A Freeze-Dried Cranberry Powder Consistently Enhances SCFA Production and Lowers Abundance of Opportunistic Pathogens In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
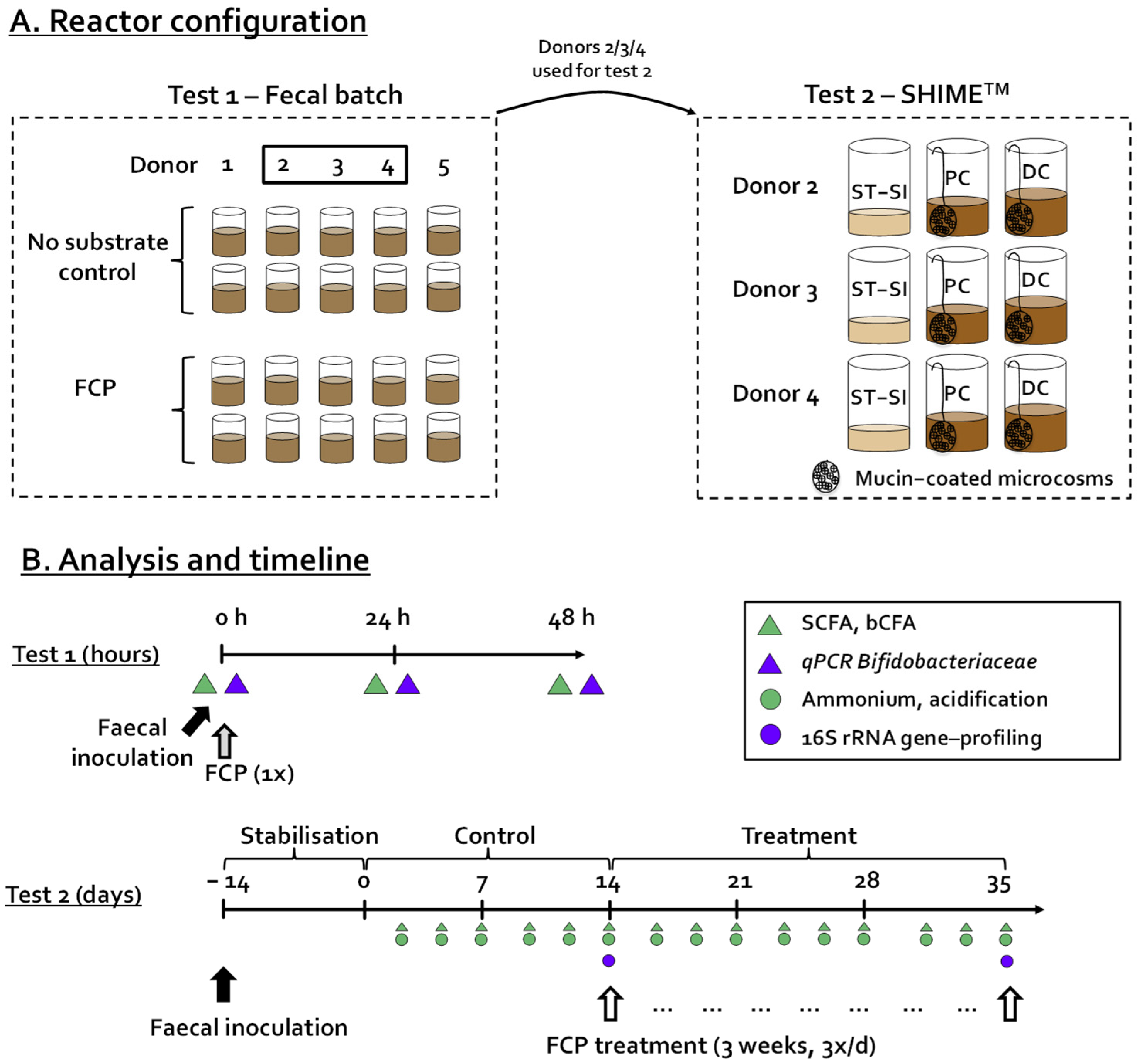
2.2. Incubation Strategy—Test 1
2.3. Incubation Strategy—Test 2
2.4. Microbial Metabolic Activity
