In Silico and In Vitro Evaluation of the Antifungal Activity of a New Chromone Derivative against Candida spp.
Abstract
1. Introduction
2. Materials and Methods
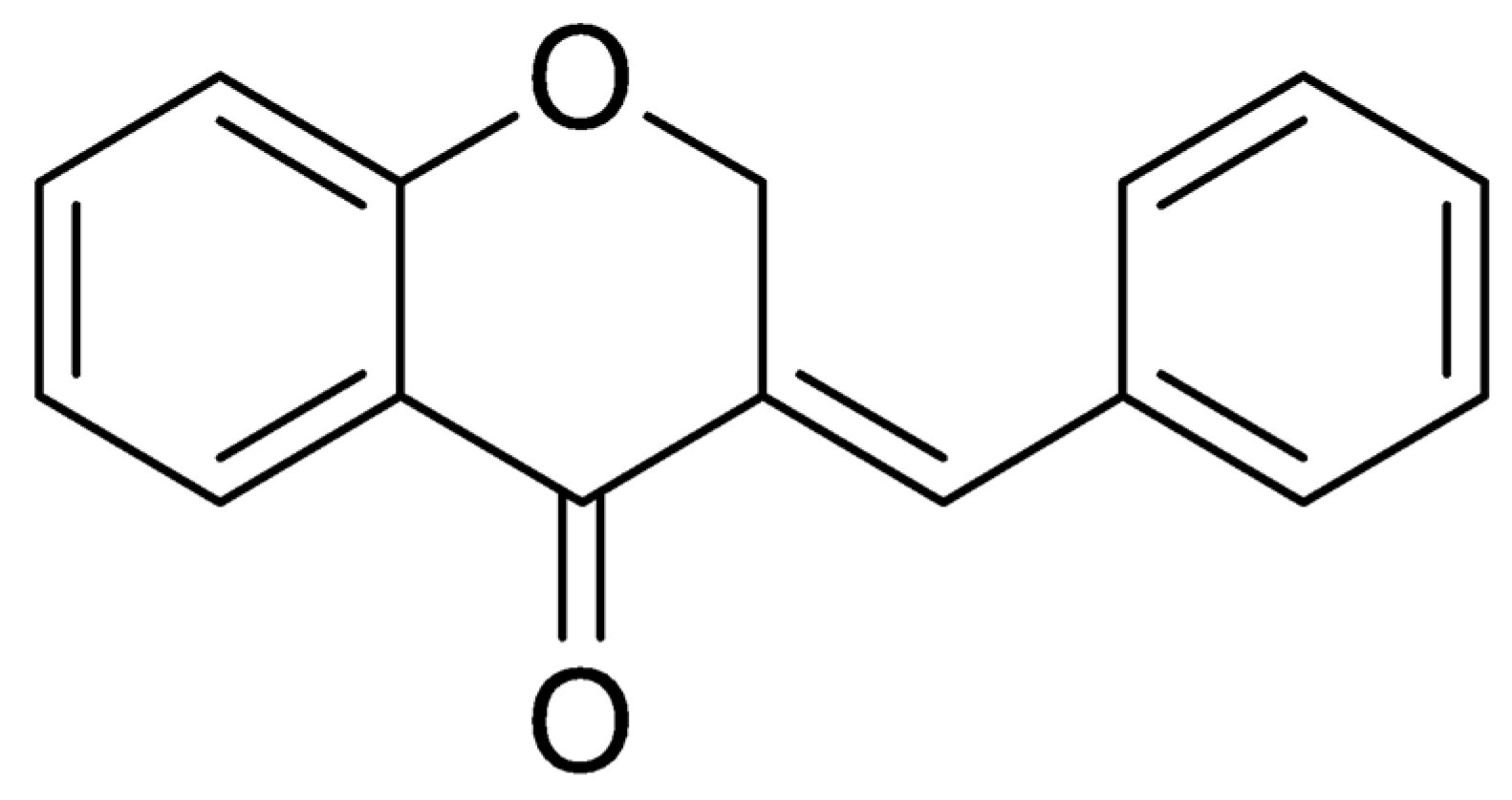
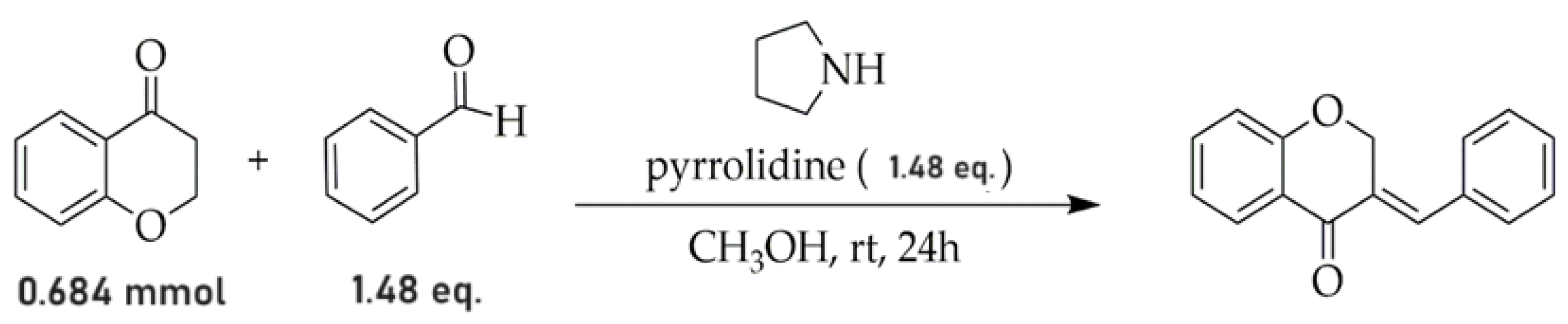
2.1. Procedure for Obtaining the AR25 Derivative
Structure Analysis of Compound AR25 Is as Follows
2.2. In Silico Analysis
2.2.1. Molecular Docking
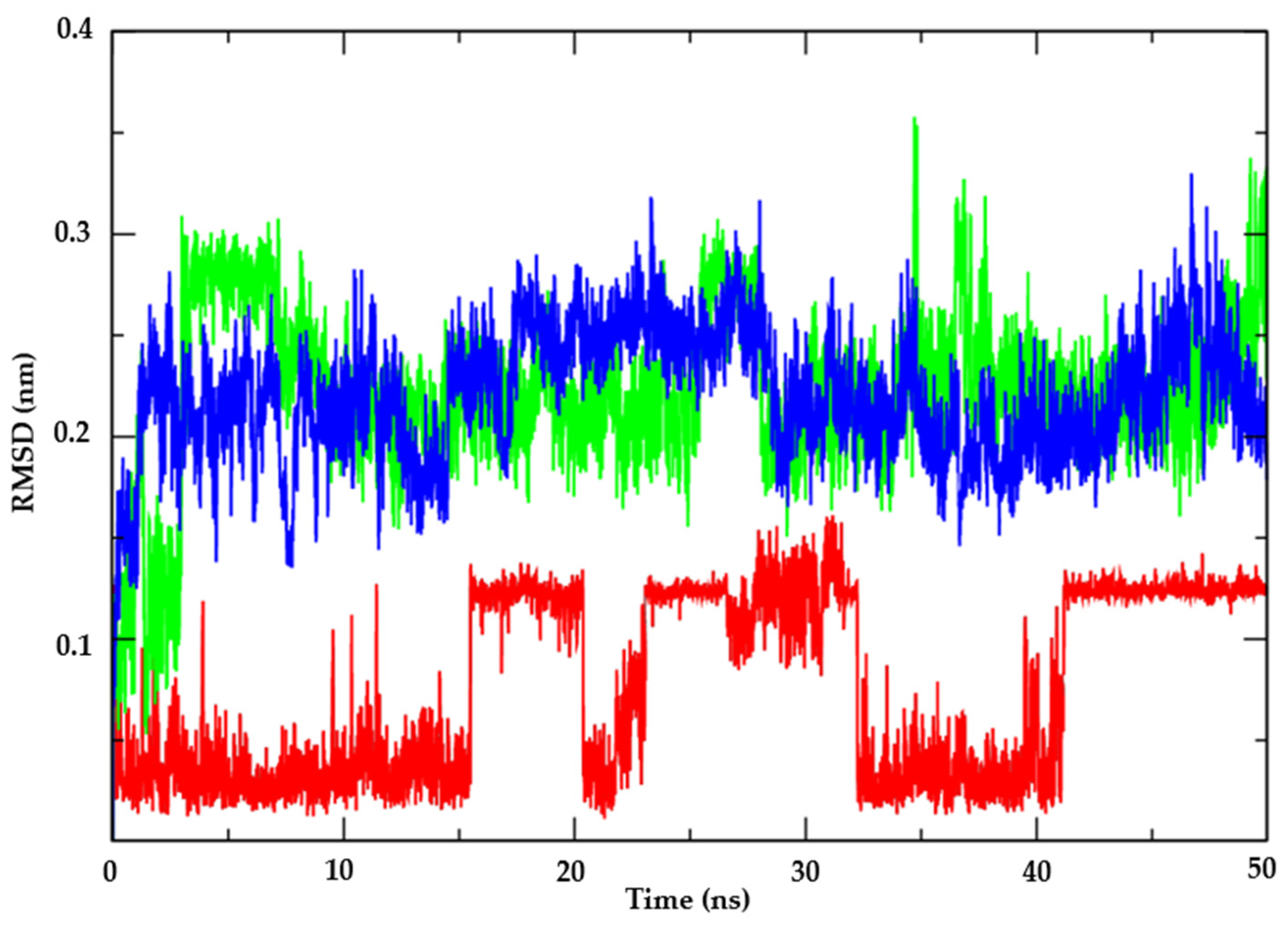
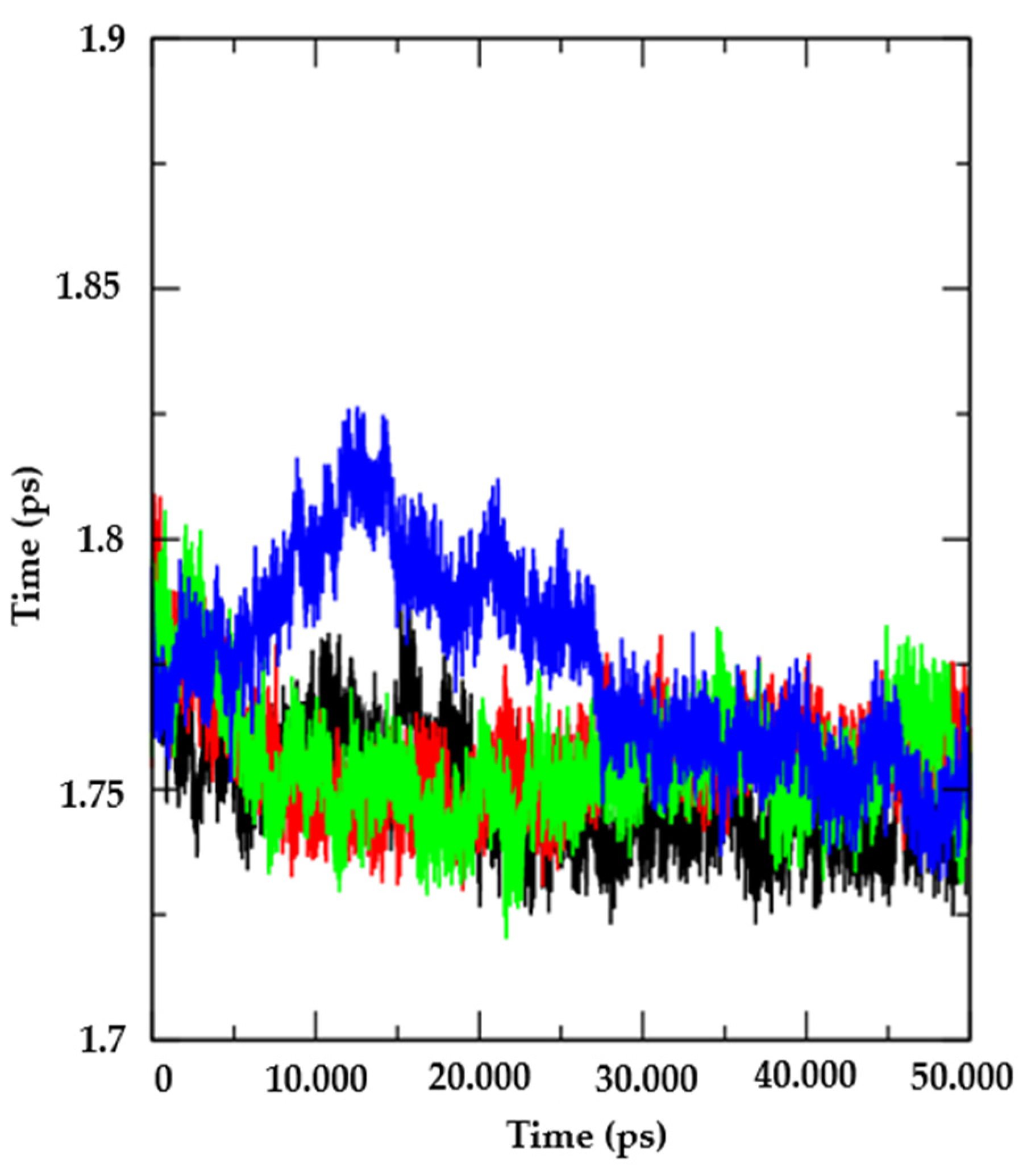
2.2.2. Molecular Dynamics
2.2.3. ADMET Predictions
2.3. In Vitro Analysis
2.3.1. Assessing the Minimum Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC)
2.3.2. Mode of Action
Ergosterol Assay
Sorbitol Assay (Cell-Wall-Related Effects)
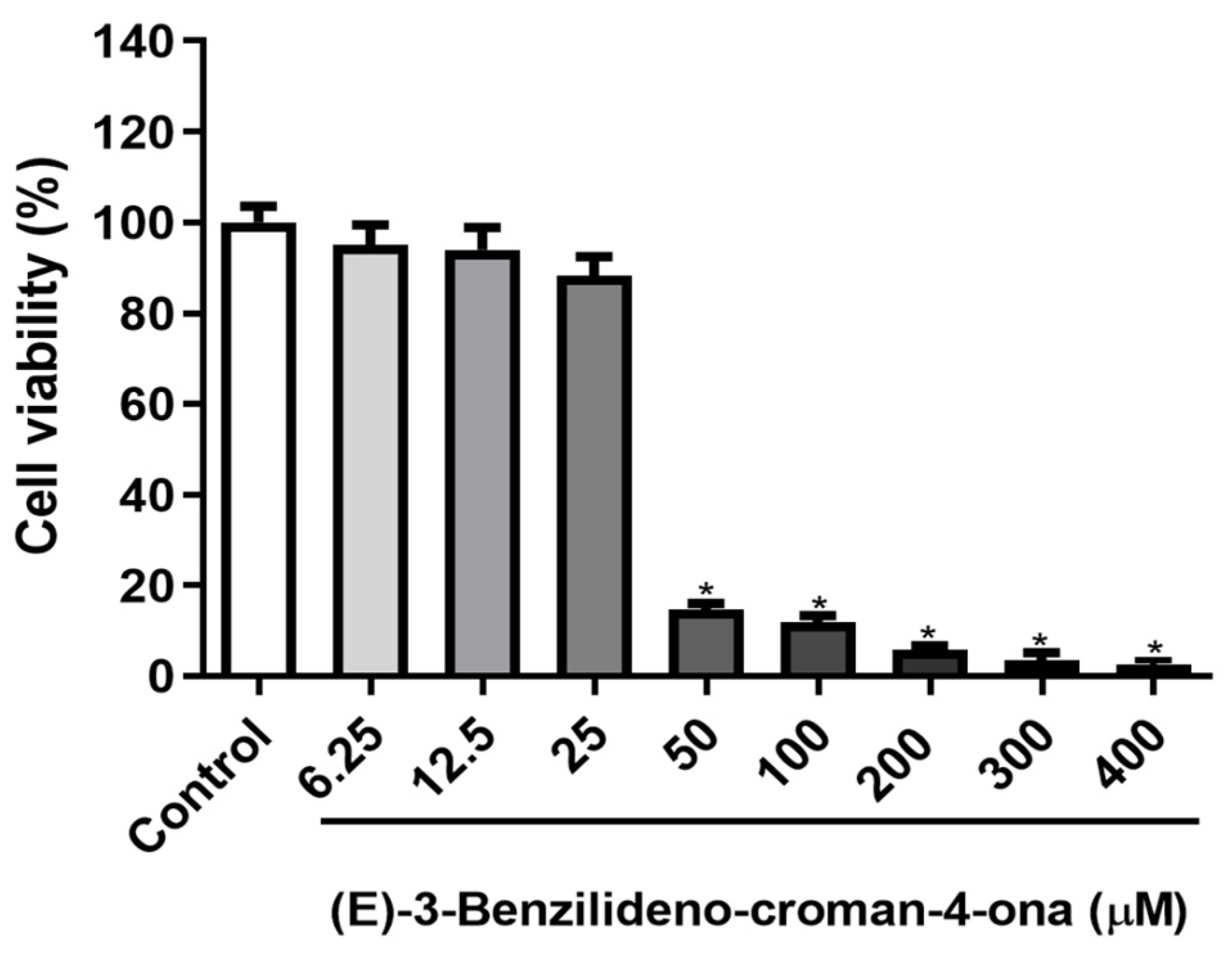
2.3.3. MTT Cell Viability Assay
3. Results
3.1. In Silico Analysis
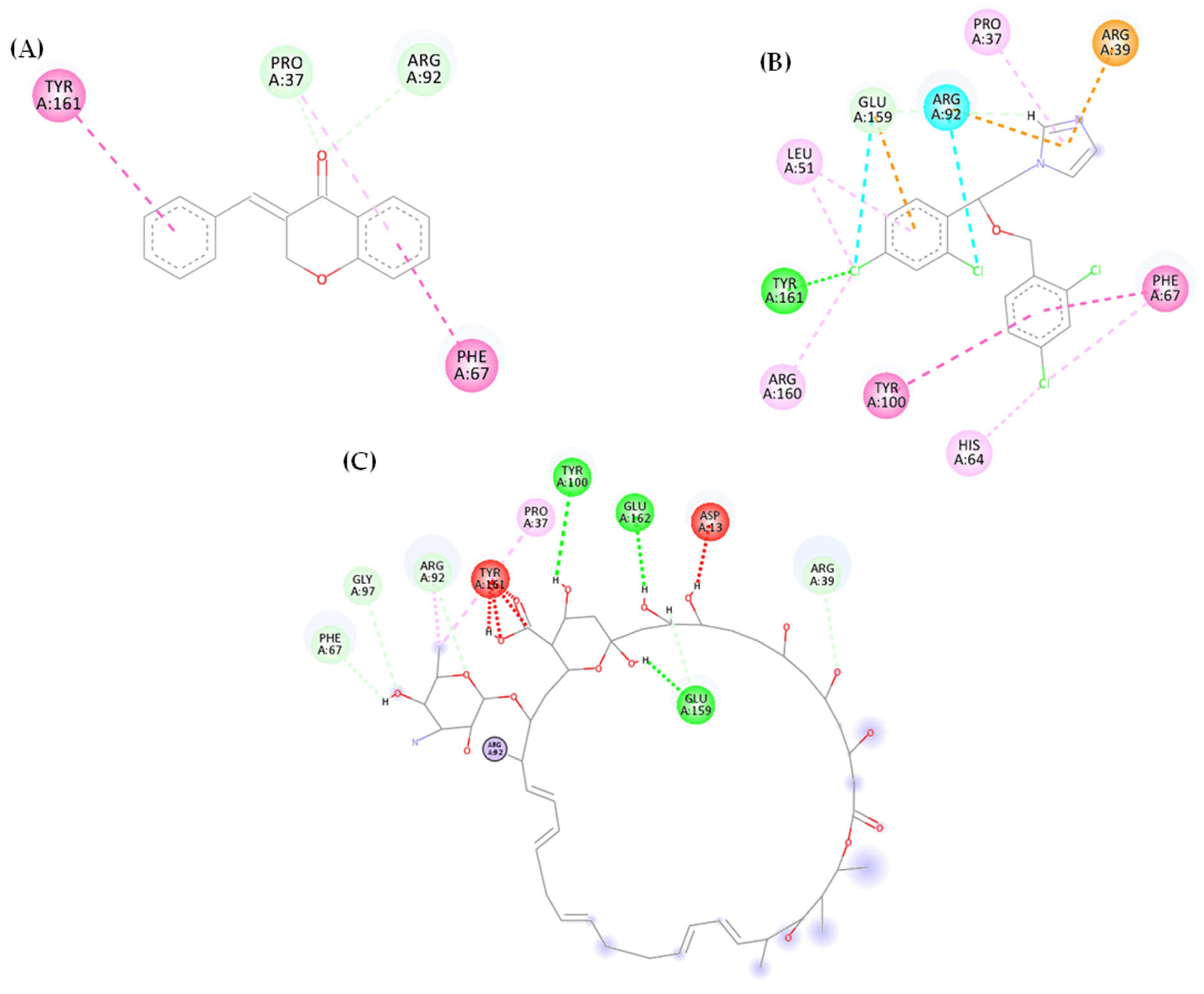
3.1.1. Molecular Docking
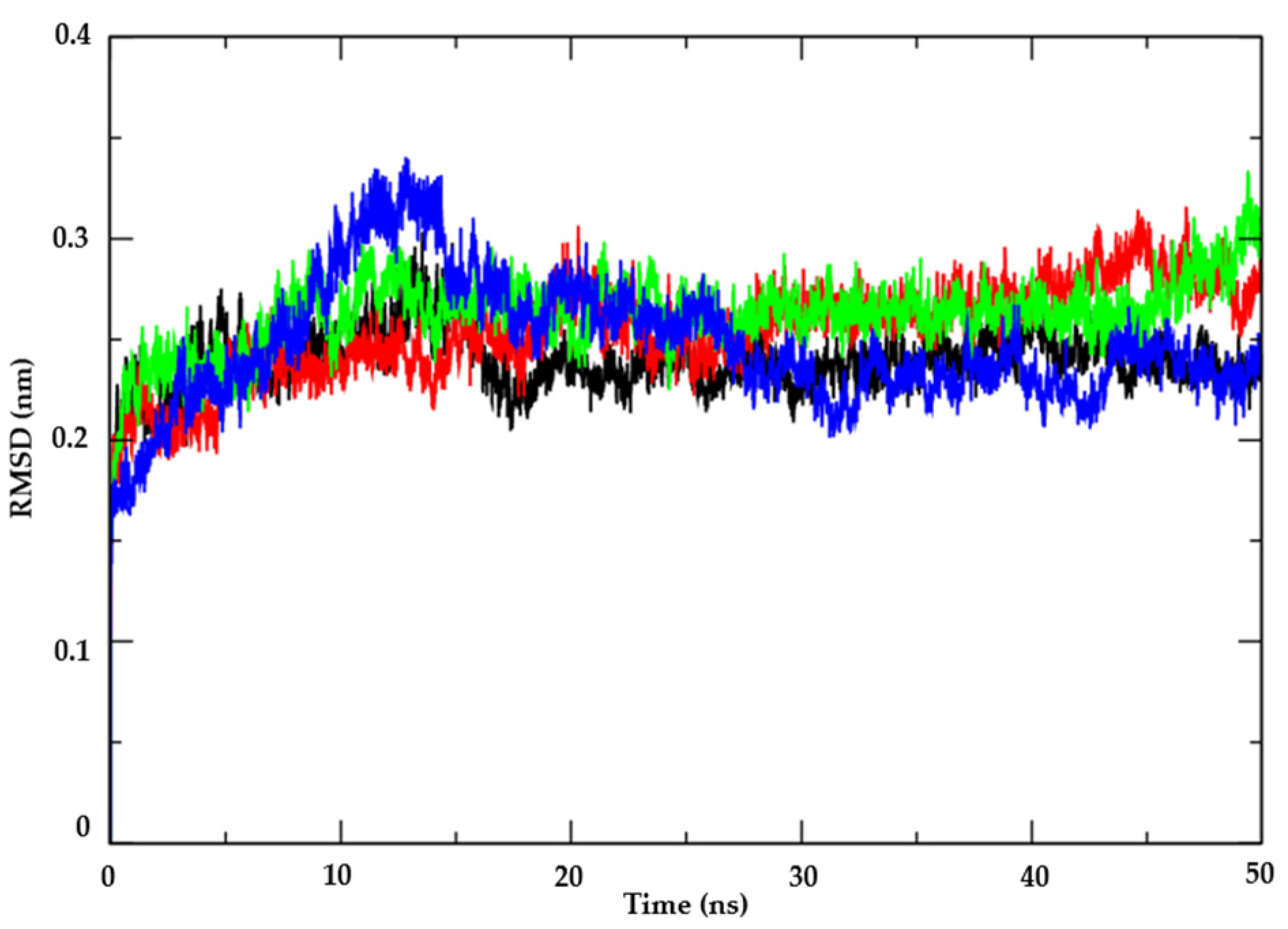
3.1.2. Molecular Dynamics
3.1.3. ADMET Predictions
3.2. In Vitro Analysis
3.2.1. Determination of MIC and MFC
3.2.2. Mechanism of Action on the Fungal Membrane and Cell Wall
Ergosterol Assay (Cell-Membrane-Related Effects)
Sorbitol Assay (Cell-Wall-Related Effects)
3.2.3. MTT Cell Viability Assay
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Colombo, A.L.; Guimarães, T.; Camargo, L.F.A.; Richtmann, R.; de Queiroz-Telles, F.; Salles, M.J.C.; da Cunha, C.A.; Yasuda, M.A.S.; Moretti, M.L.; Nucci, M. Brazilian Guidelines for the Management of Candidiasis—A Joint Meeting Report of Three Medical Societies: Sociedade Brasileira de Infectologia, Sociedade Paulista de Infectologia and Sociedade Brasileira de Medicina Tropical. Brazilian J. Infect. Dis. 2013, 17, 283–312. [Google Scholar] [CrossRef] [PubMed]
- Umme, H.; Hosakote, G.S.; Rudra, V.; Riyaz, A.M.O.; Atul, S. Candidiasis: A Fungal Infection-Current Challenges and Progress in Prevention and Treatment. Infect. Disord.—Drug Targets 2015, 15, 42–52. [Google Scholar] [CrossRef]
- Robbins, N.; Wright, G.D.; Cowen, L.E. Antifungal Drugs: The Current Armamentarium and Development of New Agents. In The Fungal Kingdom; American Society for Microbiology: Washington, DC, USA, 2017; Volume 4, pp. 903–922. ISBN 9781683670827. [Google Scholar]
- da Rocha, W.R.V.; Nunes, L.E.; Neves, M.L.R.; de Azevedo Ximenes, E.C.P.; de Azevedo Albuquerque, M.C.P. Gênero Candida-Fatores de Virulência, Epidemiologia, Candidíase e Mecanismos de Resistência. Res. Soc. Dev. 2021, 10, e43910414283. [Google Scholar] [CrossRef]
