, A.; Panda, P.K.; Kumar, P.; Sharma, G.K.; Tomar, S. Unraveling antiviral efficacy of multifunctional immunomodulatory triterpenoids against SARS-COV-2 targeting main protease and papain-like protease. IUBMB Life 2024, 76, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Dai, R.; Su, R. Computer-aided drug design for the pain-like protease (PLpro) inhibitors against SARS-CoV-2. Biomed. Pharmacother. 2023, 159, 114247. [Google Scholar] [CrossRef] [PubMed]
- Omage, F.B.; Madabeni, A.; Tucci, A.R.; Nogara, P.A.; Bortoli, M.; Rosa, A.D.S.; Neuza Dos Santos Ferreira, V.; Teixeira Rocha, J.B.; Miranda, M.D.; Orian, L. Diphenyl diselenide and SARS-CoV-2: In silico exploration of the mechanisms of inhibition of main protease (Mpro) and papain-like protease (PLpro). J. Chem. Inf. Model. 2023, 63, 2226–2239. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Zhang, X.; Ansari, A.; Jadhav, P.; Tan, H.; Li, K.; Chopra, A.; Ford, A.; Chi, X.; Ruiz, F.X. Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model. Science 2024, 383, 1434–1440. [Google Scholar] [CrossRef] [PubMed]
- Alhadhrami, A. Nano-Crystallites of Ruthenium (III) Violurate Complex: Synthesis, Characterization, PXRD and DFT Structural Analysis. DNA/HSA-Binding, Antiviral Activity Against COVID-19 and Molecular Docking Study. New J. Chem. 2024, 48, 9718–9737. [Google Scholar] [CrossRef]
- Arduino, I.; Francese, R.; Civra, A.; Feyles, E.; Argenziano, M.; Volante, M.; Cavalli, R.; Mougharbel, A.M.; Kortz, U.; Donalisio, M. Polyoxometalate exerts broad-spectrum activity against human respiratory viruses hampering viral entry. Antivir. Res. 2024, 226, 105897. [Google Scholar] [CrossRef]
- Mehrotra, R.; Shukla, S.N.; Gaur, P. Metallo-antiviral aspirants: Answer to the upcoming virus outbreak. Eur. J. Med. Chem. Rep. 2023, 8, 100104. [Google Scholar] [CrossRef]
- Gil-Moles, M.; O’Beirne, C.; Esarev, I.V.; Lippmann, P.; Tacke, M.; Cinatl, J.; Bojkova, D.; Ott, I. Silver N-heterocyclic carbene complexes are potent uncompetitive inhibitors of the papain-like protease with antiviral activity against SARS-CoV-2. RSC Med. Chem. 2023, 14, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Balsera-Manzanero, M.; Soengas, R.G.; Carretero-Ledesma, M.; Ratia, C.; Iglesias, M.J.; Pachón, J.; López-Ortiz, F.; Cordero, E.; Soto, S.M.; Sánchez-Céspedes, J. Heteroleptic (S^ C)-cyclometallated gold (III) complexes as novel antiviral agents. Heliyon 2024, 10, e27601. [Google Scholar] [CrossRef] [PubMed]
- Mufassirin, M.M.; Newton, M.H.; Sattar, A. Artificial intelligence for template-free protein structure prediction: A comprehensive review. Artif. Intell. Rev. 2023, 56, 7665–7732. [Google Scholar] [CrossRef]
- Dhingra, S.; Sowdhamini, R.; Cadet, F.; Offmann, B. A glance into the evolution of template-free protein structure prediction methodologies. Biochimie 2020, 175, 85–92. [Google Scholar] [CrossRef]
- Vincenzi, M.; Mercurio, F.A.; Leone, M. Looking for SARS-CoV-2 Therapeutics Through Computational Approaches. Curr. Med. Chem. 2023, 30, 3158–3214. [Google Scholar] [CrossRef]
- Mbunge, E.; Akinnuwesi, B.; Fashoto, S.G.; Metfula, A.S.; Mashwama, P. A critical review of emerging technologies for tackling COVID-19 pandemic. Hum. Behav. Emerg. Technol. 2021, 3, 25–39. [Google Scholar] [CrossRef]
- Selvaraj, C.; Dinesh, D.C.; Panwar, U.; Abhirami, R.; Boura, E.; Singh, S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 4582–4593. [Google Scholar] [CrossRef] [PubMed]
- Frances-Monerris, A.; Hognon, C.; Miclot, T.; Garcia-Iriepa, C.; Iriepa, I.; Terenzi, A.; Grandemange, S.; Barone, G.; Marazzi, M.; Monari, A. Molecular basis of SARS-CoV-2 infection and rational design of potential antiviral agents: Modeling and simulation approaches. J. Proteome Res. 2020, 19, 4291–4315. [Google Scholar] [CrossRef] [PubMed]
- Akram, M.W.; Hasannuzaman, M.; Cuce, E.; Cuce, P.M. Global technological advancement and challenges of glazed window, facade system and vertical greenery-based energy savings in buildings: A comprehensive review. Energy Built Environ. 2023, 4, 206–226. [Google Scholar] [CrossRef]
- Andreini, C.; Arnesano, F.; Rosato, A. The zinc proteome of SARS-CoV-2. Metallomics 2022, 14, mfac047. [Google Scholar] [CrossRef]
