Abstract
Background: Hematologic malignancies are a group of heterogeneous neoplasms which originate from hematopoietic cells. The most common among them are leukemia, lymphoma, and multiple myeloma. Machine learning (ML) is a subfield of artificial intelligence that enables the analysis of large amounts of data, possibly finding hidden patterns. Methods: We performed a narrative review about recent applications of ML in the most common hematological malignancies. We focused on the most recent scientific literature about this topic. Results: ML tools have proved useful in the most common hematological malignancies, in particular to enhance diagnostic work-up and guide treatment. Conclusions: Although ML has multiple possible applications in this field, there are some issue that have to be fixed before they can be used in daily clinical practice.
1. Introduction
Hematologic malignancies are a group of heterogeneous neoplasms which originate from hematopoietic cells [1]. Clinically, they manifest with symptoms related to bone marrow insufficiency or suppression, such as bleeding, infections, and anemia [2]. Leukemia, lymphoma, and plasma cell neoplasms are the most common tumors of this group [3].
Leukemia is composed of different hematologic malignancies, caused by the uncontrolled proliferation of a clonal hematopoietic stem cell in the bone marrow [4]. It can be classified, according to its rate of proliferation, as acute or chronic or as myelocytic or lymphocytic, based on the cell of origin. The most common subtypes are acute myeloid leukemia (AML), chronic myeloid leukemia (CML), acute lymphoblastic leukemia (ALL), and chronic lymphocytic leukemia (CLL) [3]. ALL is the most common pediatric tumor, whereas chronic types of leukemia are typical in adults [5,6]. Clinically, they cause non-specific symptoms such as fever, fatigue, weight loss, bone pain, and bleeding [7]. Complete blood count, coagulation and metabolic panel, peripheral blood smear, and a biopsy and bone marrow aspiration are usually required to make the correct diagnosis [8].
Lymphoma is a group of malignant blood tumors that develop from lymphocytes [9]. They account for about half of all newly diagnosed hematologic malignancies and constitute the sixth most common group of tumors worldwide, with peak age in the second decade of life [10].
Lymphomas have traditionally been classified as Hodgkin lymphoma (HL) or non-Hodgkin lymphoma (NHL), based on the presence or absence of Reid–Sternberg cells on histology [11]. Diagnosis involves the removal and histopathological analysis of a lymph node or the biopsy of another affected organ. Fine-needle biopsy alone can be sufficient for diagnosis if enough material can be obtained for histopathological diagnosis [12].
Multiple myeloma (MM) is a hematologic malignancy affecting the bone marrow, characterized by monoclonal proliferation of mature plasma cells [13]. MM is one of the most common hematologic malignancies and it represents about 1% of all human cancers [14]. MM typically evolves from precursor stages called monoclonal gammopathy of undetermined significance (MGUS) and smoldering MM (SMM) [15]. MM is diagnosed in patients who meet one of the CRAB criteria (calcium elevation, renal insufficiency, anemia, and bone disease) according to the consensus of the International Myeloma Working Group (IMWG) [14].
Artificial intelligence (AI) can be defined as a set of advanced computational algorithms that can identify and learn hidden patterns in huge quantities of data, possibly making evaluations and predictions [16]. In medicine, the primary purpose of AI is to extract and analyze meaningful hidden quantitative data which can aid medical decision-making [17]. AI encompasses several learning algorithms, such as machine learning (ML) [18]. ML is a method of data science that provides computers with the ability to learn without being programmed with explicit rules and enables the creation of algorithms that can learn and make predictions [19]. ML algorithms can be organized into different categories depending on the type of task: supervised, unsupervised, and reinforcement learning algorithms [20]. Supervised ML refers to techniques in which a model is trained on a range of inputs (or features) which are associated with a known outcome [21]. Data labels are provided to the algorithm in the training phase and the expected outputs are usually labeled by human experts and serve as a ground truth for the algorithm. The goal of the algorithm is usually to learn a general rule that maps inputs to outputs [20]. Once the algorithm is successfully trained, it will be capable of making outcome predictions when applied to new data [21]. In unsupervised learning, patterns are sought by algorithms without any input from the user and hidden structures in large data are automatically identified [20]. The main purpose of unsupervised learning is to discover relationships between samples or reveal the latent variables hidden in data [19]. In reinforcement learning, a computer program performs a certain task in which it receives positive and/or negative reinforcement in a dynamic environment [20]. These algorithms learn from the consequences of interactions with an environment without being explicitly taught. These different types of learning paradigms can be used in combination [20]. Deep learning (DL) is a subfield of ML, which comprises multi-layered networks composed of nodes, like neurons in the brain, thus enabling high-level abstraction from data analysis [22]. ML-based tools have numerous promising applications in several fields of medicine, especially in the field of oncology, where multiple algorithms have been proposed for oncological risk assessment, automated segmentation, lesion detection, characterization, grading and staging, prediction of prognosis, and therapy response [23].