- Vila, T.; Sultan, A.S.; Montelongo-Jauregui, D.; Jabra-Rizk, M.A. Oral Candidiasis: A Disease of Opportunity. J. Fungi 2020, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Nobile, C.J.; Johnson, A.D. Candida Albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Carvalho, A.; Hong Nguyen, M.; Hedayati, M.T.; Netea, M.G.; Perlin, D.S.; Hoenigl, M. COVID-19-Associated Candidiasis (Cac): An Underestimated Complication in the Absence of Immunological Predispositions? J. Fungi 2020, 6, 211. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Posteraro, B.; Lass-Flörl, C. Antifungal Drug Resistance among Candida Species: Mechanisms and Clinical Impact. Mycoses 2015, 58, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S.; Rautemaa-Richardson, R.; Alastruey-Izquierdo, A. The Global Problem of Antifungal Resistance: Prevalence, Mechanisms, and Management. Lancet Infect. Dis. 2017, 17, e383–e392. [Google Scholar] [CrossRef] [PubMed]
- Marquez, L.; Quave, C.L. Prevalence and Therapeutic Challenges of Fungal Drug Resistance: Role for Plants in Drug Discovery. Antibiotics 2020, 9, 150. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, J.; Hua, H.; Yan, Z. Efficacy and Safety of Miconazole for Oral Candidiasis: A Systematic Review and Meta-analysis. Oral Dis. 2016, 22, 185–195. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Kontoyiannis, D.P. Resistance to Antifungal Drugs. Infect. Dis. Clin. N. Am. 2021, 35, 279–311. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, N.P. Emerging Fungal Infections: New Species, New Names, and Antifungal Resistance. Clin. Chem. 2022, 68, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Campoy, S.; Adrio, J.L. Antifungals. Biochem. Pharmacol. 2017, 133, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Van Daele, R.; Spriet, I.; Wauters, J.; Maertens, J.; Mercier, T.; Van Hecke, S.; Brüggemann, R. Antifungal Drugs: What Brings the Future? In Medical Mycology; Oxford University Press: New York, NY, USA, 2019; Volume 57, pp. S328–S343. [Google Scholar]
- Xavier, P.A.; Carvalho, J.S.; de Castro Penteado, R.; Lemos Filho, E.F.; Pereira, R.L.; de Almeida, B.A.D. Evidências Cientificas Sobre o Uso de Cromonas (Cromoglicatos) No Tratamento Da Asma Scientific Evidence on the Use of Chromones (Cromoglycates) in the Treatment of Asthma. Brazilian J. Health Rev. 2022, 5, 12519–12527. [Google Scholar] [CrossRef]
