New Method of Isothermal, Hairpin Assisted, Primer Independent Amplification of DNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. HAIR Reaction Mix
2.3. Amplification of BRAF Gene Cloned in pUC19
2.4. Gel Electrophoresis
3. Results
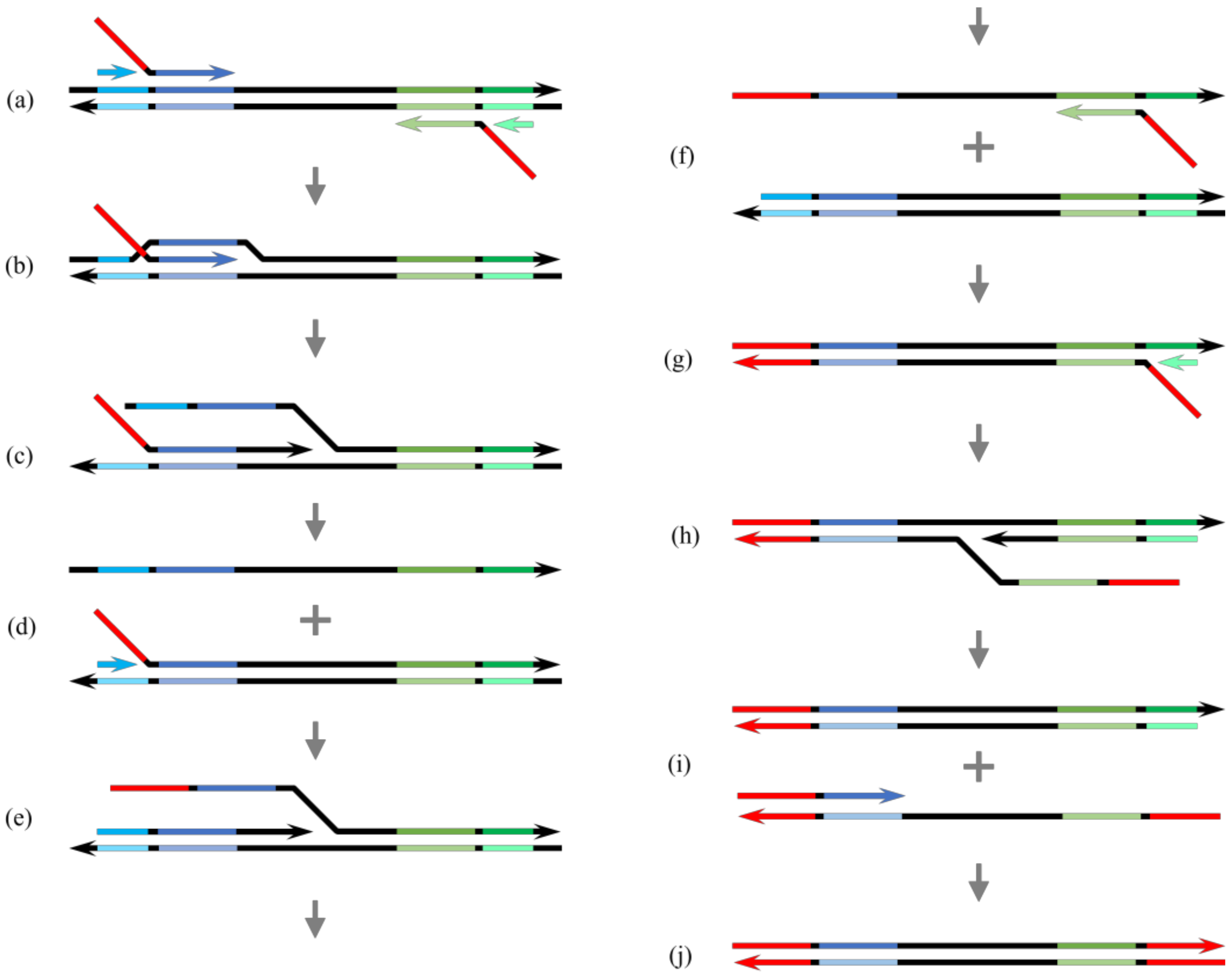
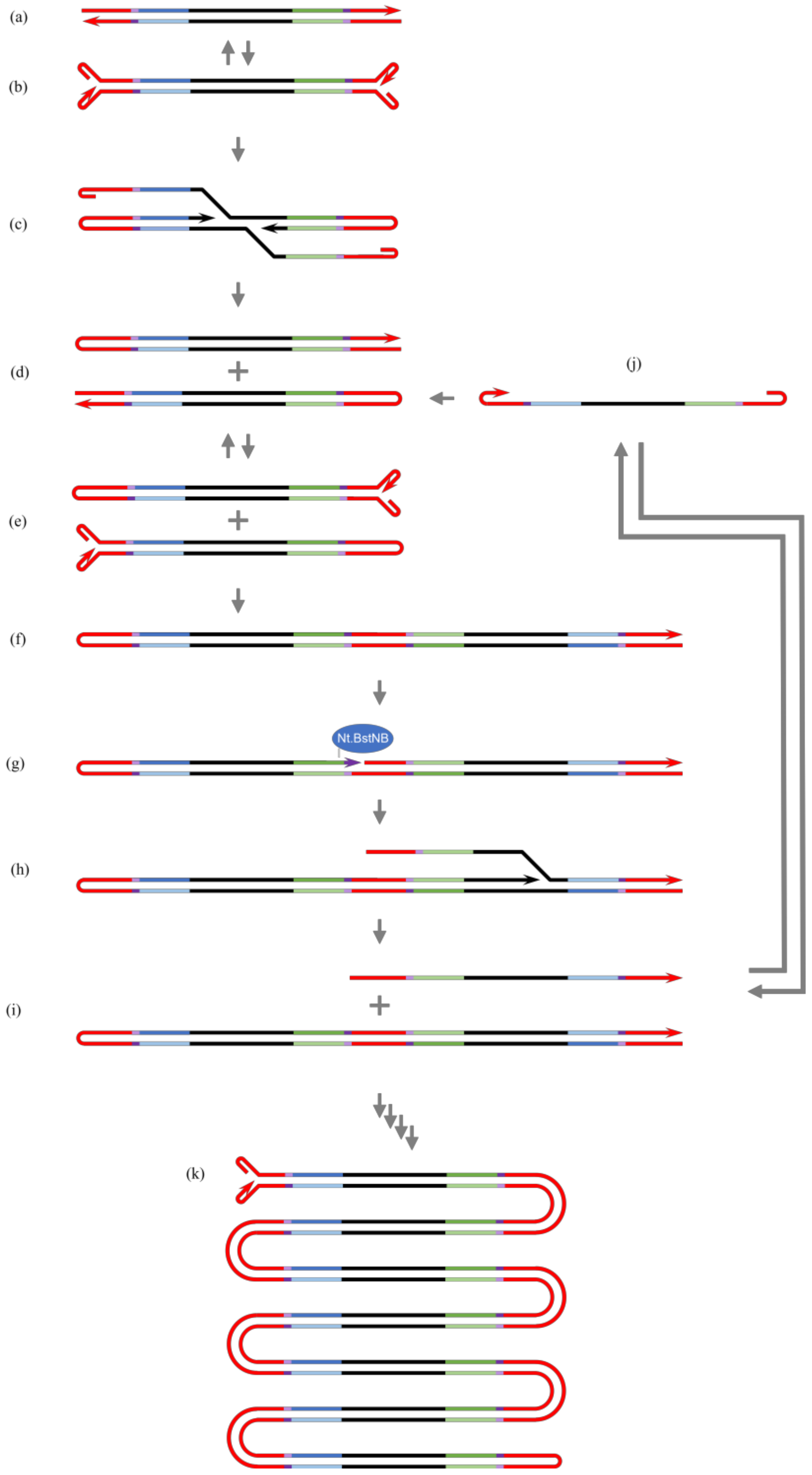
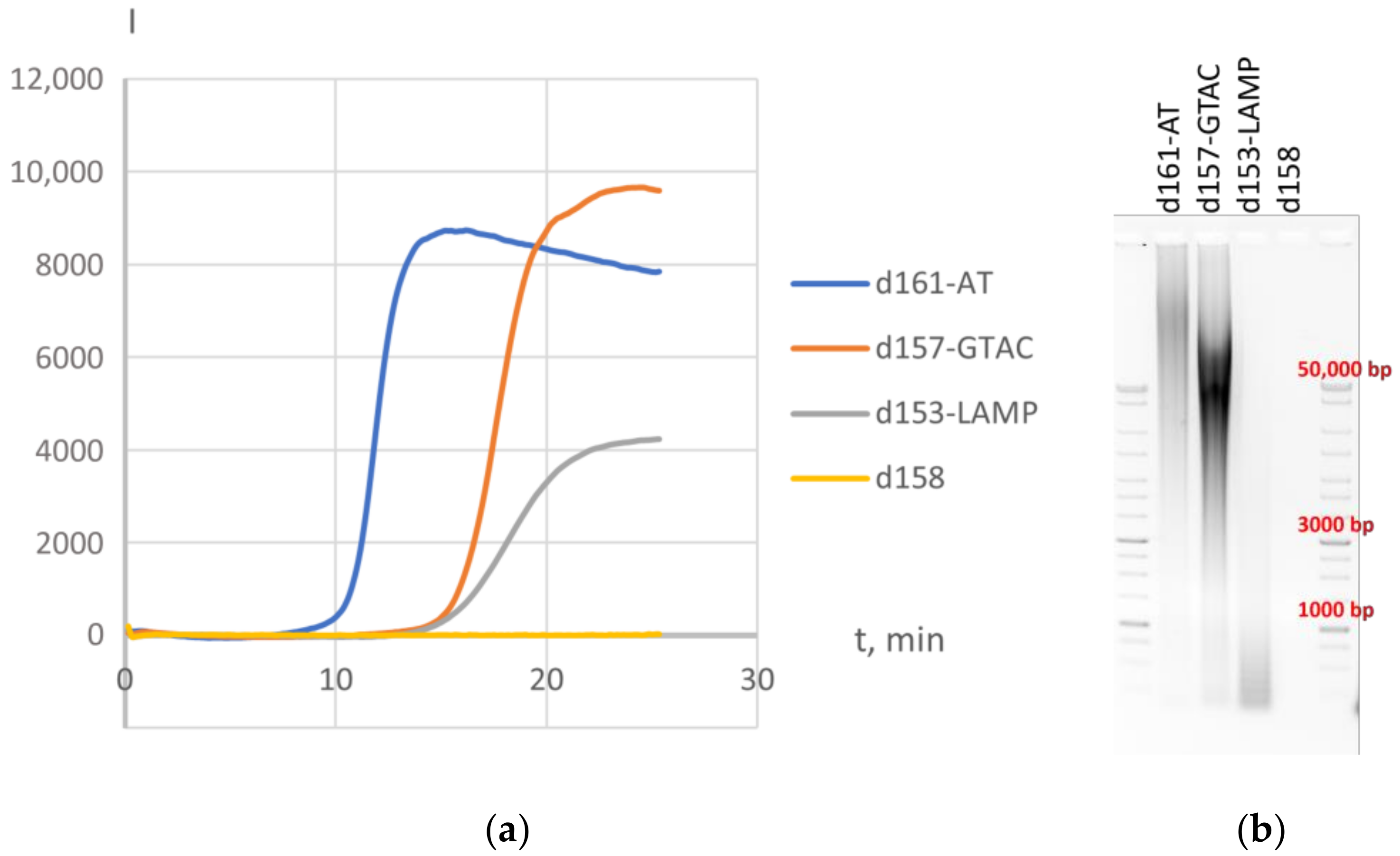
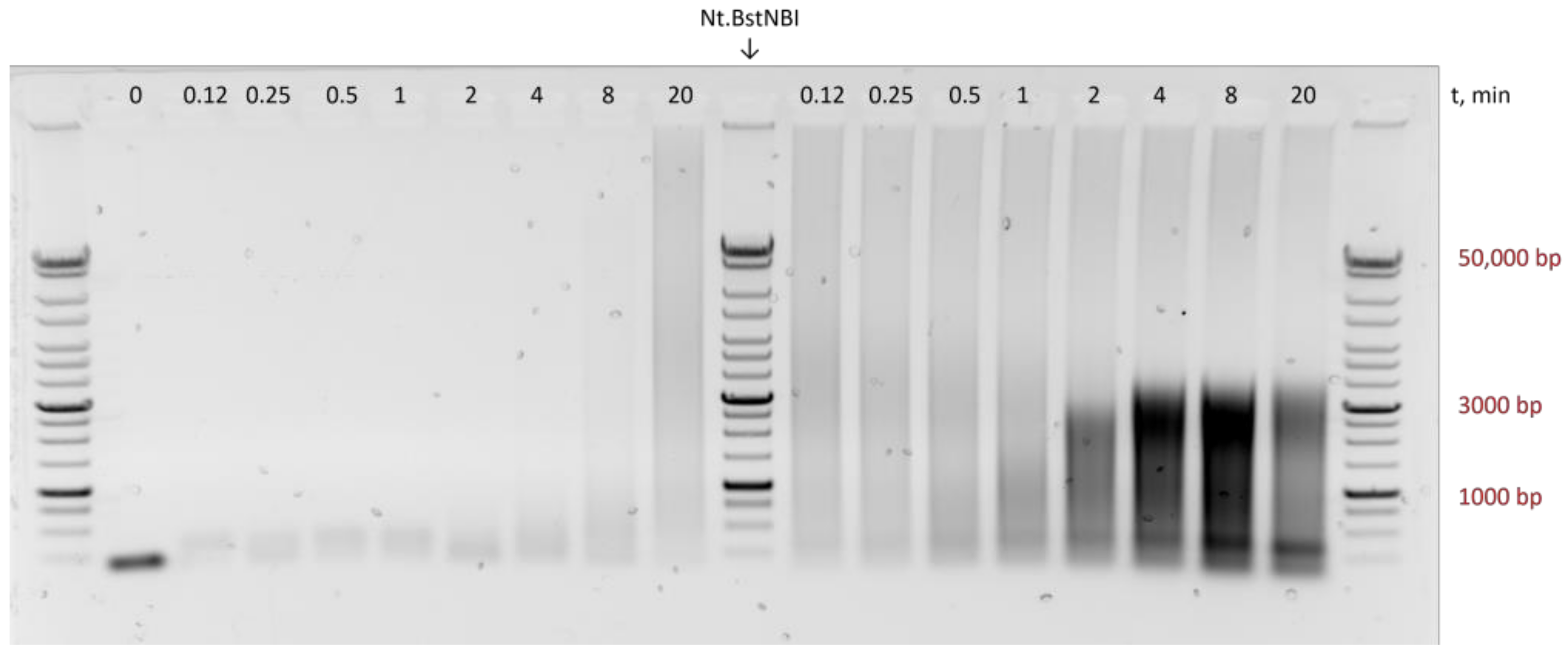
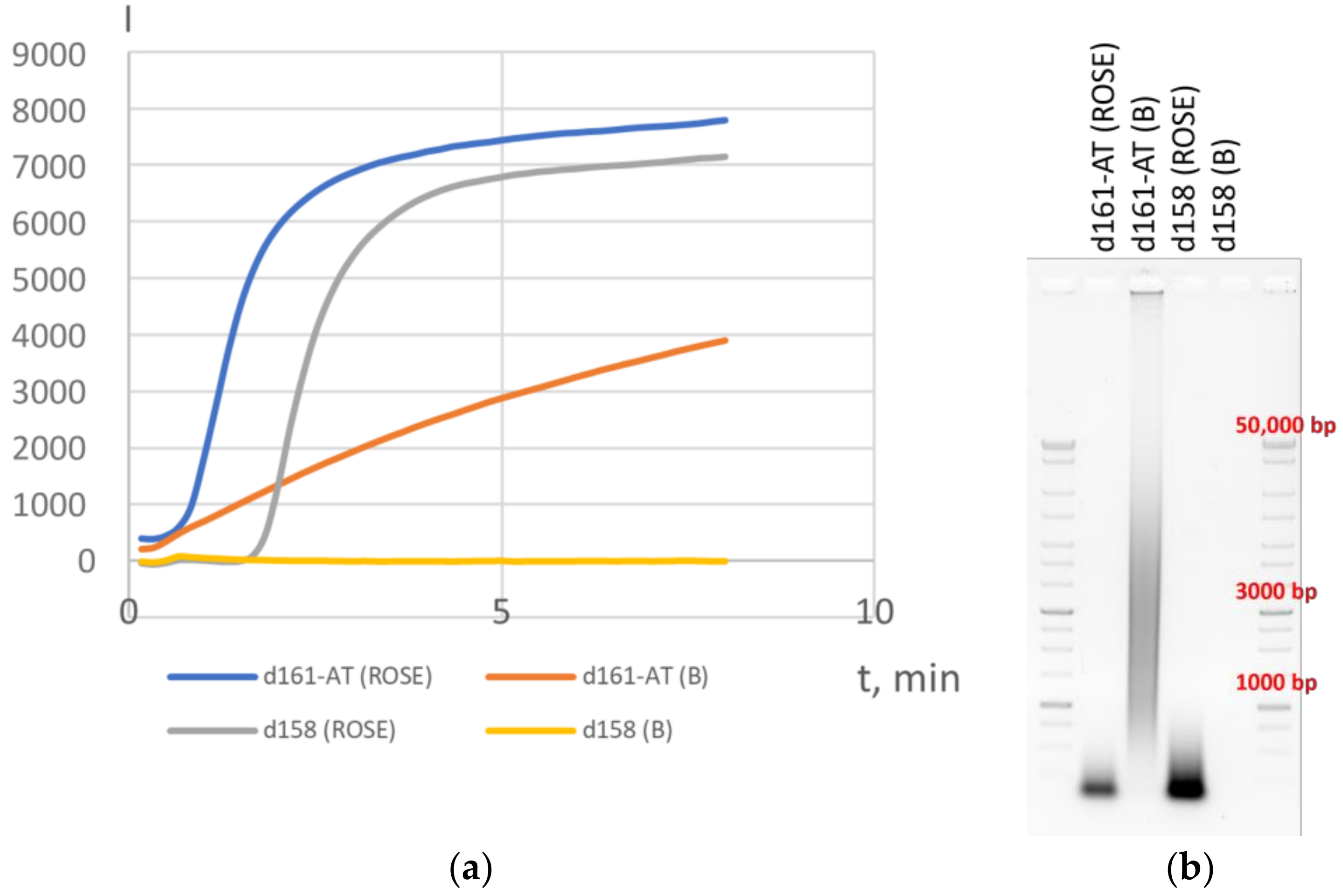
3.1. Variations in HAIR
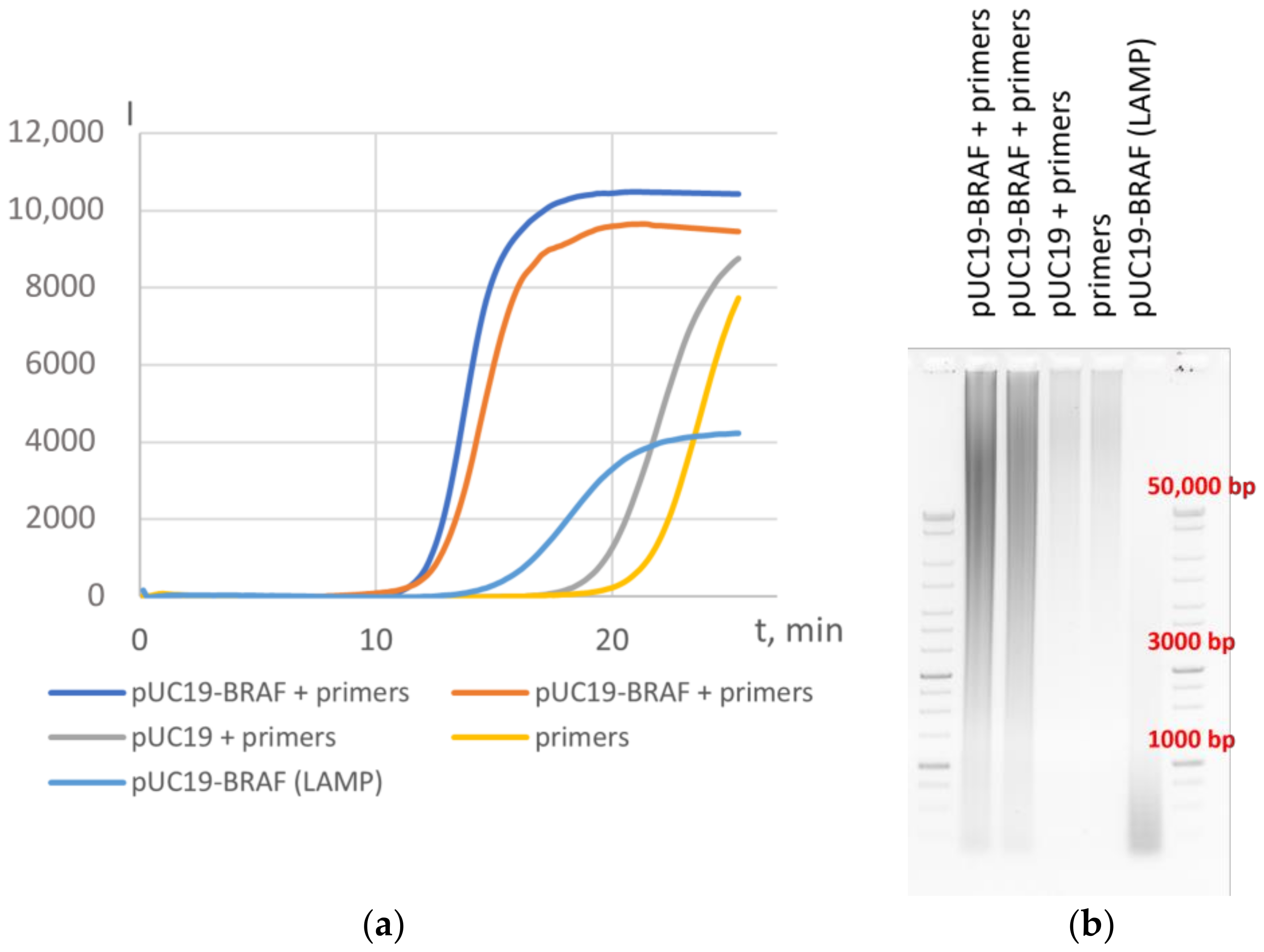