The aim of this narrative review is to depict the most recent applications of ML in the most common hematological malignancies.
2. Leukemia
Leukemia is a major hematological malignancy, with high prevalence and incidence [24]. The diagnosis of leukemia is complex and time-consuming, thus ML could be helpful in this stage [25]. ML is a promising tool for the management of leukemia, as it could identify disease-specific genetic alterations (Table 1) [26]. In this setting, the most useful ML algorithms are the least absolute shrinkage and selection operator (LASSO), random forest graph (RF), support vector machine (SVM), and decision tree [27].
In the scientific literature, there are multiple articles that use ML algorithms to improve diagnosis, predict prognosis, and suggest therapeutic strategies in patients with leukemia [28].
The first studies using ML techniques in leukemia were focused on the recognition of leukemic cells from blood samples, flow cytometry, and the evaluation of genetic data. ML can be used to accurately identify the subtypes of leukemia. For instance, Ahmed et al. tested a convolutional neural network (CNN), a subtype of DL, in the automatic differentiation of AML, CML, ALL, and CLL. The CNN algorithm showed a high accuracy in the diagnosis of leukemia (88.25%) and its classification (81.74%) (Figure 1) [29]. Similarly, Huang et al. used three types of CNN tools for the diagnosis of AML, ALL, and CML [30]. Their algorithm could identify CML with a high accuracy (>95%). Abhishek et al. developed an ML tool to automatically classify different types of leukemia (CML, ALL, CML, and ALL) using peripheral blood smear images [31]. CNN models were used for feature extraction [32]. Subsequently, the extracted features were used to train the ML tools (based on the SVM and RF models) to classify the images into normal, CML, ALL, CML, and ALL. Dese et al. used an ML algorithm based on SVM to detect and classify the major subtypes of leukemia (AML, CML, ALL, and CLL) using blood smear images [33].
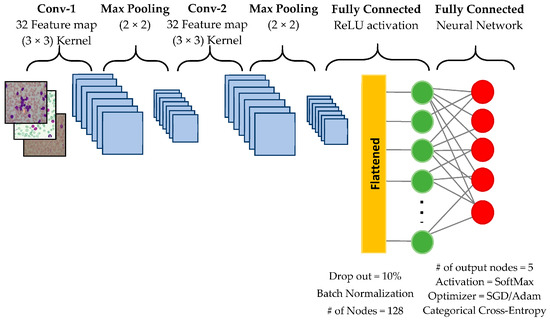
Figure 1.
An example of a convolutional neural network architecture. The figure is taken from [29].
Regarding CML, Zhang et al. used a DL model to differentiate megakaryocytes from myeloid cells [34]. The best-performing model (SVM) showed an area under the curve (AUC) of 84.93%.
ML can also be used to predict the 5-year survival of leukemia patients. In a retrospective study, Shanbehzadeh et al. developed a model for the prediction of the 5-year survival of CML patients [35]. The dataset with full and selected variables was fed into the ML models separately. The SVM model achieved the highest performance with a specificity of 85%, a sensitivity of 86%, and an AUC of 0.85. ML can also provide promising results in the prediction of a risk of Asparaginase-associated pancreatitis, which a side effect of a drug for ALL therapy.
In the setting of AML, Shaikh et al. used supervised ML to identify NRAS mutation and the absence of mutations in ASXL2, RAD21, KIT, and FLT3 genes in patients with AML, as well as low mutation levels that are associated with favorable outcomes [36]. Poor genetic risk emerged as an independent risk factor predictive of inferior outcomes. ML methods can be used to identify key genes significantly associated with leukemia prognosis. In a recent study, Cheng et al. identified genes DNM1, MEIS1, and SUSD3 to be closely related to the prognosis of AML based on ML analysis [37].