- Puhl, M.C.M.N.; Cortez, D.A.G.; Ueda-Nakamura, T.; Nakamura, C.V.; Filho, B.P.D. Antimicrobial Activity of Piper Gaudichaudianum Kuntze and Its Synergism with Different Antibiotics. Molecules 2011, 16, 9925–9938. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, A.; Matos, M.J.; Garrido, J.; Uriarte, E.; Borges, F. Chromone: A Valid Scaffold in Medicinal Chemistry. Chem. Rev. 2014, 114, 4960–4992. [Google Scholar] [CrossRef] [PubMed]
- Keri, R.S.; Budagumpi, S.; Pai, R.K.; Balakrishna, R.G. Chromones as a Privileged Scaffold in Drug Discovery: A Review. Eur. J. Med. Chem. 2014, 78, 340–374. [Google Scholar] [CrossRef] [PubMed]
- Emami, S.; Ghanbarimasir, Z. Recent Advances of Chroman-4-One Derivatives: Synthetic Approaches and Bioactivities. Eur. J. Med. Chem. 2015, 93, 539–563. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.R.; Alves, D.d.N.; de Castro, R.D.; Perez-Castillo, Y.; de Sousa, D.P. Synthesis of Coumarin and Homoisoflavonoid Derivatives and Analogs: The Search for New Antifungal Agents. Pharmaceuticals 2022, 15, 712. [Google Scholar] [CrossRef]
- da Silva Rivas, A.C.; Lopes, P.M.; de Azevedo Barros, M.M.; Costa Machado, D.C.; Alviano, C.S.; Alviano, D.S. Biological Activities of α-Pinene and β-Pinene Enantiomers. Molecules 2012, 17, 6305–6316. [Google Scholar]
- Hung, L.-H.; Guerquin, M.; Samudrala, R. GPU-QJ, a Fast Method for Calculating Root Mean Square Deviation (RMSD) after Optimal Superposition. BMC Res. Notes 2011, 4, 97. [Google Scholar] [CrossRef] [PubMed]
- Schneider, N.; Hindle, S.; Lange, G.; Klein, R.; Albrecht, J.; Briem, H.; Beyer, K.; Claußen, H.; Gastreich, M.; Lemmen, C. Substantial Improvements in Large-Scale Redocking and Screening Using the Novel HYDE Scoring Function. J. Comput. Aided. Mol. Des. 2012, 26, 701–723. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX. 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Popiołek, Ł.; Biernasiuk, A. Design, Synthesis, and in Vitro Antimicrobial Activity of Hydrazide–Hydrazones of 2-substituted Acetic Acid. Chem. Biol. Drug Des. 2016, 88, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, Z.N.; Farooq, F.; Musthafa, T.N.M.; Ahmad, A.; Khan, A.U. Synthesis, Characterization and Antimicrobial Evaluation of Novel Halopyrazole Derivatives. J. Saudi Chem. Soc. 2013, 17, 237–243. [Google Scholar] [CrossRef]
- Sousa, J.; Costa, A.; Leite, M.; Guerra, F.; Silva, V.; Menezes, C.; Pereira, F.; Lima, E. Antifungal Activity of Citral by Disruption of Ergosterol Biosynthesis in Fluconazole Resistant Candida Tropicalis. Int. J. Trop. Dis. Health 2016, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pantella, F.; Jesus, K. De ISSN 0102-5716 Veterinária e Zootecnia 175. Veterinária e Zootec. 2011, 18, 175–186. [Google Scholar]
