Effect of Chitosan Degradation Products, Glucosamine and Chitosan Oligosaccharide, on Osteoclastic Differentiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Osteoclastic Differentiation
2.3. Cell Viability Assay
2.4. TRAP Enzyme Activity Assay
2.5. TRAP Staining
2.6. Actin Staining
2.7. Statistical Analysis
3. Results
3.1. Effects of GlcN and COS on Cell Viability of Human PBMCs and Murine RAW264 Cells
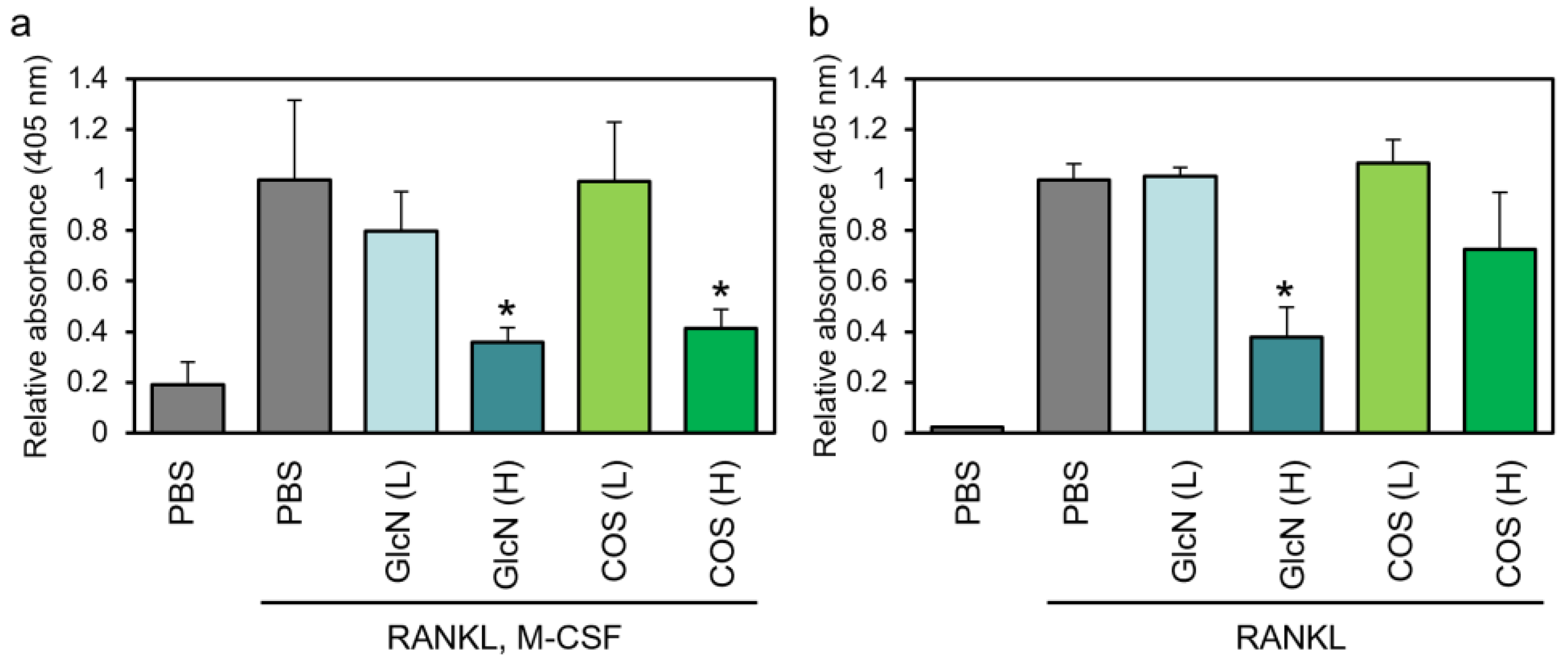
3.2. Effects of GlcN and COS on TRAP Enzyme Activity of Human PBMCs and Murine RAW264 Cells
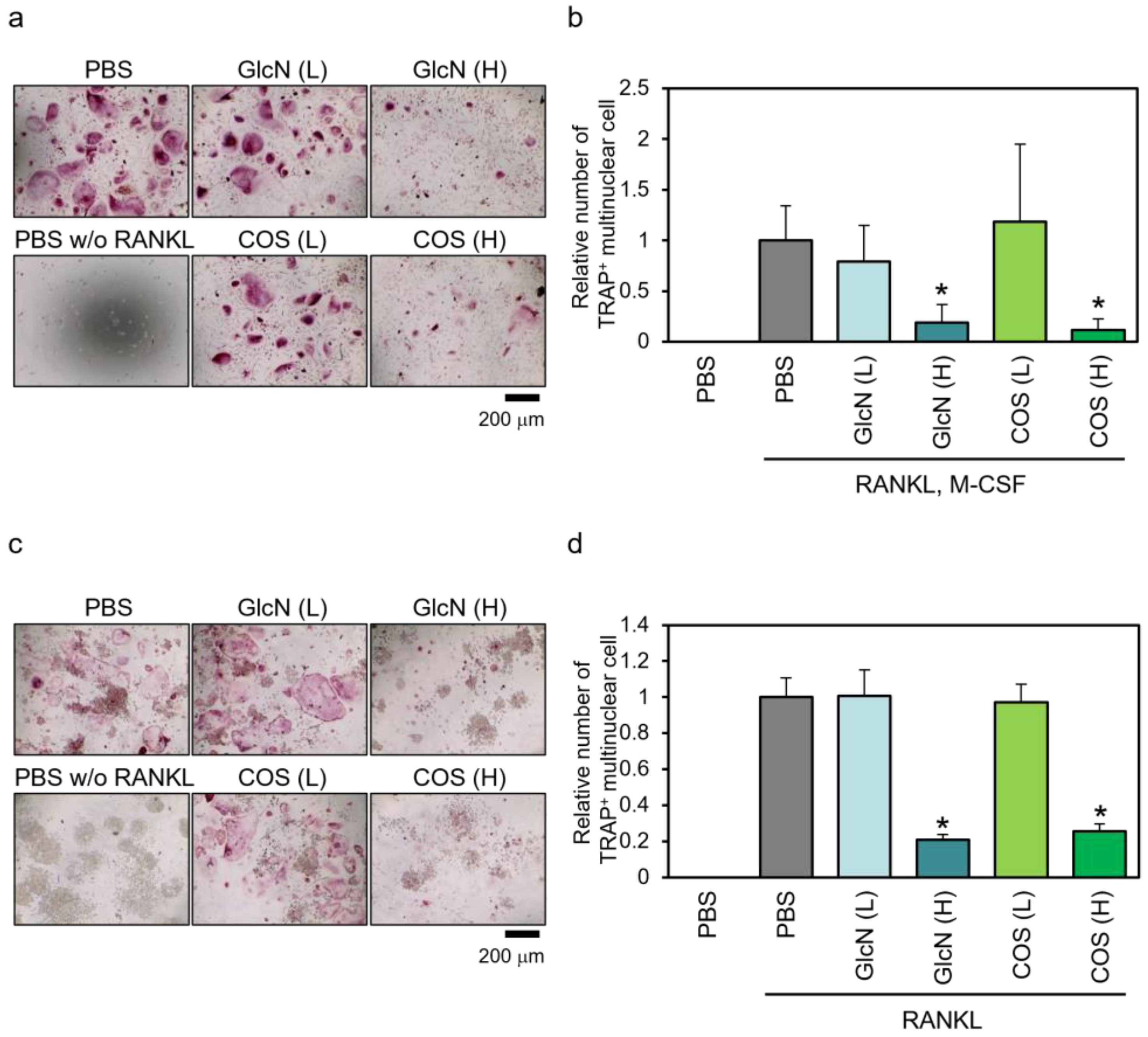
3.3. Effects of GlcN and COS on TRAP-Positive Multinuclear Cell Formation of Human PBMCs and Murine RAW264 Cells
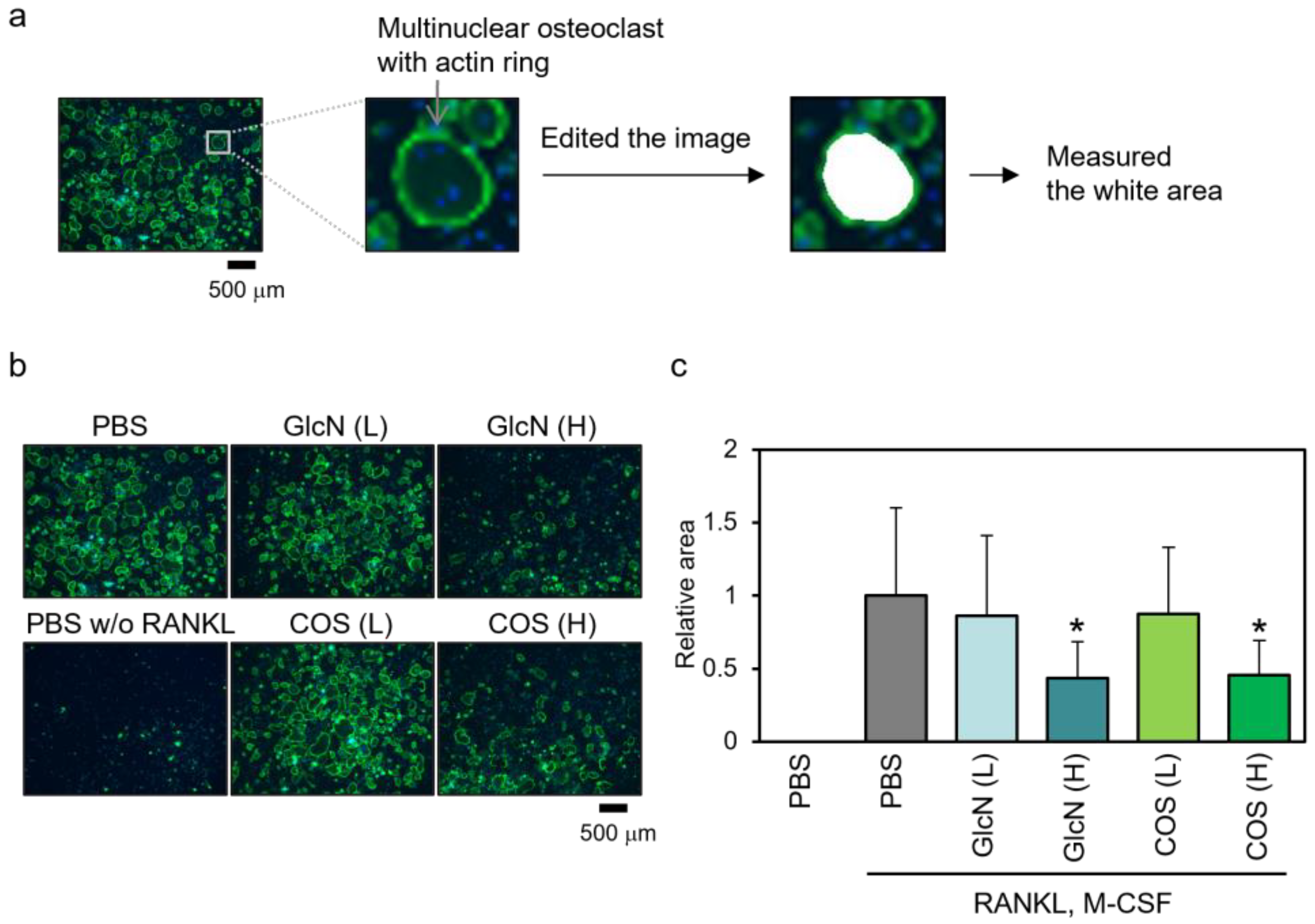
3.4. Effects of GlcN and COS on Actin Ring Formation of Human PBMCs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Valachova, K.; Soltes, L. Versatile Use of Chitosan and Hyaluronan in Medicine. Molecules 2021, 26, 1195. [Google Scholar] [CrossRef]
- Desai, N.; Rana, D.; Salave, S.; Gupta, R.; Patel, P.; Karunakaran, B.; Sharma, A.; Giri, J.; Benival, D.; Kommineni, N. Chitosan: A Potential Biopolymer in Drug Delivery and Biomedical Applications. Pharmaceutics 2023, 15, 1313. [Google Scholar] [CrossRef]
- Gholap, A.D.; Rojekar, S.; Kapare, H.S.; Vishwakarma, N.; Raikwar, S.; Garkal, A.; Mehta, T.A.; Jadhav, H.; Prajapati, M.K.; Annapure, U. Chitosan scaffolds: Expanding horizons in biomedical applications. Carbohydr. Polym. 2024, 323, 121394. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Tian, X.; Fan, J.; Tong, H.; Ao, Q.; Wang, X. Chitosans for Tissue Repair and Organ Three-Dimensional (3D) Bioprinting. Micromachines 2019, 10, 765. [Google Scholar] [CrossRef]