Finally, Nielsen et al. used ML strategies, including regression, RF, AdaBoost, and artificial neural networks, to predict individual-level risk of Asparaginase-associated pancreatitis in children with ALL [38].
3. Lymphoma
Over the past decade, ML tools have improved the accuracy of diagnosis, genomics, proteomics, and histopathology of Lymphoma, leading to personalized medicine and improving survival rates [39].
The application of ML in lymphoma management has been shown to be promising concerning clinical studies, demonstrating great potential in different aspects of clinical practice [40]. In particular, ML can improve diagnosis, classification, genetic evaluation, and prognosis. Although biopsy is the gold standard for diagnosis, ML can also be used to infer the type of lymphoma through the analysis of mediastinal bulky masses via imaging [41].
Can et al. developed a DL tool to diagnose cervical lymphadenopathies without the need for radiologist consultation and histopathological examinations, enabling automatic distinction of granulomatous diseases, lymphoma, squamous cell tumor, and reactive hyperplasia [42]. To train the model, which achieved an accuracy higher than 92.5%, they used a database of the computed tomography (CT) of 400 patients who had undergone neck surgery.
Regarding diffuse large B-cell lymphoma (DLBCL), De Jesus et al. developed an ML tool which showed promising results in differentiating it from follicular lymphoma (FL) [43]. They trained the classifier on a database of 120 [18F] Fluorodeoxyglucose (FDG) positron emission tomography (PET)/CT results. Their best-performing model showed an AUC of 0.86, significantly higher than the performance of the SUVmax-based model (AUC, 0.79). These results might have important clinical implications in monitoring for aggressive transformation. Sarcoidosis and lymphoma often share common features on 18 F-FDG PET/CT, thus Lovinfosse et al. developed radiomic signatures to differentiate them from DLBCL and HL. The characterization of sarcoidosis and lymphoma lesions using ML and radiomics was found to be equivalent to or better than characterization by physicians [44]. Irshaid et al. demonstrated the association between specific morphologic findings in bone marrow and the clinical outcome of lymphoma transformation, and investigated ML correlates that may aid in this endeavor [45]. During prognosis assessment, ML models have proved valuable in predicting disease progression from FL and CLL to DLBCL and treatment outcomes, helping clinicians in the formulation of management plans tailored to individual patients. Finally, Carreras et al. used ML to automatically quantify the expression of the TNFAIP8 protein in patients with DLBCL, which is related to a bad prognosis [46].
ML can also be used to guide treatment and develop new targeted drugs for lymphoma [47]. In particular, ML could enhance the process of drug production, especially in the context of tailored medicine [48]. For instance, Liu and colleagues used an ML tool to develop new inhibitors of B-cell lymphoma [49].
In a recent study, Hill et al. incorporated baseline clinicopathologic, molecular, and cytogenetic features in ML models to predict outcomes in patients with mantle cell lymphoma [50]. They used clinicopathologic, cytogenetic, and genomic information from a large database of 862 patients to train the model, which could discriminate patients with indolent from responsive mantle cell lymphoma with an AUC of 0.83.
Finally, the total metabolic tumor volume is considered as an independent prognostic factor in both HL and NHL. Capobianco et al. demonstrated that the DL method can estimate it, measuring the body FDG-PET/CT [51].
4. Multiple Myeloma
MM is a malignant tumor of plasma cells that predominantly affects elderly patients. Serological, genetic, morphological, immunophenotypic, histological, and radiological information is important for diagnosis and staging. Multiple studies evaluated the potential role of ML in identification of early markers of MM, which can aid the selection of the appropriate therapy, impacting recurrence and survival. Imaging techniques are extremely relevant for the recognition of bone lesions and the identification of lytic bone lesions is crucial for diagnosis, prognosis, and therapy, and ML can be useful in this setting [52]. For instance, Xiong et al. developed an ML algorithm to automatically differentiate MM bony lesions from metastasis of the lumbar spine using MRI [53]. In this retrospective analysis, 107 patients were enrolled, and 10 ML algorithms were tested. Among them, the artificial neural network showed the best performance, with an accuracy of 0.81. In MM, undetectable measurable residual disease correlates with better survival. Thus, Guerrero et al. used ML to identify elements which could predict this condition [54]. They retrospectively enrolled 487 patients with newly diagnosed MM, and trained their model based on cytogenetic, tumor burden, and immune-related biomarkers. The model proved to be effective as it could identify a subgroup of patients with active MM with 80% and 93% progression-free and overall survival rates at 5 years. Bi and colleagues examined the prognostic value of radiomic features extracted from 18F-FDG-PET/CT images, integrating them with clinical features and conventional PET/CT metrics with an ML model, in patients with newly diagnosed MM. They demonstrated that the combination of the radiomic features of PET/CT with clinical data provides better prognostic performance than models with radiomic features or clinical data only.