- Alves, D.D.N.; Ferreira, A.R.; Duarte, A.B.S.; Melo, A.K.V.; De Sousa, D.P.; Castro, R.D. De Breakpoints for the Classification of Anti- Candida Compounds in Antifungal Screening. Biomed Res. Int. 2021, 2021, 3311. [Google Scholar] [CrossRef]
- Oliveira, A.P.; Guimarães, A.L.; Pacheco, A.G.M.; Araújo, C.S.; Oliveira Júnior, R.G.; Lavor, É.M.; Silva, M.G.; Araújo, E.C.C.; Mendes, R.L.; Rolim, L.A. Estudo Fitoquímico, Atividade Antimicrobiana e Citotóxica de Espécimes de Leonotis Nepetifolia LR (Br). Quim. Nova 2016, 39, 32–37. [Google Scholar]
- Guedes, I.A.; de Magalhães, C.S.; Dardenne, L.E. Receptor–Ligand Molecular Docking. Biophys. Rev. 2014, 6, 75–87. [Google Scholar] [CrossRef] [PubMed]
- da Nóbrega Alves, D.; Monteiro, A.F.M.; Andrade, P.N.; Lazarini, J.G.; Abílio, G.M.F.; Guerra, F.Q.S.; Scotti, M.T.; Scotti, L.; Rosalen, P.L.; de Castro, R.D. Docking Prediction, Antifungal Activity, Anti-Biofilm Effects on Candida Spp., and Toxicity against Human Cells of Cinnamaldehyde. Molecules 2020, 25, 5969. [Google Scholar] [CrossRef] [PubMed]
- Canteiro, G.D.; Camargo, C.C.; da Silva, E.G.; Fernandes, J.R.S.; do Vale, M.C.S.; Montesino, A.C. O Uso Da Terapia Fotodinâmica Na Candidíase Oral: Uma Revisão de Literatura. E-Acadêmica 2021, 2, e322377. [Google Scholar] [CrossRef]
- Silva, F.D.; Alves, F. Docking Molecular Como Processo de Verificação da Afinidade Entre Receptores Farmacológicos no Tratamento de Adenocarcinoma Gástrico. NBC-Periódico Científico Do Núcleo Biociências 2019, 9, 17. [Google Scholar]
- Silva, D.R.; Sardi, J.d.C.O.; Freires, I.A.; Silva, A.C.B.; Rosalen, P.L. In Silico Approaches for Screening Molecular Targets in Candida Albicans: A Proteomic Insight into Drug Discovery and Development. Eur. J. Pharmacol. 2019, 842, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, B.G.; Araújo, R.C.M.U. SAPT: Ligação de Hidrogênio Ou Interação de van Der Waals? Quim. Nova 2012, 35, 2002–2012. [Google Scholar] [CrossRef]
- Sinha, K.; Rule, G.S. The Structure of Thymidylate Kinase from Candida Albicans Reveals a Unique Structural Element. Biochemistry 2017, 56, 4360–4370. [Google Scholar] [CrossRef] [PubMed]