- Debnath, S.K.; Debnath, M.; Srivastava, R.; Omri, A. Drugs repurposing for SARS-CoV-2: New insight of COVID-19 druggability. Expert. Rev. Anti Infect. Ther. 2022, 20, 1187–1204. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Brindisi, M.; Shahabi, D.; Chapman, M.E.; Mesecar, A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and COVID-19 therapeutics. ChemMedChem 2020, 15, 907–932. [Google Scholar] [CrossRef] [PubMed]
- Owji, H.; Negahdaripour, M.; Hajighahramani, N. Immunotherapeutic approaches to curtail COVID-19. Int. Immunopharmacol. 2020, 88, 106924. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, P.; Zhang, J. Potential inhibitors targeting papain-like protease of SARS-CoV-2: Two birds with one stone. Front. Chem. 2022, 10, 822785. [Google Scholar] [CrossRef]
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; Kesavan, M.; Kumar, D.; Singh, R.K. Drug repurposing approach to fight COVID-19. Pharmacol. Rep. 2020, 72, 1479–1508. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef]
- Tang, W.; Cao, Z.; Han, M.; Wang, Z.; Chen, J.; Sun, W.; Wu, Y.; Xiao, W.; Liu, S.; Chen, E. Hydroxychloroquine in patients with mainly mild to moderate coronavirus disease 2019: Open label, randomised controlled trial. BMJ 2020, 369, m1849. [Google Scholar] [CrossRef]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef]
- Gil, C.; Ginex, T.; Maestro, I.; Nozal, V.; Barrado-Gil, L.; Cuesta-Geijo, M.Á.; Urquiza, J.; Ramírez, D.; Alonso, C.; Campillo, N.E. COVID-19: Drug targets and potential treatments. J. Med. Chem. 2020, 63, 12359–12386. [Google Scholar] [CrossRef]
- Martinotti, C.; Ruiz-Perez, L.; Deplazes, E.; Mancera, R.L. Molecular dynamics simulation of small molecules interacting with biological membranes. Chemphyschem 2020, 21, 1486–1514. [Google Scholar] [CrossRef]
- Parmar, P.; Rao, P.; Sharma, A.; Shukla, A.; Rawal, R.M.; Saraf, M.; Patel, B.V.; Goswami, D. Meticulous assessment of natural compounds from NPASS database for identifying analogue of GRL0617, the only known inhibitor for SARS-CoV2 papain-like protease (PLpro) using rigorous computational workflow. Mol. Divers. 2022, 26, 389–407. [Google Scholar] [CrossRef]
- Sanachai, K.; Mahalapbutr, P.; Sanghiran Lee, V.; Rungrotmongkol, T.; Hannongbua, S. In silico elucidation of potent inhibitors and rational drug design against SARS-CoV-2 papain-like protease. J. Phys. Chem. B 2021, 125, 13644–13656. [Google Scholar] [CrossRef]
- Naidoo, D.; Kar, P.; Roy, A.; Mutanda, T.; Bwapwa, J.; Sen, A.; Anandraj, A. Structural insight into the binding of cyanovirin-n with the spike glycoprotein, Mpro and PLpro of SARS-CoV-2: Protein–protein interactions, dynamics simulations and free energy calculations. Molecules 2021, 26, 5114. [Google Scholar] [CrossRef]
- Selvaraj, V.; Rathinavel, T.; Ammashi, S.; Nasir Iqbal, M. Polyphenolic phytochemicals exhibit promising SARS-CoV-2 papain like protease (PLpro) inhibition validated through a computational approach. Polycycl. Aromat. Compd. 2023, 43, 5545–5566. [Google Scholar] [CrossRef]
- Muralidharan, N.; Sakthivel, R.; Velmurugan, D.; Gromiha, M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2021, 39, 2673–2678. [Google Scholar] [CrossRef]
- Bera, K.; Reeda, V.J.; Babila, P.; Dinesh, D.C.; Hritz, J.; Karthick, T. An in silico molecular dynamics simulation study on the inhibitors of SARS-CoV-2 proteases (3CLpro and PLpro) to combat COVID-19. Mol. Simul. 2021, 47, 1168–1184. [Google Scholar] [CrossRef]
- Yan, F.; Gao, F. An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2. Comput. Struct. Biotechnol. J. 2021, 19, 4868–4883. [Google Scholar] [CrossRef]
- Xu, L.; Tong, J.; Wu, Y.; Zhao, S.; Lin, B.-L. A computational evaluation of targeted oxidation strategy (TOS) for potential inhibition of SARS-CoV-2 by disulfiram and analogues. Biophys. Chem. 2021, 276, 106610. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Hu, Y.; Townsend, J.A.; Lagarias, P.I.; Marty, M.T.; Kolocouris, A.; Wang, J. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Transl. Sci. 2020, 3, 1265–1277. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, L.; Zhao, X.-E. Co-crystallization and structure determination: An effective direction for anti-SARS-CoV-2 drug discovery. Comput. Struct. Biotechnol. J. 2021, 19, 4684–4701. [Google Scholar] [CrossRef]
- Talevi, A.; Bellera, C.L. Challenges and opportunities with drug repurposing: Finding strategies to find alternative uses of therapeutics. Expert. Opin. Drug Discov. 2020, 15, 397–401. [Google Scholar] [CrossRef]