A further target is to provide a stratification of treatment sensitivity at the time of MM diagnosis. Povoa et al. developed an ML algorithm to predict treatment sensitivity with an AUC of 68.7%. This tool could help treatment choice in newly diagnosed MM patients, improving survival.
5. Discussion
ML is a branch of AI that relies on algorithms that automatically learn from available data, without previous programming, with the aim of creating decision models [23]. In medicine, it can have multiple possible applications, thanks also to technological advances in terms of production and extrapolation, and the digitalization of medical data, especially in the subfields of radiology and pathology. Multiple algorithms have also been proposed in oncohematology, with a role in risk assessment, segmentation, lesion detection, characterization, grading and staging, prediction of prognosis, and response to therapy.
Although ML has shown multiple promising applications, ML has several limitations, which should be addressed before these tools are introduced into daily clinical practice. Firstly, the reproducibility of ML in multicenter studies is still poor due to the different acquisition parameters and the use of non-standard pipelines [55]. Recently, some online platforms have been developed to offer standardized ML pipelines [56]. Interpretation and elaboration of data also requires specific expertise, and it is not always possible to fully integrate the ML components under consideration [57]. Technical variables still affect the quantification of radiomic characteristics. In particular, large public databases are required in the training step to reduce overfitting, which consists of poor generalization of ML models to future observations [58]. In the future, large open datasets will be required to improve ML tool performance. Finally, in the past few years many articles using ML in medicine have been published; therefore, it is important to identify those articles which are lacking in terms of scientific rigorousness. Radiomics quality scores and metrics could help evaluate this feature [59].
6. Future Directions
The diagnosis and treatment of hematological malignancies are continuously evolving, and ML tools could be used to empower them in clinical practice. For instance, they can be used to identify genetic mutations which could correlate with treatment response and prognosis. This could lead to tailored treatments, thus improving clinical outcomes. ML can also be used to identify features which could predict the evolution of hematological malignancies from indolent to aggressive forms, possibly revealing the pathophysiology underlying this process.
Although ML has multiple possible applications in this field, it is crucial to acknowledge some issues. For example, large multicenter open databases are required for the training phase, to improve the ability of ML tools to generalize. The use of standard ML pipelines could also enhance this task. Furthermore, in the past few years there has been an enormous number of scientific articles regarding ML tools, thus it is important to evaluate their scientific rigor. Multiple scientific scores have been created to evaluate the scientific rigor of these original articles, which is crucial to introduce them into clinical practice [60].
7. Conclusions
Recently, multiple ML tools have been developed to improve diagnosis, prognosis, and treatment in the most common hematological malignancies. Although the results are promising, there are still multiple limitations that should be addressed before ML could be routinely used in daily clinical practice.