- de Barros, D.B.; e Lima, L.d.O.; Silva, L.A.; Fonseca, M.C.; Diniz-Neto, H.; da Silva Rocha, W.P.; de Medeiros Beltrão, G.V.; Castellano, L.R.C.; Guerra, F.Q.S.; da Silva, M.V. Efeito Antifúngico de α-Pineno Isolado e Em Associação Com Antifúngicos Frente Às Cepas de Candida Albicans. Res. Soc. Dev. 2022, 11, e58711427748. [Google Scholar]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Iwashima, M.; Tako, N.; Hayakawa, T.; Matsunaga, T.; Mori, J.; Saito, H. New Chromane Derivatives Isolated from the Brown Alga, Sargassum Micracanthum. Chem. Pharm. Bull. 2008, 56, 124–128. [Google Scholar] [CrossRef]
- Tamayo, L.V.; Santos, A.F.; Ferreira, I.P.; Santos, V.G.; Lopes, M.T.P.; Beraldo, H. Silver(I) Complexes with Chromone-Derived Hydrazones: Investigation on the Antimicrobial and Cytotoxic Effects. BioMetals 2017, 30, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Malefo, M.S.; Ramadwa, T.E.; Famuyide, I.M.; McGaw, L.J.; Eloff, J.N.; Sonopo, M.S.; Selepe, M.A. Synthesis and Antifungal Activity of Chromones and Benzoxepines from the Leaves of Ptaeroxylon Obliquum. J. Nat. Prod. 2020, 83, 2508–2517. [Google Scholar] [CrossRef] [PubMed]
- Chohan, Z.H.; Rauf, A.; Naseer, M.M.; Somra, M.A.; Supuran, C.T. Antibacterial, Antifungal and Cytotoxic Properties of Some Sulfonamide-Derived Chromones. J. Enzyme Inhib. Med. Chem. 2006, 21, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Prakash, O.; Kumar, R.; Parkash, V. Synthesis and Antifungal Activity of Some New 3-Hydroxy-2-(1-Phenyl-3-Aryl-4-Pyrazolyl) Chromones. Eur. J. Med. Chem. 2008, 43, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.Y.; Chang, C.-Y.; Liau, H.-H.; Lu, P.-J.; Chen, H.-L.; Yang, C.-N.; Li, H.-Y. Synthesis of 2-Styrylchromones as a Novel Class of Antiproliferative Agents Targeting Carcinoma Cells. Eur. J. Med. Chem. 2009, 44, 2552–2562. [Google Scholar] [CrossRef] [PubMed]
- Resende, E.A.; Pereira, R.J. CROMONAS E XANTONAS. Compost. Bioativos Veg. 2014, 82, 82–139. [Google Scholar]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal Drug Resistance: Evolution, Mechanisms and Impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Rezende, C.; Segura, R.; RIVA, S.B.M.; da Cruz Oliveira Castro, V. Mecanismos de Ação Dos Antifúngicos. Rev. unifev ciência Tecnol. 2017, 2, 316. [Google Scholar]
- Cerdeira, C.D.; Pereira de Araújo, M.; Ribeiro Jorge Ferreira, C.B.; Tranches Dias, A.L.; Pereira Lima Brigagão, M.R. Explorando Os Estresses Oxidativo e Nitrosativo Contra Fungos: Um Mecanismo Subjacente à Ação de Tradicionais Antifúngicos e Um Potencial Novo Alvo Terapêutico Na Busca Por Indutores Oriundos de Fontes Naturais. Rev. Colomb. Ciencias Químico-Farmacéuticas 2021, 50, 100–157. [Google Scholar] [CrossRef]