- Aguilar, A.; Zein, N.; Harmouch, E.; Hafdi, B.; Bornert, F.; Offner, D.; Clauss, F.; Fioretti, F.; Huck, O.; Benkirane-Jessel, N.; et al. Application of Chitosan in Bone and Dental Engineering. Molecules 2019, 24, 3009. [Google Scholar] [CrossRef]
- Lauritano, D.; Limongelli, L.; Moreo, G.; Favia, G.; Carinci, F. Nanomaterials for Periodontal Tissue Engineering: Chitosan-Based Scaffolds. A Systematic Review. Nanomaterials 2020, 10, 605. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Ren, Q.; Li, Z.; Zhang, L. Chitosan-Based Biomimetically Mineralized Composite Materials in Human Hard Tissue Repair. Molecules 2020, 25, 4785. [Google Scholar] [CrossRef] [PubMed]
- Ressler, A. Chitosan-Based Biomaterials for Bone Tissue Engineering Applications: A Short Review. Polymers 2022, 14, 3430. [Google Scholar] [CrossRef]
- Robling, A.G.; Castillo, A.B.; Turner, C.H. Biomechanical and molecular regulation of bone remodeling. Annu. Rev. Biomed. Eng. 2006, 8, 455–498. [Google Scholar] [CrossRef]
- Ono, T.; Nakashima, T. Recent advances in osteoclast biology. Histochem. Cell Biol. 2018, 149, 325–341. [Google Scholar] [CrossRef]
- Gyori, D.S.; Mocsai, A. Osteoclast Signal Transduction During Bone Metastasis Formation. Front. Cell Dev. Biol. 2020, 8, 507. [Google Scholar] [CrossRef]
- Goncalves, C.; Ferreira, N.; Lourenco, L. Production of Low Molecular Weight Chitosan and Chitooligosaccharides (COS): A Review. Polymers 2021, 13, 2466. [Google Scholar] [CrossRef]
- Rochet, N.; Balaguer, T.; Boukhechba, F.; Laugier, J.P.; Quincey, D.; Goncalves, S.; Carle, G.F. Differentiation and activity of human preosteoclasts on chitosan enriched calcium phosphate cement. Biomaterials 2009, 30, 4260–4267. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Miyahara, T.; Tezuka, Y.; Watanabe, M.; Nemoto, N.; Seto, H.; Kadota, S. The effect of low molecular weight Chitosan on bone resorption in vitro and in vivo. Phytomedicine 1999, 6, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.L.; Xie, Z.J.; Weng, S.J.; Wu, Z.Y.; Li, H.; Tao, Z.S.; Boodhun, V.; Yan, D.Y.; Shen, Z.J.; Tang, J.H.; et al. Chitosan oligosaccharide promotes osteoclast formation by stimulating the activation of MAPK and AKT signaling pathways. J. Biomater. Sci. Polym. Ed. 2018, 29, 1207–1218. [Google Scholar] [CrossRef]
- Takeuchi, T.; Sugimoto, A.; Imazato, N.; Tamura, M.; Nakatani, S.; Kobata, K.; Arata, Y. Glucosamine Suppresses Osteoclast Differentiation through the Modulation of Glycosylation Including O-GlcNAcylation. Biol. Pharm. Bull. 2017, 40, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Smith, W.; Hao, D. Overview of RAW264.7 for osteoclastogensis study: Phenotype and stimuli. J. Cell Mol. Med. 2019, 23, 3077–3087. [Google Scholar] [CrossRef]
- Lampiasi, N.; Russo, R.; Kireev, I.; Strelkova, O.; Zhironkina, O.; Zito, F. Osteoclasts Differentiation from Murine RAW 264.7 Cells Stimulated by RANKL: Timing and Behavior. Biology 2021, 10, 117. [Google Scholar] [CrossRef]
- Raschke, W.C.; Baird, S.; Ralph, P.; Nakoinz, I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 1978, 15, 261–267. [Google Scholar] [CrossRef]
- Takeuchi, T.; Nagasaka, M.; Shimizu, M.; Tamura, M.; Arata, Y. N-acetylglucosamine suppresses osteoclastogenesis in part through the promotion of O-GlcNAcylation. Bone Rep. 2016, 5, 15–21. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Guo, K.; Liu, Z.L.; Wang, W.C.; Xu, W.F.; Yu, S.Q.; Zhang, S.Y. Chitosan oligosaccharide inhibits skull resorption induced by lipopolysaccharides in mice. BMC Oral Health 2019, 19, 263. [Google Scholar] [CrossRef]