Author Contributions
Conceptualization, T.P.; methodology, T.P.; writing—original draft preparation, C.G. and M.d.G.; writing—review and editing, T.P.; supervision, R.C. and A.P.; project administration, C.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zierhut, M.; Haen, S.P.; Moehle, R.; Chan, C.-C. Hematological Neoplasms. In Intraocular Inflammation; Zierhut, M., Pavesio, C., Ohno, S., Orefice, F., Rao, N.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 1493–1510. [Google Scholar]
- Button, E.; Chan, R.J.; Chambers, S.; Butler, J.; Yates, P. A systematic review of prognostic factors at the end of life for people with a hematological malignancy. BMC Cancer 2017, 17, 213. [Google Scholar] [CrossRef]
- Rodriguez-Abreu, D.; Bordoni, A.; Zucca, E. Epidemiology of hematological malignancies. Ann. Oncol. 2007, 18, i3–i8. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Li, S.; Li, L.; Liang, S.; Zhang, B.; Meng, Y.; Zhang, X.; Zhang, Y.; Zhao, S. Differentiation of paediatric posterior fossa tumours by the multiregional and multiparametric MRI radiomics approach: A study on the selection of optimal multiple sequences and multiregions. Br. J. Radiol. 2022, 95, 20201302. [Google Scholar] [CrossRef] [PubMed]
- Ekpa, Q.L.; Akahara, P.C.; Anderson, A.M.; O Adekoya, O.; O Ajayi, O.; O Alabi, P.; E Okobi, O.; Jaiyeola, O.; Ekanem, M.S. A Review of Acute Lymphocytic Leukemia (ALL) in the Pediatric Population: Evaluating Current Trends and Changes in Guidelines in the Past Decade. Cureus 2023, 15, e49930. [Google Scholar] [CrossRef] [PubMed]
- Juliusson, G.; Hough, R. Leukemia. In Progress in Tumor Research; Stark, D.P., Vassal, G., Eds.; Karger AG: Basel, Switzerland, 2016; pp. 87–100. [Google Scholar]
- Clarke, R.T.; Van den Bruel, A.; Bankhead, C.; Mitchell, C.D.; Phillips, B.; Thompson, M.J. Clinical presentation of childhood leukaemia: A systematic review and meta-analysis. Arch. Dis. Child. 2016, 101, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.A.; Grimwade, D.; Tallman, M.S.; Lowenberg, B.; Fenaux, P.; Estey, E.H.; Naoe, T.; Lengfelder, E.; Büchner, T.; Döhner, H.; et al. Management of acute promyelocytic leukemia: Recommendations from an expert panel on behalf of the European LeukemiaNet. Blood 2009, 113, 1875–1891. [Google Scholar] [CrossRef]
- Huh, J. Epidemiologic overview of malignant lymphoma. Korean J. Hematol. 2012, 47, 92–104. [Google Scholar] [CrossRef]
- Roman, E.; Smith, A.G. Epidemiology of lymphomas: Epidemiology and lymphomas. Histopathology 2011, 58, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Parente, P.; Zanelli, M.; Sanguedolce, F.; Mastracci, L.; Graziano, P. Hodgkin Reed–Sternberg-Like Cells in Non-Hodgkin Lymphoma. Diagnostics 2020, 10, 1019. [Google Scholar] [CrossRef]
- Momotow, J.; Borchmann, S.; Eichenauer, D.A.; Engert, A.; Sasse, S. Hodgkin Lymphoma—Review on Pathogenesis, Diagnosis, Current and Future Treatment Approaches for Adult Patients. J. Clin. Med. 2021, 10, 1125. [Google Scholar] [CrossRef]
- The International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: A report of the International Myeloma Working Group. Br. J. Haematol. 2003, 121, 749–757. [Google Scholar] [CrossRef]
- Fend, F.; Dogan, A.; Cook, J.R. Plasma cell neoplasms and related entities—Evolution in diagnosis and classification. Virchows Arch. 2023, 482, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Hameed, M.; Sandhu, A.; Soneji, N.; Amiras, D.; Rockall, A.; Messiou, C.; Wallitt, K.; Barwick, T.D. Pictorial review of whole body MRI in myeloma: Emphasis on diffusion-weighted imaging. Br. J. Radiol. 2020, 93, 20200312. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Ates, E.; Kilickesmez, O. Radiomics with artificial intelligence: A practical guide for beginners. Diagn. Interv. Radiol. 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Thrall, J.H.; Li, X.; Li, Q.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. J. Am. Coll. Radiol. 2018, 15, 504–508. [Google Scholar] [CrossRef]