- Oliveira-Tavares, N.N.; Lemos, A.S.; Pereira, A.P.O.; Fabri, R.L.; Chedier, L.M. Atividade Antifúngica Do Látex de Jatropha Multifida L.(Euphorbiaceae) e de Lupenona Isolada de Suas Folhas. Rev. Virtual Química 2019, 11, 1579–1590. [Google Scholar] [CrossRef]
- Vieira, A.J.H.; Santos, J.I. Mecanismos de Resistência de Candida Albicans Aos Antifúngicos Anfotericina B, Fluconazol e Caspofungina. RBAC 2017, 49, 235–239. [Google Scholar] [CrossRef]
- Guerra, F.Q.S.; Araújo, R.S.A.d.; Sousa, J.P.d.; Pereira, F.d.O.; Mendonça-Junior, F.J.B.; Barbosa-Filho, J.M.; de Oliveira Lima, E. Evaluation of Antifungal Activity and Mode of Action of New Coumarin Derivative, 7-Hydroxy-6-Nitro-2h-1-Benzopyran-2-One, against Aspergillus Spp. Evid.-Based Complement. Altern. Med. 2015, 2015, 925096. [Google Scholar] [CrossRef]
- Vieira, F.; Nascimento, T. Resistência a Fármacos Antifúngicos Por Candida e Abordagem Terapêutica. Rev. Port. Farmacoter. 2017, 9, 29–36. [Google Scholar]
- de Oliveira, L.; Silva, L.A.; Fonseca, M.C.; Diniz-Neto, H.; de Oliveira Lima, E.; Barbosa Filho, J.M.; Tavares, J.F.; da Silva-Rocha, W.P.; Guerra, F.Q.S. Efeito Inibitório de Di-Hidrojasmona Frente Cepas de Candida Spp. Fluconazol Resistentes. Res. Soc. Dev. 2021, 10, e440101523110. [Google Scholar]
- Deberaldini, M.G.; Santos, J.L. Infecções Fúngicas Invasivas: Aspectos Gerais e Tratamento: Invasive Fungal Infections: An Overview and Treatment. Ulakes J. Med. 2021, 1, 209–221. [Google Scholar]
- De Souza, M.M.; Pereira, M.A.; Ardenghi, J.V.; Mora, T.C.; Bresciani, L.F.; Yunes, R.A.; Delle Monache, F.; Cechinel-Filho, V. Filicene Obtained from Adiantum Cuneatum Interacts with the Cholinergic, Dopaminergic, Glutamatergic, GABAergic, and Tachykinergic Systems to Exert Antinociceptive Effect in Mice. Pharmacol. Biochem. Behav. 2009, 93, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Langer, L.T.A.; Staudt, K.J.; do Carmo, R.L.; Alves, I.A. Biofilmes Em Infecção Por Candida: Uma Revisão Da Literatura Biofilmes in Infection by Candida: A Review of the Literature. Rev. Interdiscip. Ciências Saúde Biológicas 2018, 2, 1–15. [Google Scholar]
- Katopodi, A.; Tsotsou, E.; Iliou, T.; Deligiannidou, G.-E.; Pontiki, E.; Kontogiorgis, C.; Tsopelas, F.; Detsi, A. Synthesis, Bioactivity, Pharmacokinetic and Biomimetic Properties of Multi-Substituted Coumarin Derivatives. Molecules 2021, 26, 5999. [Google Scholar] [CrossRef]
- Olejarz, W.; Wrzosek, M.; Jóźwiak, M.; Grosicka-Maciąg, E.; Roszkowski, P.; Filipek, A.; Cychol, A.; Nowicka, G.; Struga, M. Synthesis and Anticancer Effects of α-Lipoic Ester of Alloxanthoxyletin. Med. Chem. Res. 2019, 28, 788–796. [Google Scholar] [CrossRef]