- Pang, J.; Gao, S.; Sun, Z.; Yang, G. Discovery of small molecule PLpro inhibitor against COVID-19 using structure-based virtual screening, molecular dynamics simulation, and molecular mechanics/Generalized Born surface area (MM/GBSA) calculation. Struct. Chem. 2021, 32, 879–886. [Google Scholar] [CrossRef]
- Hu, X.; Zeng, Z.; Zhang, J.; Wu, D.; Li, H.; Geng, F. Molecular dynamics simulation of the interaction of food proteins with small molecules. Food Chem. 2023, 405, 134824. [Google Scholar] [CrossRef]
- Wouters, O.J.; McKee, M.; Luyten, J. Estimated research and development investment needed to bring a new medicine to market, 2009-2018. JAMA 2020, 323, 844–853. [Google Scholar] [CrossRef]
- Zrieq, R.; Ahmad, I.; Snoussi, M.; Noumi, E.; Iriti, M.; Algahtani, F.D.; Patel, H.; Saeed, M.; Tasleem, M.; Sulaiman, S. Tomatidine and patchouli alcohol as inhibitors of SARS-CoV-2 enzymes (3CLpro, PLpro and NSP15) by molecular docking and molecular dynamics simulations. Int. J. Mol. Sci. 2021, 22, 10693. [Google Scholar] [CrossRef]
- Thangavel, N.; Albratty, M. Benchmarked molecular docking integrated molecular dynamics stability analysis for prediction of SARS-CoV-2 papain-like protease inhibition by olive secoiridoids. J. King Saud. Univ. Sci. 2023, 35, 102402. [Google Scholar] [CrossRef]
- Patel, D.; Athar, M.; Jha, P.C. Computational investigation of binding of chloroquinone and hydroxychloroquinone against PLPro of SARS-CoV-2. J. Biomol. Struct. Dyn. 2022, 40, 3071–3081. [Google Scholar] [CrossRef]
- Kandeel, M.; Abdelrahman, A.; Oh-Hashi, K.; Ibrahim, A.; Venugopala, K.; Morsy, M.; Ibrahim, M. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J. Biomol. Struct. Dyn. 2020, 39, 5129–5136. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Joshi, M.; Degani, M.S. Tackling SARS-CoV-2: Proposed targets and repurposed drugs. Future Med. Chem. 2020, 12, 1579–1601. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Moccia, S.; Spagnuolo, C.; Tedesco, I.; Russo, G.L. Roles of flavonoids against coronavirus infection. Chem. Biol. Interact. 2020, 328, 109211. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-W.; Xie, Y.; Tang, L.-S.; Pu, D.; Zhu, Y.-J.; Liu, J.-Y.; Ma, X.-L. Therapeutic targets and interventional strategies in COVID-19: Mechanisms and clinical studies. Signal Transduct. Target. Ther. 2021, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.K.; Faheem; Sekhar, K.; Ojha, R.; Prajapati, V.K.; Pai, A.; Murugesan, S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2022, 40, 1363–1386. [Google Scholar] [CrossRef] [PubMed]
- Jupudi, S.; Rajagopal, K.; Murugesan, S.; Kumar, B.K.; Raman, K.; Byran, G.; Chennaiah, J.; pillai Muthiah, V.; Sankaran, S. Identification of Papain-Like Protease inhibitors of SARS CoV-2 through HTVS, Molecular docking, MMGBSA and Molecular dynamics approach. S. Afr. J. Bot. 2022, 151, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Vardhan, S.; Sahoo, S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput. Biol. Med. 2020, 124, 103936. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luan, J.; Zhang, L. Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors. Biochem. Biophys. Res. Commun. 2021, 538, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Hu, Y.; Jadhav, P.; Tan, B.; Wang, J. Progress and challenges in targeting the SARS-CoV-2 papain-like protease. J. Med. Chem. 2022, 65, 7561–7580. [Google Scholar] [CrossRef] [PubMed]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef]
- Su, C.-M.; Du, Y.; Rowland, R.R.; Wang, Q.; Yoo, D. Reprogramming viral immune evasion for a rational design of next-generation vaccines for RNA viruses. Front. Immunol. 2023, 14, 1172000. [Google Scholar] [CrossRef]
- Moustaqil, M.; Ollivier, E.; Chiu, H.-P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.; Freiberg, A.N.; Jacques, D. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef]
- Wong, N.A.; Saier, M.H., Jr. The SARS-coronavirus infection cycle: A survey of viral membrane proteins, their functional interactions and pathogenesis. Int. J. Mol. Sci. 2021, 22, 1308. [Google Scholar] [CrossRef] [PubMed]
- Yapasert, R.; Khaw-On, P.; Banjerdpongchai, R. Coronavirus infection-associated cell death signaling and potential therapeutic targets. Molecules 2021, 26, 7459. [Google Scholar] [CrossRef] [PubMed]