- Lambertini, E.; Penolazzi, L.; Pandolfi, A.; Mandatori, D.; Sollazzo, V.; Piva, R. Human osteoclasts/osteoblasts 3D dynamic co-culture system to study the beneficial effects of glucosamine on bone microenvironment. Int. J. Mol. Med. 2021, 47, 57. [Google Scholar] [CrossRef]
- Mourya, V.K.; Inamdar, N.N.; Choudhari, Y.M. Chitooligosaccharides: Synthesis, characterization and applications. Polym. Sci. Ser. A 2011, 53, 583–612. [Google Scholar] [CrossRef]
- Garrett, I.R.; Boyce, B.F.; Oreffo, R.O.; Bonewald, L.; Poser, J.; Mundy, G.R. Oxygen-derived free radicals stimulate osteoclastic bone resorption in rodent bone in vitro and in vivo. J. Clin. Investig. 1990, 85, 632–639. [Google Scholar] [CrossRef]
- Suda, N.; Morita, I.; Kuroda, T.; Murota, S. Participation of oxidative stress in the process of osteoclast differentiation. Biochim. Biophys. Acta 1993, 1157, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Yu, G.; Hirata, T.; Terao, J.; Lin, H. Antioxidative activities of several marine polysaccharides evaluated in a phosphatidylcholine-liposomal suspension and organic solvents. Biosci. Biotechnol. Biochem. 1998, 62, 206–209. [Google Scholar] [CrossRef] [PubMed]
- Xing, R.; Liu, S.; Guo, Z.; Yu, H.; Wang, P.; Li, C.; Li, Z.; Li, P. Relevance of molecular weight of chitosan and its derivatives and their antioxidant activities in vitro. Bioorg. Med. Chem. 2005, 13, 1573–1577. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shu, R.; Shao, J.; Xu, G.; Gu, X. Radical scavenging activity of chitooligosaccharide with different molecular weights. Eur. Food Res. Technol. 2005, 222, 36–40. [Google Scholar] [CrossRef]
- Kulig, D.; Krol-Kilinska, Z.; Bobak, L.; Zarowska, B.; Jarmoluk, A.; Zimoch-Korzycka, A. Functional Properties of Chitosan Oligomers Obtained by Enzymatic Hydrolysis. Polymers 2023, 15, 3801. [Google Scholar] [CrossRef] [PubMed]