| Substances | Molecular Targets | ||||
|---|---|---|---|---|---|
| 1EQP | 4MAI | 4QUV | 5TZ1 | 5UIV | |
| (E)-benzylidene-chroman-4-one | −66.4624 | −62.0636 | −67.2416 | −56.1563 | −102.589 |
| Miconazole | −106.367 | −91.5091 | −98.4458 | −82.8594 | −133.473 |
| Nystatin | −174.305 | −157.953 | −210.923 | −75.3106 | −91.3965 |
| PDB Ligand | - | - | - | −85.3911 | - |
| Energy | Thymidylate Kinase (PDB: 5UIV) | ||
|---|---|---|---|
| AR25 | Miconazole | Nystatin | |
| Coulomb (C) | −51.4842 | −48.0814 | −223.375 |
| Lennard-Jones (LJ) | −97.2097 | −188.998 | −182.996 |
| ID | GI 1 | BBB 2 | Lipinski Violations | Mutagenicity | Tumorigenicity | Reproductive Effect | Irritant Skin Effect |
|---|---|---|---|---|---|---|---|
| AR25 | High | Yes | 0 | None | None | None | None |
| ID | CYP | ||||
|---|---|---|---|---|---|
| CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 | |
| AR25 | Yes | Yes | No | No | No |
| (E)-3-Benzylidene-Chroman-4-One | Nystatin | |||||
|---|---|---|---|---|---|---|
| Strains | MIC | MFC | MFC/ MIC | MIC | MFC | MFC/ MIC |
| C. albicans ATCC 10231 | 62.5 (264.52) | 62.5 (264.52) | 1 | 3 (3.23) | 3 (3.23) | 1 |
| C. krusei ATCC 6258 | 62.5 (264.52) | 62.5 (264.52) | 1 | 3 (3.23) | 3 (3.23) | 1 |
| C. glabrata ATCC 90030 | 250.0 (1058.11) | 250.0 (1058.11) | 1 | 1.5 (1.61) | 1.5 (1.61) | 1 |
| C. tropicalis ATCC 750 | 250.0 (1058.11) | 250.0 (1058.11) | 1 | 1.5 (1.61) | 1.5 (1.61) | 1 |
| C. parapsilosis ATCC 22019 | 500.0 (2116.22) | 1000.0 (423.44) | 2 | 1.5 (1.61) | 1.5 (1.61) | 1 |
| C. albicans ATCC 60193 | 1000.0 (4232.44) | 1000.0 (4232.44) | 1 | 1.5 (1.61) | 1.5 (1.61) | 1 |
| C. albicans ATCC 90028 | 500.0 (2116.22) | 500.0 (2116.22) | 1 | 1.5 (1.61) | 1.5 (1.61) | 1 |
| (E)-3-Benzylidene-Chroman-4-One | Nystatin | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Concentration (μg/mL) | C. albicans ATCC 90028 | C. krusei ATCC 6258 | Concentration (μg/mL) | C. albicans ATCC 90028 | C. krusei ATCC 6258 | ||||
| Absence of Ergosterol | Presence of Ergosterol | Absence of Ergosterol | Presence of Ergosterol | Absence of Ergosterol | Presence of Ergosterol | Absence of Ergosterol | Presence of Ergosterol | ||
| 1000 | − | − | − | − | 48 | − | − | − | − |
| 500 | − | + | − | + | 24 | − | + | − | + |
| 250 | + | + | − | + | 12 | − | + | − | + |
| 125 | + | + | − | + | 6 | − | + | − | + |
| 62.5 | + | + | − | + | 3 | − | + | − | + |
| 31.25 | + | + | + | + | 1.5 | − | + | + | + |