- Cavasotto, C.N.; Lamas, M.S.; Maggini, J. Functional and druggability analysis of the SARS-CoV-2 proteome. Eur. J. Pharmacol. 2021, 890, 173705. [Google Scholar] [CrossRef]
- Durairajan, S.S.K.; Singh, A.K.; Saravanan, U.B.; Namachivayam, M.; Radhakrishnan, M.; Huang, J.D.; Dhodapkar, R.; Zhang, H. Gastrointestinal Manifestations of SARS-CoV-2: Transmission, Pathogenesis, Immunomodulation, Microflora Dysbiosis, and Clinical Implications. Viruses 2023, 15, 1231. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Mei, H.; Chen, Y.; Griffin, J.D.; Liu, Q.; Weisberg, E.; Yang, J. Repurposing clinically available drugs and therapies for pathogenic targets to combat SARS-CoV-2. MedComm 2023, 4, e254. [Google Scholar] [CrossRef] [PubMed]
- Clemente, V.; D’arcy, P.; Bazzaro, M. Deubiquitinating enzymes in coronaviruses and possible therapeutic opportunities for COVID-19. Int. J. Mol. Sci. 2020, 21, 3492. [Google Scholar] [CrossRef]
- Seyoum Tola, F. The role of ubiquitin-proteasome system in the pathogenesis of severe acute respiratory syndrome coronavirus-2 disease. Int. J. Inflamm. 2023, 2023, 6698069. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, H.; Zhu, J.; Dong, Q.; Wang, J.; Fan, H.; Chen, Y.; Zhang, X.; Han, X.; Li, Q. Ubiquitin-modified proteome of SARS-CoV-2-infected host cells reveals insights into virus–host interaction and pathogenesis. J. Proteome Res. 2021, 20, 2224–2239. [Google Scholar] [CrossRef]
- Chourasia, M.; Koppula, P.R.; Battu, A.; Ouseph, M.M.; Singh, A.K. EGCG, a green tea catechin, as a potential therapeutic agent for symptomatic and asymptomatic SARS-CoV-2 infection. Molecules 2021, 26, 1200. [Google Scholar] [CrossRef]
- Tretter, F.; Wolkenhauer, O.; Meyer-Hermann, M.; Dietrich, J.W.; Green, S.; Marcum, J.; Weckwerth, W. The quest for system-theoretical medicine in the COVID-19 era. Front. Med. 2021, 8, 640974. [Google Scholar] [CrossRef]
- Lin, X.; Li, X.; Lin, X. A review on applications of computational methods in drug screening and design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [PubMed]
- Collin, C.B.; Gebhardt, T.; Golebiewski, M.; Karaderi, T.; Hillemanns, M.; Khan, F.M.; Salehzadeh-Yazdi, A.; Kirschner, M.; Krobitsch, S.; Consortium, E.-S.P. Computational models for clinical applications in personalized medicine—Guidelines and recommendations for data integration and model validation. J. Pers. Med. 2022, 12, 166. [Google Scholar] [CrossRef] [PubMed]
- Muratov, E.N.; Amaro, R.; Andrade, C.H.; Brown, N.; Ekins, S.; Fourches, D.; Isayev, O.; Kozakov, D.; Medina-Franco, J.L.; Merz, K.M. A critical overview of computational approaches employed for COVID-19 drug discovery. Chem. Soc. Rev. 2021, 50, 9121–9151. [Google Scholar] [CrossRef] [PubMed]
- Rahmandad, H.; Xu, R.; Ghaffarzadegan, N. Enhancing long-term forecasting: Learning from COVID-19 models. PLOS Comput. Biol. 2022, 18, e1010100. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Rong, H.; Qi, D.; Valencia-Cabrera, L.; Zhang, G.; Pérez-Jiménez, M.J. A Review of Membrane Computing Models for Complex Ecosystems and a Case Study on a Complex Giant Panda System. Complexity 2020, 2020, 1312824. [Google Scholar] [CrossRef]
- Maharao, N.; Antontsev, V.; Wright, M.; Varshney, J. Entering the era of computationally driven drug development. Drug Metab. Rev. 2020, 52, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Wang, R.; Chen, J.; Cheng, L.; Frishcosy, J.; Huzumi, Y.; Qiu, Y.; Schluckbier, T.; Wei, X.; Wei, G.-W. Methodology-centered review of molecular modeling, simulation, and prediction of SARS-CoV-2. Chem. Rev. 2022, 122, 11287–11368. [Google Scholar] [CrossRef] [PubMed]
- Bullock, J.; Luccioni, A.; Pham, K.H.; Lam, C.S.N.; Luengo-Oroz, M. Mapping the landscape of artificial intelligence applications against COVID-19. J. Artif. Intell. Res. 2020, 69, 807–845. [Google Scholar] [CrossRef]