- Chartrand, G.; Cheng, P.M.; Vorontsov, E.; Drozdzal, M.; Turcotte, S.; Pal, C.J.; Kadoury, S.; Tang, A. Deep Learning: A Primer for Radiologists. RadioGraphics 2017, 37, 2113–2131. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Summers, R.M. Machine learning and radiology. Med. Image Anal. 2012, 16, 933–951. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Synho, D.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64. [Google Scholar] [CrossRef]
- Zaharchuk, G.; Gong, E.; Wintermark, M.; Rubin, D.; Langlotz, C. Deep Learning in Neuroradiology. Am. J. Neuroradiol. 2018, 39, 1776–1784. [Google Scholar] [CrossRef]
- Cuocolo, R.; Caruso, M.; Perillo, T.; Ugga, L.; Petretta, M. Machine Learning in oncology: A clinical appraisal. Cancer Lett. 2020, 481, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.D. Leukaemia prevalence worldwide: Raising aetiology questions. Lancet Haematol. 2018, 5, e2–e3. [Google Scholar] [CrossRef] [PubMed]
- Salah, H.T.; Muhsen, I.N.; Salama, M.E.; Owaidah, T.; Hashmi, S.K. Machine learning applications in the diagnosis of leukemia: Current trends and future directions. Int. J. Lab. Hematol. 2019, 41, 717–725. [Google Scholar] [CrossRef]
- Eckardt, J.-N.; Bornhäuser, M.; Wendt, K.; Middeke, J.M. Application of machine learning in the management of acute myeloid leukemia: Current practice and future prospects. Blood Adv. 2020, 4, 6077–6085. [Google Scholar] [CrossRef]
- McEligot, A.J.; Poynor, V.; Sharma, R.; Panangadan, A. Logistic LASSO Regression for Dietary Intakes and Breast Cancer. Nutrients 2020, 12, 2652. [Google Scholar] [CrossRef]
- Elhadary, M.; Elsabagh, A.A.; Ferih, K.; Elsayed, B.; Elshoeibi, A.M.; Kaddoura, R.; Akiki, S.; Ahmed, K.; Yassin, M. Applications of Machine Learning in Chronic Myeloid Leukemia. Diagnostics 2023, 13, 1330. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Yigit, A.; Isik, Z.; Alpkocak, A. Identification of Leukemia Subtypes from Microscopic Images Using Convolutional Neural Network. Diagnostics 2019, 9, 104. [Google Scholar] [CrossRef]
- Huang, F.; Guang, P.; Li, F.; Liu, X.; Zhang, W.; Huang, W. AML, ALL, and CML classification and diagnosis based on bone marrow cell morphology combined with convolutional neural network: A STARD compliant diagnosis research. Medicine 2020, 99, e23154. [Google Scholar] [CrossRef] [PubMed]
- Iman, M.; Arabnia, H.R.; Rasheed, K. A Review of Deep Transfer Learning and Recent Advancements. Technologies 2023, 11, 40. [Google Scholar] [CrossRef]
- Abhishek, A.; Jha, R.K.; Sinha, R.; Jha, K. Automated detection and classification of leukemia on a subject-independent test dataset using deep transfer learning supported by Grad-CAM visualization. Biomed. Signal Process. Control. 2023, 83, 104722. [Google Scholar] [CrossRef]
- Dese, K.; Raj, H.; Ayana, G.; Yemane, T.; Adissu, W.; Krishnamoorthy, J.; Kwa, T. Accurate Machine-Learning-Based classification of Leukemia from Blood Smear Images. Clin. Lymphoma Myeloma Leuk. 2021, 21, e903–e914. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, X.; Yan, Q.; Lin, Y.; Liu, E.; Mi, Y.; Liang, S.; Wang, H.; Xu, J.; Ru, K. The Diagnosis of Chronic Myeloid Leukemia with Deep Adversarial Learning. Am. J. Pathol. 2022, 192, 1083–1091. [Google Scholar] [CrossRef]