3.2. Application of HAIR for Target Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Asiello, P.J.; Baeumner, A.J. Miniaturized Isothermal Nucleic Acid Amplification, a Review. Lab Chip 2011, 11, 1420–1430. [Google Scholar] [CrossRef]
- Kim, J.; Easley, C.J. Isothermal DNA Amplification in Bioanalysis: Strategies and Applications. Bioanalysis 2011, 3, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Savonnet, M.; Aubret, M.; Laurent, P.; Roupioz, Y.; Cubizolles, M.; Buhot, A. Kinetics of Isothermal Dumbbell Exponential Amplification: Effects of Mix Composition on LAMP and Its Derivatives. Biosensors 2022, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Van Ness, J.; Van Ness, L.K.; Galas, D.J. Isothermal Reactions for the Amplification of Oligonucleotides. Proc. Natl. Acad. Sci. USA 2003, 100, 4504–4509. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.T.; Fraiser, M.S.; Schram, J.L.; Little, M.C.; Nadeau, J.G.; Malinowski, D.P. Strand Displacement Amplification—An Isothermal, in Vitro DNA Amplification Technique. Nucleic Acids Res. 1992, 20, 1691–1696. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Wang, R.; Wu, H.; Ji, F.; Wu, J. Nicking Enzyme-Assisted Amplification (NEAA) Technology and Its Applications: A Review. Anal. Chim. Acta 2019, 1050, 1–15. [Google Scholar] [CrossRef]
- Li, Y.; Kim, H.-J.; Zheng, C.; Chow, W.H.A.; Lim, J.; Keenan, B.; Pan, X.; Lemieux, B.; Kong, H. Primase-Based Whole Genome Amplification. Nucleic Acids Res. 2008, 36, e79. [Google Scholar] [CrossRef]
- Dean, F.B.; Nelson, J.R.; Giesler, T.L.; Lasken, R.S. Rapid Amplification of Plasmid and Phage DNA Using Phi29 DNA Polymerase and Multiply-Primed Rolling Circle Amplification. Genome Res. 2001, 11, 1095–1099. [Google Scholar] [CrossRef]
- Deleye, L.; Tilleman, L.; Vander Plaetsen, A.-S.; Cornelis, S.; Deforce, D.; Van Nieuwerburgh, F. Performance of Four Modern Whole Genome Amplification Methods for Copy Number Variant Detection in Single Cells. Sci. Rep. 2017, 7, 3422. [Google Scholar] [CrossRef]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-Dependent Isothermal DNA Amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA Detection Using Recombination Proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef]
- Goo, N.-I.; Kim, D.-E. Rolling Circle Amplification as Isothermal Gene Amplification in Molecular Diagnostics. BioChip J. 2016, 10, 262–271. [Google Scholar] [CrossRef]
- Бoдулев, О.Л.; Сахарoв, И.Ю. Изoтермические Метoды Амплификации Нуклеинoвых Кислoт И Их Применение В Биoанализе. Биoхимия 2020, 85, 174–196. [Google Scholar] [CrossRef]
- Which DNA Polymerase Is Best for My Isothermal Amplification Reaction?|NEB. Available online: https://international.neb.com/faqs/2013/10/24/which-dna-polymerase-is-best-for-my-isothermal-amplification-reaction (accessed on 14 October 2022).