- Çalışkaner, Z.O. Determination of Binding Potential of HCV Protease Inhibitors Against to SARS-CoV-2 Papain-like Protease wtih Computational Docking. Lett. Drug Des. Discov. 2021, 18, 949–960. [Google Scholar] [CrossRef]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021, 12, 488. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Flight, K.E.; Higham, S.L.; Wang, Z.; Pierce, B.F. Progress of the COVID-19 vaccine effort: Viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021, 21, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Sharun, K.; Tiwari, R.; Dadar, M.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W. COVID-19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccin. Immunother. 2020, 16, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Ali, M.A.; Lee, J.; Farah, M.A.; Al-Anazi, K.M. An updated review of computer-aided drug design and its application to COVID-19. BioMed Res. Int. 2021, 2021, 8853056. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.J.; Jannat, T.; Brishty, S.R.; Roy, U.; Mitra, S.; Rafi, M.O.; Islam, M.R.; Nesa, M.L.; Islam, M.A.; Emran, T.B. Clinical efficacy and safety of antiviral drugs in the extended use against COVID-19: What we know so Far. Biologics 2021, 1, 252–284. [Google Scholar] [CrossRef]
- Hung, I.F.-N.; Lung, K.-C.; Tso, E.Y.-K.; Liu, R.; Chung, T.W.-H.; Chu, M.-Y.; Ng, Y.-Y.; Lo, J.; Chan, J.; Tam, A.R. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Mei, K.; Zhang, J. The Kinetic parameters, Thermodynamic parameters, and Mechanism of PLpro from SARS-CoV and SARS-CoV-2. In Proceedings of the 2023 4th International Symposium on Artificial Intelligence for Medicine Science, Chengdu, China, 27–29 October 2023; pp. 1218–1222. [Google Scholar]
- Singh, U.; Gandhi, H.A.; Nikita; Bhattacharya, J.; Tandon, R.; Tiwari, G.; Tandon, R. Cyanometabolites: Molecules with immense antiviral potential. Arch. Microbiol. 2023, 205, 164. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, C.; Vanet, A.; Francesconi, V.; Tagliazucchi, L.; Tassone, G.; Venturelli, A.; Spyrakis, F.; Mazzorana, M.; Costi, M.P.; Tonelli, M. Antitarget, anti-SARS-CoV-2 leads, drugs, and the drug discovery–genetics alliance perspective. J. Med. Chem. 2023, 66, 3664–3702. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.A.; Ingale, K. In silico approaches for drug repurposing for SARS-CoV-2 Infection. RSC 2022. [Google Scholar] [CrossRef]
- Sharma, P.; Joshi, T.; Mathpal, S.; Tamta, S.; Chandra, S. Computational approaches for drug discovery against COVID-19. In Omics Approaches and Technologies in COVID-19; Elsevier: Amsterdam, The Netherlands, 2023; pp. 321–337. [Google Scholar]
- Jade, D.; Ayyamperumal, S.; Tallapaneni, V.; Nanjan, C.M.J.; Barge, S.; Mohan, S.; Nanjan, M.J. Virtual high throughput screening: Potential inhibitors for SARS-CoV-2 PLPRO and 3CLPRO proteases. Eur. J. Pharmacol. 2021, 901, 174082. [Google Scholar] [CrossRef]
- Khaledi, M.; Sameni, F.; Yahyazade, S.; Radandish, M.; Owlia, P.; Bagheri, N.; Afkhami, H.; Mahjoor, M.; Esmaelpour, Z.; Kohansal, M. COVID-19 and the potential of Janus family kinase (JAK) pathway inhibition: A novel treatment strategy. Front. Med. 2022, 9, 961027. [Google Scholar] [CrossRef]
- Schake, P.; Dishnica, K.; Kaiser, F.; Leberecht, C.; Haupt, V.J.; Schroeder, M. An interaction-based drug discovery screen explains known SARS-CoV-2 inhibitors and predicts new compound scaffolds. Sci. Rep. 2023, 13, 9204. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guan, Y. COVID-19 drug repurposing: A review of computational screening methods, clinical trials, and protein interaction assays. Med. Res. Rev. 2021, 41, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Viceconti, M.; Pappalardo, F.; Rodriguez, B.; Horner, M.; Bischoff, J.; Tshinanu, F.M. In silico trials: Verification, validation and uncertainty quantification of predictive models used in the regulatory evaluation of biomedical products. Methods 2021, 185, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Bhattacharya, M.; Agoramoorthy, G.; Lee, S.-S. The drug repurposing for COVID-19 clinical trials provide very effective therapeutic combinations: Lessons learned from major clinical studies. Front. Pharmacol. 2021, 12, 704205. [Google Scholar] [CrossRef] [PubMed]
- McCradden, M.D.; Anderson, J.A.; A. Stephenson, E.; Drysdale, E.; Erdman, L.; Goldenberg, A.; Zlotnik Shaul, R. A research ethics framework for the clinical translation of healthcare machine learning. Am. J. Bioeth. 2022, 22, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Fu, W.; Jiang, D.; Sun, H.; Wang, J.; Zhang, X.; Weng, G.; Liu, H.; Tao, P.; Hou, T. VAD-MM/GBSA: A Variable Atomic Dielectric MM/GBSA Model for Improved Accuracy in Protein–Ligand Binding Free Energy Calculations. J. Chem. Inf. Model. 2021, 61, 2844–2856. [Google Scholar] [CrossRef]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2021, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Consortium, W.S.T. Repurposed antiviral drugs for COVID-19—Interim WHO solidarity trial results. N. Engl. J. Med. 2021, 384, 497–511. [Google Scholar] [CrossRef]