- Shanbehzadeh, M.; Afrash, M.R.; Mirani, N.; Kazemi-Arpanahi, H. Comparing machine learning algorithms to predict 5-year survival in patients with chronic myeloid leukemia. BMC Med. Inform. Decis. Mak. 2022, 22, 236. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, A.F.; Kakirde, C.; Dhamne, C.; Bhanshe, P.; Joshi, S.; Chaudhary, S.; Chatterjee, G.; Tembhare, P.; Prasad, M.; Moulik, N.R.; et al. Machine learning derived genomics driven prognostication for acute myeloid leukemia with RUNX1-RUNX1T1. Leuk. Lymphoma 2020, 61, 3154–3160. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Yang, X.; Wang, Y.; Li, Q.; Chen, W.; Dai, R.; Zhang, C. Multiple machine-learning tools identifying prognostic biomarkers for acute Myeloid Leukemia. BMC Med Inform. Decis. Mak. 2024, 24, 2. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.L.; Wolthers, B.O.; Helenius, M.M.; Albertsen, B.K.; Clemmensen, L.; Nielsen, K.; Kanerva, J.; Niinimäki, R.; Frandsen, T.L.; Attarbaschi, A.; et al. Can Machine Learning Models Predict Asparaginase-associated Pancreatitis in Childhood Acute Lymphoblastic Leukemia. J. Pediatr. Hematol. 2022, 44, E628–E636. [Google Scholar] [CrossRef]
- Moran-Sanchez, J.; Santisteban-Espejo, A.; Martin-Piedra, M.A.; Perez-Requena, J.; Garcia-Rojo, M. Translational Applications of Artificial Intelligence and Machine Learning for Diagnostic Pathology in Lymphoid Neoplasms: A Comprehensive and Evolutive Analysis. Biomolecules 2021, 11, 793. [Google Scholar] [CrossRef]
- Yuan, J.; Zhang, Y.; Wang, X. Application of machine learning in the management of lymphoma: Current practice and future prospects. Digit. Health 2024, 10, 20552076241247963. [Google Scholar] [CrossRef]
- Abenavoli, E.M.; Barbetti, M.; Linguanti, F.; Mungai, F.; Nassi, L.; Puccini, B.; Romano, I.; Sordi, B.; Santi, R.; Passeri, A.; et al. Characterization of Mediastinal Bulky Lymphomas with FDG-PET-Based Radiomics and Machine Learning Techniques. Cancers 2023, 15, 1931. [Google Scholar] [CrossRef]
- Can, S.; Türk, Ö.; Ayral, M.; Kozan, G.; Arı, H.; Akdağ, M.; Baylan, M.Y. Can deep learning replace histopathological examinations in the differential diagnosis of cervical lymphadenopathy? Eur. Arch. Oto-Rhino-Laryngol. 2024, 281, 359–367. [Google Scholar] [CrossRef]
- de Jesus, F.M.; Yin, Y.; Mantzorou-Kyriaki, E.; Kahle, X.U.; de Haas, R.J.; Yakar, D.; Glaudemans, A.W.J.M.; Noordzij, W.; Kwee, T.C.; Nijland, M. Machine learning in the differentiation of follicular lymphoma from diffuse large B-cell lymphoma with radiomic [18F]FDG PET/CT features. Eur. J. Nucl. Med. 2022, 49, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Lovinfosse, P.; Ferreira, M.; Withofs, N.; Jadoul, A.; Derwael, C.; Frix, A.-N.; Guiot, J.; Bernard, C.; Diep, A.N.; Donneau, A.-F.; et al. Distinction of Lymphoma from Sarcoidosis on18F-FDG PET/CT: Evaluation of Radiomics-Feature–Guided Machine Learning Versus Human Reader Performance. J. Nucl. Med. 2022, 63, 1933–1940. [Google Scholar] [CrossRef] [PubMed]
- Irshaid, L.; Bleiberg, J.; Weinberger, E.; Garritano, J.; Shallis, R.M.; Patsenker, J.; Lindenbaum, O.; Kluger, Y.; Katz, S.G.; Xu, M.L. Histopathologic and Machine Deep Learning Criteria to Predict Lymphoma Transformation in Bone Marrow Biopsies. Arch. Pathol. Lab. Med. 2022, 146, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Kikuti, Y.Y.; Miyaoka, M.; Hiraiwa, S.; Tomita, S.; Ikoma, H.; Kondo, Y.; Ito, A.; Shiraiwa, S.; Hamoudi, R.; et al. A Single Gene Expression Set Derived from Artificial Intelligence Predicted the Prognosis of Several Lymphoma Subtypes; and High Immunohistochemical Expression of TNFAIP8 Associated with Poor Prognosis in Diffuse Large B-Cell Lymphoma. AI 2020, 1, 342–360. [Google Scholar] [CrossRef]