| 15.62 | + | + | + | + | 0.75 | + | + | + | + |
| 7.81 | + | + | + | + | 0.37 | + | + | + | + |
| (E)-3-Benzylidene-Chroman-4-One | Caspofungin® | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Concentration (μg/mL) | C. albicans ATCC 90028 | C. krusei ATCC 6258 | Concentration (μg/mL) | C. albicans ATCC 90028 | C. krusei ATCC 6258 | ||||
| Absence of Ergosterol | Presence of Ergosterol | Absence of Ergosterol | Presence of Ergosterol | Absence of Ergosterol | Presence of Ergosterol | Absence of Ergosterol | Presence of Ergosterol | ||
| 1000 | − | − | − | − | 4 | − | − | − | − |
| 500 | − | − | − | − | 2 | − | + | − | − |
| 250 | + | + | − | − | 1 | − | + | − | + |
| 125 | + | + | − | − | 0.5 | − | + | − | + |
| 62.5 | + | + | − | − | 0.25 | − | + | − | + |
| 31.25 | + | + | + | + | 0.125 | − | + | + | + |
| 15.62 | + | + | + | + | 0.062 | + | + | + | + |
| 7.81 | + | + | + | + | 0.031 | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Araújo, G.R.; Costa, P.C.Q.G.d.; Nogueira, P.L.; Alves, D.d.N.; Ferreira, A.R.; da Silva, P.R.; de Andrade, J.C.; de Sousa, N.F.; Loureiro, P.B.A.; Sobral, M.V.; et al. In Silico and In Vitro Evaluation of the Antifungal Activity of a New Chromone Derivative against Candida spp. BioTech 2024, 13, 16. https://doi.org/10.3390/biotech13020016
Araújo GR, Costa PCQGd, Nogueira PL, Alves DdN, Ferreira AR, da Silva PR, de Andrade JC, de Sousa NF, Loureiro PBA, Sobral MV, et al. In Silico and In Vitro Evaluation of the Antifungal Activity of a New Chromone Derivative against Candida spp. BioTech. 2024; 13(2):16. https://doi.org/10.3390/biotech13020016
Chicago/Turabian StyleAraújo, Gleycyelly Rodrigues, Palloma Christine Queiroga Gomes da Costa, Paula Lima Nogueira, Danielle da Nóbrega Alves, Alana Rodrigues Ferreira, Pablo R. da Silva, Jéssica Cabral de Andrade, Natália F. de Sousa, Paulo Bruno Araujo Loureiro, Marianna Vieira Sobral, and et al. 2024. "In Silico and In Vitro Evaluation of the Antifungal Activity of a New Chromone Derivative against Candida spp." BioTech 13, no. 2: 16. https://doi.org/10.3390/biotech13020016
APA StyleAraújo, G. R., Costa, P. C. Q. G. d., Nogueira, P. L., Alves, D. d. N., Ferreira, A. R., da Silva, P. R., de Andrade, J. C., de Sousa, N. F., Loureiro, P. B. A., Sobral, M. V., Sousa, D. P., Scotti, M. T., de Castro, R. D., & Scotti, L. (2024). In Silico and In Vitro Evaluation of the Antifungal Activity of a New Chromone Derivative against Candida spp. BioTech, 13(2), 16. https://doi.org/10.3390/biotech13020016