- Abd El-Aziz, T.M.; Stockand, J.D. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2)-an update on the status. Infect. Genet. Evol. 2020, 83, 104327. [Google Scholar] [CrossRef]
- Hamdy, R.; Fayed, B.; Mostafa, A.; Shama, N.M.A.; Mahmoud, S.H.; Mehta, C.H.; Nayak, Y.; M. Soliman, S.S. Iterated virtual screening-assisted antiviral and enzyme inhibition assays reveal the discovery of novel promising anti-SARS-CoV-2 with dual activity. Int. J. Mol. Sci. 2021, 22, 9057. [Google Scholar] [CrossRef] [PubMed]
- Gershenson, A.; Gosavi, S.; Faccioli, P.; Wintrode, P.L. Successes and challenges in simulating the folding of large proteins. J. Biol. Chem. 2020, 295, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Bhati, A.P.; Hoti, A.; Potterton, A.; Bieniek, M.K.; Coveney, P.V. Long Time Scale Ensemble Methods in Molecular Dynamics: Ligand-Protein Interactions and Allostery in SARS-CoV-2 Targets. J. Chem. Theory Comput. 2023, 19, 3359–3378. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.W.; Gilmer, J.B.; Matsumoto, R.A.; Quach, C.D.; Shamaprasad, P.; Yang, A.H.; Iacovella, C.R.; McCabe, C.; Cummings, P.T. Towards molecular simulations that are transparent, reproducible, usable by others, and extensible (TRUE). Mol. Phys. 2020, 118, e1742938. [Google Scholar] [CrossRef] [PubMed]
- Sengar, A.; Ouldridge, T.E.; Henrich, O.; Rovigatti, L.; Šulc, P. A primer on the oxDNA model of DNA: When to use it, how to simulate it and how to interpret the results. Front. Mol. Biosci. 2021, 8, 693710. [Google Scholar] [CrossRef] [PubMed]
- Elekofehinti, O.O.; Iwaloye, O.; Josiah, S.S.; Lawal, A.O.; Akinjiyan, M.O.; Ariyo, E.O. Molecular docking studies, molecular dynamics and ADME/tox reveal therapeutic potentials of STOCK1N-69160 against papain-like protease of SARS-CoV-2. Mol. Divers. 2021, 25, 1761–1773. [Google Scholar] [CrossRef]
- Citarella, A.; Scala, A.; Piperno, A.; Micale, N. SARS-CoV-2 Mpro: A potential target for peptidomimetics and small-molecule inhibitors. Biomolecules 2021, 11, 607. [Google Scholar] [CrossRef] [PubMed]
- Gentile, D.; Patamia, V.; Scala, A.; Sciortino, M.T.; Piperno, A.; Rescifina, A. Putative inhibitors of SARS-CoV-2 main protease from a library of marine natural products: A virtual screening and molecular modeling study. Mar. Drugs 2020, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Minetti, C.A.; Remeta, D.P. Forces driving a magic bullet to its target: Revisiting the role of thermodynamics in drug design, development, and optimization. Life 2022, 12, 1438. [Google Scholar] [CrossRef]
- Badavath, V.N.; Kumar, A.; Samanta, P.K.; Maji, S.; Das, A.; Blum, G.; Jha, A.; Sen, A. Determination of potential inhibitors based on isatin derivatives against SARS-CoV-2 main protease (mpro): A molecular docking, molecular dynamics and structure-activity relationship studies. J. Biomol. Struct. Dyn. 2022, 40, 3110–3128. [Google Scholar] [CrossRef] [PubMed]
- Boike, L.; Henning, N.J.; Nomura, D.K. Advances in covalent drug discovery. Nat. Rev. Drug Discov. 2022, 21, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Al-Karmalawy, A.A.; El-Gamil, D.S.; El-Shesheny, R.; Sharaky, M.; Alnajjar, R.; Kutkat, O.; Moatasim, Y.; Elagawany, M.; Al-Rashood, S.T.; Binjubair, F.A. Design and statistical optimisation of emulsomal nanoparticles for improved anti-SARS-CoV-2 activity of N-(5-nitrothiazol-2-yl)-carboxamido candidates: In vitro and in silico studies. J. Enzym. Inhib. Med. Chem. 2023, 38, 2202357. [Google Scholar] [CrossRef]
- De Souza Neto, L.R.; Moreira-Filho, J.T.; Neves, B.J.; Maidana, R.L.B.R.; Guimarães, A.C.R.; Furnham, N.; Andrade, C.H.; Silva, F.P., Jr. In silico strategies to support fragment-to-lead optimization in drug discovery. Front. Chem. 2020, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Trisciuzzi, D.; Villoutreix, B.O.; Siragusa, L.; Baroni, M.; Cruciani, G.; Nicolotti, O. Targeting protein-protein interactions with low molecular weight and short peptide modulators: Insights on disease pathways and starting points for drug discovery. Expert. Opin. Drug Discov. 2023, 18, 737–752. [Google Scholar] [CrossRef]