- Yuan, B.; Xie, H.; Wang, Z.; Xu, Y.; Zhang, H.; Liu, J.; Chen, L.; Li, C.; Tan, S.; Lin, Z.; et al. The domain-separation language network dynamics in resting state support its flexible functional segregation and integration during language and speech processing. NeuroImage 2023, 274, 120132. [Google Scholar] [CrossRef]
- Nayarisseri, A.; Khandelwal, R.; Tanwar, P.; Madhavi, M.; Sharma, D.; Thakur, G.; Speck-Planche, A.; Singh, S.K. Artificial Intelligence, Big Data and Machine Learning Approaches in Precision Medicine & Drug Discovery. Curr. Drug Targets 2021, 22, 631–655. [Google Scholar] [CrossRef]
- Liu, L.; Na, R.; Yang, L.; Liu, J.; Tan, Y.; Zhao, X.; Huang, X.; Chen, X. A Workflow Combining Machine Learning with Molecular Simulations Uncovers Potential Dual-Target Inhibitors against BTK and JAK3. Molecules 2023, 28, 7140. [Google Scholar] [CrossRef]
- Hill, H.A.; Jain, P.; Ok, C.Y.; Sasaki, K.; Chen, H.; Wang, M.L.; Chen, K. Integrative Prognostic Machine-Learning Models in Mantle Cell Lymphoma. Cancer Res. Commun. 2023, 3, 1435–1446. [Google Scholar] [CrossRef]
- Capobianco, N.; Meignan, M.A.; Cottereau, A.-S.; Vercellino, L.; Sibille, L.; Spottiswoode, B.; Zuehlsdorff, S.; Casasnovas, O.; Thieblemont, C.; Buvat, I. Deep-Learning 18F-FDG Uptake Classification Enables Total Metabolic Tumor Volume Estimation in Diffuse Large B-Cell Lymphoma. J. Nucl. Med. 2021, 62, 30–36. [Google Scholar] [CrossRef]
- Gangemi, S.; Allegra, A.; Alonci, A.; Cristani, M.; Russo, S.; Speciale, A.; Penna, G.; Spatari, G.; Cannavò, A.; Bellomo, G.; et al. Increase of novel biomarkers for oxidative stress in patients with plasma cell disorders and in multiple myeloma patients with bone lesions. Inflamm. Res. 2012, 61, 1063–1067. [Google Scholar] [CrossRef]
- Xiong, X.; Wang, J.; Hu, S.; Dai, Y.; Zhang, Y.; Hu, C. Differentiating Between Multiple Myeloma and Metastasis Subtypes of Lumbar Vertebra Lesions Using Machine Learning–Based Radiomics. Front. Oncol. 2021, 11, 601699. [Google Scholar] [CrossRef]
- Guerrero, C.; Puig, N.; Cedena, M.-T.; Goicoechea, I.; Perez, C.; Garcés, J.-J.; Botta, C.; Calasanz, M.-J.; Gutierrez, N.C.; Martin-Ramos, M.-L.; et al. A Machine Learning Model Based on Tumor and Immune Biomarkers to Predict Undetectable MRD and Survival Outcomes in Multiple Myeloma. Clin. Cancer Res. 2022, 28, 2598–2609. [Google Scholar] [CrossRef] [PubMed]
- Perillo, T.; Somma, C.; de Giorgi, M.; Papace, U.M.; Perillo, S.; Serino, A.; Manto, A.; Cuocolo, R. Radiomics and radiogenomics of central nervous system metastatic lesions. In Radiomics and Radiogenomics in Neuro-Oncology; Elsevier: Amsterdam, The Netherlands, 2024; pp. 235–249. [Google Scholar]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Kazerooni, A.F.; Arif, S.; Madhogarhia, R.; Khalili, N.; Haldar, D.; Bagheri, S.; Familiar, A.M.; Anderson, H.; Haldar, S.; Tu, W.; et al. Automated tumor segmentation and brain tissue extraction from multiparametric MRI of pediatric brain tumors: A multi-institutional study. Neuro-Oncol. Adv. 2023, 5, vdad027. [Google Scholar] [CrossRef]
- Lasocki, A.; Abdalla, G.; Chow, G.; Thust, S.C. Imaging features associated with H3 K27-altered and H3 G34-mutant gliomas: A narrative systematic review. Cancer Imaging 2022, 22, 63. [Google Scholar] [CrossRef] [PubMed]
- Kocak, B.; D’aNtonoli, T.A.; Mercaldo, N.; Alberich-Bayarri, A.; Baessler, B.; Ambrosini, I.; Andreychenko, A.E.; Bakas, S.; Beets-Tan, R.G.H.; Bressem, K.; et al. METhodological RadiomICs Score (METRICS): A quality scoring tool for radiomics research endorsed by EuSoMII. Insights Imaging 2024, 15, 8. [Google Scholar] [CrossRef]
- Cosgun, E.; Oh, M. Exploring the Consistency of the Quality Scores with Machine Learning for Next-Generation Sequencing Experiments. BioMed Res. Int. 2020, 2020, 8531502. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).