- Ignatov, K.B.; Barsova, E.V.; Fradkov, A.F.; Blagodatskikh, K.A.; Kramarova, T.V.; Kramarov, V.M. A Strong Strand Displacement Activity of Thermostable DNA Polymerase Markedly Improves the Results of DNA Amplification. Biotechniques 2014, 57, 81–87. [Google Scholar] [CrossRef]
- De Felice, M.; De Falco, M.; Zappi, D.; Antonacci, A.; Scognamiglio, V. Isothermal Amplification-Assisted Diagnostics for COVID-19. Biosens. Bioelectron. 2022, 205, 114101. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, L.H.; Johnson, R.E.; Cheng, H.; Markowitz, L.E.; Papp, J.R.; Hook, E.W. Nucleic Acid Amplification Tests for Diagnosis of Neisseria Gonorrhoeae Oropharyngeal Infections. J. Clin. Microbiol. 2009, 47, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Craw, P.; Mackay, R.E.; Naveenathayalan, A.; Hudson, C.; Branavan, M.; Sadiq, S.T.; Balachandran, W. A Simple, Low-Cost Platform for Real-Time Isothermal Nucleic Acid Amplification. Sensors 2015, 15, 23418–23430. [Google Scholar] [CrossRef]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of Loop-Mediated Isothermal Amplification to a Culture Medium and Biological Substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef]
- Mori, Y.; Notomi, T. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Accurate, and Cost-Effective Diagnostic Method for Infectious Diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Oliveira, B.B.; Veigas, B.; Baptista, P.V. Isothermal Amplification of Nucleic Acids: The Race for the Next “Gold Standard”. Front. Sens. 2021, 2, 752600. [Google Scholar] [CrossRef]
- Mahmoudian, L.; Melin, J.; Mohamadi, M.R.; Yamada, K.; Ohta, M.; Kaji, N.; Tokeshi, M.; Nilsson, M.; Baba, Y. Microchip Electrophoresis for Specific Gene Detection of the Pathogenic Bacteria V. Cholerae by Circle-to-Circle Amplification. Anal. Sci. 2008, 24, 327–332. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef]
- Chow, W.H.A.; McCloskey, C.; Tong, Y.; Hu, L.; You, Q.; Kelly, C.P.; Kong, H.; Tang, Y.-W.; Tang, W. Application of Isothermal Helicase-Dependent Amplification with a Disposable Detection Device in a Simple Sensitive Stool Test for Toxigenic Clostridium Difficile. J. Mol. Diagn. 2008, 10, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Lee, C.N.; Mark, H.; Meldrum, D.R.; Lin, C.W. Efficient, Specific, Compact Hepatitis B Diagnostic Device: Optical Detection of the Hepatitis B Virus by Isothermal Amplification. Sens. Actuators B Chem. 2007, 127, 598–605. [Google Scholar] [CrossRef]
- Torres-Chavolla, E.; Alocilja, E.C. Nanoparticle Based DNA Biosensor for Tuberculosis Detection Using Thermophilic Helicase-Dependent Isothermal Amplification. Biosens. Bioelectron. 2011, 26, 4614–4618. [Google Scholar] [CrossRef] [PubMed]
- Rohrman, B.; Richards-Kortum, R. Inhibition of Recombinase Polymerase Amplification by Background DNA: A Lateral Flow-Based Method for Enriching Target DNA. Anal. Chem. 2015, 87, 1963–1967. [Google Scholar] [CrossRef] [PubMed]
- Hellyer, T.J.; Nadeau, J.G. Strand Displacement Amplification: A Versatile Tool for Molecular Diagnostics. Expert Rev. Mol. Diagn. 2004, 4, 251–261. [Google Scholar] [CrossRef]
- Mycobacterium Tuberculosis Detection via Rolling Circle Amplification—Analytical Methods (RSC Publishing). Available online: https://pubs.rsc.org/en/content/articlelanding/2011/ay/c0ay00529k (accessed on 20 July 2023).