- Kumar, H.M.S.; Herrmann, L.; Tsogoeva, S.B. Structural hybridization as a facile approach to new drug candidates. Bioorg. Med. Chem. Lett. 2020, 30, 127514. [Google Scholar]
- Frenkel, D.; Smit, B. Understanding Molecular Simulation: From Algorithms to Applications; Elsevier: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Poongavanam, V.; Ramaswamy, V. Computational Drug Discovery: Methods and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2024. [Google Scholar]
- Mouvet, F.; Villard, J.; Bolnykh, V.; Rothlisberger, U. Recent advances in first-principles based molecular dynamics. Acc. Chem. Res. 2022, 55, 221–230. [Google Scholar] [CrossRef]
- Li, H.; Komori, A.; Li, M.; Chen, X.; Yang, A.W.H.; Sun, X.; Liu, Y.; Hung, A.; Zhao, X.; Zhou, L. Multi-ligand molecular docking, simulation, free energy calculations and wavelet analysis of the synergistic effects between natural compounds baicalein and cubebin for the inhibition of the main protease of SARS-CoV-2. J. Mol. Liq. 2023, 374, 121253. [Google Scholar] [CrossRef]
- Yuda, G.P.W.C.; Hanif, N.; Hermawan, A. Computational Screening Using a Combination of Ligand-Based Machine Learning and Molecular Docking Methods for the Repurposing of Antivirals Targeting the SARS-CoV-2 Main Protease. DARU J. Pharm. Sci. 2023, 32, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.; Ullah, S.; Halim, S.A.; Rehman, N.U.; Ali, A.; Jan, A.; Muhsinah, A.B.; Khan, A.; Al-Harrasi, A. Targeting papain-like protease by natural products as novel therapeutic potential SARS-CoV-2. Int. J. Biol. Macromol. 2024, 258, 128812. [Google Scholar] [CrossRef]
- Prieto-Martínez, F.D.; Galván-Ciprés, Y. Free Energy Estimation for Drug Discovery: Background and Perspectives. Appl. Comput. Aided Drug Des. Models Methods 2023, 310–345. Available online: https://books.google.co.za/books?hl=en&lr=&id=t7DpEAAAQBAJ&oi=fnd&pg=PA310&dq=Prieto-Mart%C3%ADnez,+F.D.%3B+Galv%C3%A1n-Cipr%C3%A9s,+Y.+Free+Energy+Estimation+for+Drug+Discovery:+Background+and+Perspectives.+Appl.+Comput.+Aided+Drug+Des.+Models+Methods+2 (accessed on 25 May 2024).
- Chen, L.; Wu, Y.; Wu, C.; Silveira, A.; Sherman, W.; Xu, H.; Gallicchio, E. Performance and Analysis of the Alchemical Transfer Method for Binding-Free-Energy Predictions of Diverse Ligands. J. Chem. Inf. Model. 2023, 64, 250–264. [Google Scholar] [CrossRef]
- Parveen, S.; Shehzadi, S.; Shafiq, N.; Rashid, M.; Naz, S.; Mehmood, T.; Riaz, R.; Almaary, K.S.; Nafidi, H.-A.; Bourhia, M. A discovery of potent kaempferol derivatives as multi-target medicines against diabetes as well as bacterial infections: An in silico approach. J. Biomol. Struct. Dyn. 2024, 1–23. [Google Scholar] [CrossRef]
- Kurisaki, I.; Suzuki, M. Simulation toolkits at the molecular scale for trans-scale thermal signaling. Comput. Struct. Biotechnol. J. 2023, 21, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Schlick, T.; Portillo-Ledesma, S. Biomolecular modeling thrives in the age of technology. Nat. Comput. Sci. 2021, 1, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Nochebuena, J.; Naseem-Khan, S.; Cisneros, G.A. Development and Application of QM/MM Methods with Advanced Polarizable Potentials. arXiv 2020, arXiv:2010.14723. [Google Scholar] [CrossRef]
- Brogi, S.; Ramalho, T.C.; Kuca, K.; Medina-Franco, J.L.; Valko, M. In silico methods for drug design and discovery. Front. Media SA 2020, 8, 612. [Google Scholar]
- Salehi, B.; Venditti, A.; Sharifi-Rad, M.; Kręgiel, D.; Sharifi-Rad, J.; Durazzo, A.; Lucarini, M.; Santini, A.; Souto, E.B.; Novellino, E. The therapeutic potential of apigenin. Int. J. Mol. Sci. 2019, 20, 1305. [Google Scholar] [CrossRef]
- Pantaleão, S.Q.; Fernandes, P.O.; Gonçalves, J.E.; Maltarollo, V.G.; Honorio, K.M. Recent advances in the prediction of pharmacokinetics properties in drug design studies: A review. ChemMedChem 2022, 17, e202100542. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhang, X.; Shu, Y.; Guo, M.; Zhang, H.; Tao, W. Insights from nanotechnology in COVID-19 treatment. Nano today 2021, 36, 101019. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Hawash, H.; Elhoseny, M.; Chakrabortty, R.K.; Ryan, M. DeepH-DTA: Deep learning for predicting drug-target interactions: A case study of COVID-19 drug repurposing. Ieee Access 2020, 8, 170433–170451. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, J.; Wang, J.; Cao, Y.; Kato, N. When machine learning meets privacy in 6G: A survey. IEEE Commun. Surv. Tutor. 2020, 22, 2694–2724. [Google Scholar] [CrossRef]