- Paneque, A.; Fortus, H.; Zheng, J.; Werlen, G.; Jacinto, E. The Hexosamine Biosynthesis Pathway: Regulation and Function. Genes 2023, 14, 933. [Google Scholar] [CrossRef]
- Nakajima, K.; Ito, E.; Ohtsubo, K.; Shirato, K.; Takamiya, R.; Kitazume, S.; Angata, T.; Taniguchi, N. Mass isotopomer analysis of metabolically labeled nucleotide sugars and N- and O-glycans for tracing nucleotide sugar metabolisms. Mol. Cell. Proteom. 2013, 12, 2468–2480. [Google Scholar] [CrossRef]
- Li, Y.N.; Chen, C.W.; Trinh-Minh, T.; Zhu, H.; Matei, A.E.; Gyorfi, A.H.; Kuwert, F.; Hubel, P.; Ding, X.; Manh, C.T.; et al. Dynamic changes in O-GlcNAcylation regulate osteoclast differentiation and bone loss via nucleoporin 153. Bone Res. 2022, 10, 51. [Google Scholar] [CrossRef]
- Takeuchi, T.; Horimoto, Y.; Oyama, M.; Nakatani, S.; Kobata, K.; Tamura, M.; Arata, Y.; Hatanaka, T. Osteoclast Differentiation Is Suppressed by Increased O-GlcNAcylation Due to Thiamet G Treatment. Biol. Pharm. Bull. 2020, 43, 1501–1505. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, H.S.; Lee, S.; Min, K.Y.; Choi, W.S.; You, J.S. Hexosamine Biosynthetic Pathway-Derived O-GlcNAcylation Is Critical for RANKL-Mediated Osteoclast Differentiation. Int. J. Mol. Sci. 2021, 22, 8888. [Google Scholar] [CrossRef]
- Block, J.A.; Oegema, T.R.; Sandy, J.D.; Plaas, A. The effects of oral glucosamine on joint health: Is a change in research approach needed? Osteoarthr. Cartil. 2010, 18, 5–11. [Google Scholar] [CrossRef]
- Reginster, J.Y.; Neuprez, A.; Lecart, M.P.; Sarlet, N.; Bruyere, O. Role of glucosamine in the treatment for osteoarthritis. Rheumatol. Int. 2012, 32, 2959–2967. [Google Scholar] [CrossRef]