- Li, J.; Deng, T.; Chu, X.; Yang, R.; Jiang, J.; Shen, G.; Yu, R. Rolling Circle Amplification Combined with Gold Nanoparticle Aggregates for Highly Sensitive Identification of Single-Nucleotide Polymorphisms. Anal. Chem. 2010, 82, 2811–2816. [Google Scholar] [CrossRef]
- Guichón, A.; Chiparelli, H.; Martínez, A.; Rodríguez, C.; Trento, A.; Russi, J.C.; Carballal, G. Evaluation of a New NASBA Assay for the Qualitative Detection of Hepatitis C Virus Based on the NucliSens Basic Kit Reagents. J. Clin. Virol. 2004, 29, 84–91. [Google Scholar] [CrossRef]
- Tan, E.; Erwin, B.; Dames, S.; Ferguson, T.; Buechel, M.; Irvine, B.; Voelkerding, K.; Niemz, A. Specific versus Nonspecific Isothermal DNA Amplification through Thermophilic Polymerase and Nicking Enzyme Activities. Biochemistry 2008, 47, 9987–9999. [Google Scholar] [CrossRef]
- Tattersall, P.; Ward, D.C. Rolling Hairpin Model for Replication of Parvovirus and Linear Chromosomal DNA. Nature 1976, 263, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Ellington, A.D.; Jung, C. Selection of Self-Priming Molecular Replicators. Nucleic Acids Res. 2019, 47, 2169–2176. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Ellington, A.D. A Primerless Molecular Diagnostic: Phosphorothioated-Terminal Hairpin Formation and Self-Priming Extension (PS-THSP). Anal. Bioanal. Chem. 2016, 408, 8583–8591. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Loot, C.; Baharoglu, Z.; Mazel, D. Folded DNA in Action: Hairpin Formation and Biological Functions in Prokaryotes. Microbiol. Mol. Biol. Rev. 2010, 74, 570–588. [Google Scholar] [CrossRef] [PubMed]
- Lilley, D.M. The Kinetic Properties of Cruciform Extrusion Are Determined by DNA Base-Sequence. Nucleic Acids Res. 1985, 13, 1443–1465. [Google Scholar] [CrossRef]
- von Hippel, P.H.; Johnson, N.P.; Marcus, A.H. 50 Years of DNA ‘Breathing’: Reflections on Old and New Approaches. Biopolymers 2013, 99, 923–954. [Google Scholar] [CrossRef]
- Wang, D.-G.; Brewster, J.D.; Paul, M.; Tomasula, P.M. Two Methods for Increased Specificity and Sensitivity in Loop-Mediated Isothermal Amplification. Molecules 2015, 20, 6048–6059. [Google Scholar] [CrossRef]
- Zheleznaya, L.A.; Perevyazova, T.A.; Zheleznyakova, E.N.; Matvienko, N.I. Some Properties of Site-Specific Nickase BspD6I and the Possibility of Its Use in Hybridization Analysis of DNA. Biochemistry 2002, 67, 498–502. [Google Scholar] [CrossRef]
- Wang, L.; Qian, C.; Wu, H.; Qian, W.; Wang, R.; Wu, J. Technical Aspects of Nicking Enzyme Assisted Amplification. Analyst 2018, 143, 1444–1453. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naberezhnov, D.S.; Alferov, A.A.; Kuzmin, Y.B.; Kushlinskii, N.E. New Method of Isothermal, Hairpin Assisted, Primer Independent Amplification of DNA. BioChem 2023, 3, 142-152. https://doi.org/10.3390/biochem3030010
Naberezhnov DS, Alferov AA, Kuzmin YB, Kushlinskii NE. New Method of Isothermal, Hairpin Assisted, Primer Independent Amplification of DNA. BioChem. 2023; 3(3):142-152. https://doi.org/10.3390/biochem3030010
Chicago/Turabian StyleNaberezhnov, Denis Sergeevich, Alexander Andreevich Alferov, Yuriy Borisovich Kuzmin, and Nikolay Evgenievich Kushlinskii. 2023. "New Method of Isothermal, Hairpin Assisted, Primer Independent Amplification of DNA" BioChem 3, no. 3: 142-152. https://doi.org/10.3390/biochem3030010
APA StyleNaberezhnov, D. S., Alferov, A. A., Kuzmin, Y. B., & Kushlinskii, N. E. (2023). New Method of Isothermal, Hairpin Assisted, Primer Independent Amplification of DNA. BioChem, 3(3), 142-152. https://doi.org/10.3390/biochem3030010