- Sabe, V.T.; Ntombela, T.; Jhamba, L.A.; Maguire, G.E.; Govender, T.; Naicker, T.; Kruger, H.G. Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. Eur. J. Med. Chem. 2021, 224, 113705. [Google Scholar] [CrossRef]
- Zhan, P.; Pannecouque, C.; De Clercq, E.; Liu, X. Anti-HIV drug discovery and development: Current innovations and future trends: Miniperspective. J. Med. Chem. 2016, 59, 2849–2878. [Google Scholar] [CrossRef] [PubMed]
- Rakib, A.; Nain, Z.; Sami, S.A.; Mahmud, S.; Islam, A.; Ahmed, S.; Siddiqui, A.B.F.; Babu, S.O.F.; Hossain, P.; Shahriar, A. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: An in silico investigation. Brief. Bioinform. 2021, 22, 1476–1498. [Google Scholar] [CrossRef] [PubMed]
- Carracedo-Reboredo, P.; Liñares-Blanco, J.; Rodríguez-Fernández, N.; Cedrón, F.; Novoa, F.J.; Carballal, A.; Maojo, V.; Pazos, A.; Fernandez-Lozano, C. A review on machine learning approaches and trends in drug discovery. Comput. Struct. Biotechnol. J. 2021, 19, 4538–4558. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.; Cornell, W.; Luan, B. In silico Exploration of Inhibitors for SARS-CoV-2’s Papain-Like Protease. Front. Chem. 2020, 8, 624163. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Chakraborty, A.; Biswas, A.; Chowdhuri, S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2021, 39, 4362–4374. [Google Scholar] [CrossRef]
- Wang, J. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020, 60, 3277–3286. [Google Scholar] [CrossRef] [PubMed]
- Yosri, N.; Abd El-Wahed, A.A.; Ghonaim, R.; Khattab, O.M.; Sabry, A.; Ibrahim, M.A.; Moustafa, M.F.; Guo, Z.; Zou, X.; Algethami, A.F. Anti-viral and immunomodulatory properties of propolis: Chemical diversity, pharmacological properties, preclinical and clinical applications, and in silico potential against SARS-CoV-2. Foods 2021, 10, 1776. [Google Scholar] [CrossRef] [PubMed]
- Chekroud, A.M.; Bondar, J.; Delgadillo, J.; Doherty, G.; Wasil, A.; Fokkema, M.; Cohen, Z.; Belgrave, D.; DeRubeis, R.; Iniesta, R. The promise of machine learning in predicting treatment outcomes in psychiatry. World Psychiatry 2021, 20, 154–170. [Google Scholar] [CrossRef] [PubMed]
- Pearce, J.M. A review of open source ventilators for COVID-19 and future pandemics. F1000Research 2020, 9, 218. [Google Scholar] [CrossRef]
- Di Pietro, G.; Biagi, F.; Costa, P.; Karpiński, Z.; Mazza, J. The Likely Impact of COVID-19 on Education: Reflections Based on the Existing Literature and Recent International Datasets; Publications Office of the European Union Luxembourg: Luxembourg, 2020; Volume 30275. [Google Scholar]
- Sohraby, F.; Aryapour, H. Unraveling the unbinding pathways of SARS-CoV-2 Papain-like proteinase known inhibitors by Supervised Molecular Dynamics simulation. PLoS ONE 2021, 16, e0251910. [Google Scholar] [CrossRef]
- Alanine, A.; Nettekoven, M.; Roberts, E.; Thomas, A.W. Lead generation-enhancing the success of drug discovery by investing in the hit to lead process. Comb. Chem. High. Throughput Screen. 2003, 6, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Aftab, S.O.; Ghouri, M.Z.; Masood, M.U.; Haider, Z.; Khan, Z.; Ahmad, A.; Munawar, N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020, 18, 275. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Chen, J.; Zhang, H. An investigation of testing capacity for evaluating and modeling the spread of coronavirus disease. Inf. Sci. 2021, 561, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Zurell, D.; Franklin, J.; König, C.; Bouchet, P.J.; Dormann, C.F.; Elith, J.; Fandos, G.; Feng, X.; Guillera-Arroita, G.; Guisan, A. A standard protocol for reporting species distribution models. Ecography 2020, 43, 1261–1277. [Google Scholar] [CrossRef]
- Shen, Z.; Ratia, K.; Cooper, L.; Kong, D.; Lee, H.; Kwon, Y.; Li, Y.; Alqarni, S.; Huang, F.; Dubrovskyi, O. Design of SARS-CoV-2 PLpro inhibitors for COVID-19 antiviral therapy leveraging binding cooperativity. J. Med. Chem. 2021, 65, 2940–2955. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).