- Grigorian, A.; Araujo, L.; Naidu, N.N.; Place, D.J.; Choudhury, B.; Demetriou, M. N-acetylglucosamine inhibits T-helper 1 (Th1)/T-helper 17 (Th17) cell responses and treats experimental autoimmune encephalomyelitis. J. Biol. Chem. 2011, 286, 40133–40141. [Google Scholar] [CrossRef] [PubMed]
- Sy, M.; Newton, B.L.; Pawling, J.; Hayama, K.L.; Cordon, A.; Yu, Z.; Kuhle, J.; Dennis, J.W.; Brandt, A.U.; Demetriou, M. N-acetylglucosamine inhibits inflammation and neurodegeneration markers in multiple sclerosis: A mechanistic trial. J. Neuroinflamm. 2023, 20, 209. [Google Scholar] [CrossRef] [PubMed]
- Asai, H.; Nakatani, S.; Kato, T.; Shimizu, T.; Mano, H.; Kobata, K.; Wada, M. Glucosamines Attenuate Bone Loss Due to Menopause by Regulating Osteoclast Function in Ovariectomized Mice. Biol. Pharm. Bull. 2016, 39, 1035–1041. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takeuchi, T.; Oyama, M.; Hatanaka, T. Effect of Chitosan Degradation Products, Glucosamine and Chitosan Oligosaccharide, on Osteoclastic Differentiation. BioTech 2024, 13, 6. https://doi.org/10.3390/biotech13010006
Takeuchi T, Oyama M, Hatanaka T. Effect of Chitosan Degradation Products, Glucosamine and Chitosan Oligosaccharide, on Osteoclastic Differentiation. BioTech. 2024; 13(1):6. https://doi.org/10.3390/biotech13010006
Chicago/Turabian StyleTakeuchi, Tomoharu, Midori Oyama, and Tomomi Hatanaka. 2024. "Effect of Chitosan Degradation Products, Glucosamine and Chitosan Oligosaccharide, on Osteoclastic Differentiation" BioTech 13, no. 1: 6. https://doi.org/10.3390/biotech13010006
APA StyleTakeuchi, T., Oyama, M., & Hatanaka, T. (2024). Effect of Chitosan Degradation Products, Glucosamine and Chitosan Oligosaccharide, on Osteoclastic Differentiation. BioTech, 13(1), 6. https://doi.org/10.3390/biotech13010006