2.5. Microbial Composition Analysis
2.6. Statistics
3. Results
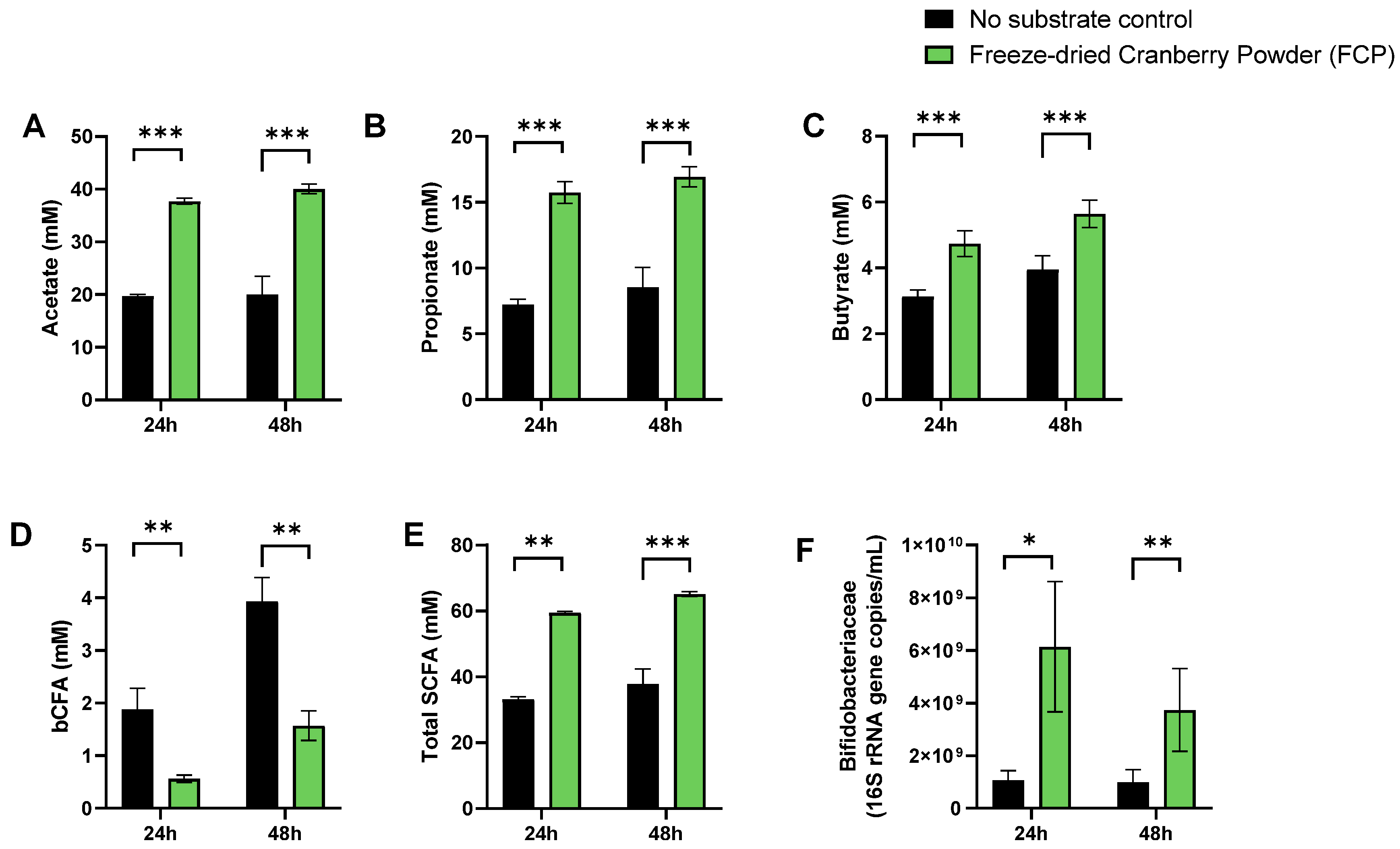
3.1. A Single Dose of FCP Consistently Boosted SCFA and Lowered bCFA Levels during Fecal Batch Incubations of Five Human Adults (Test 1)
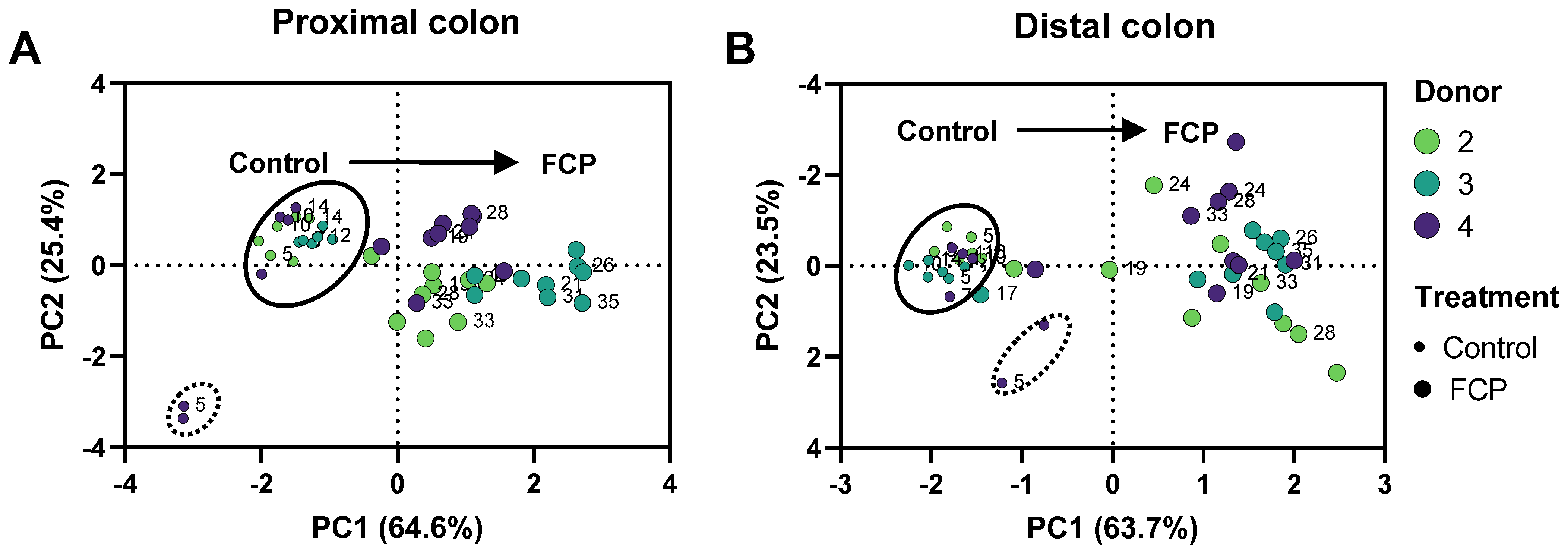
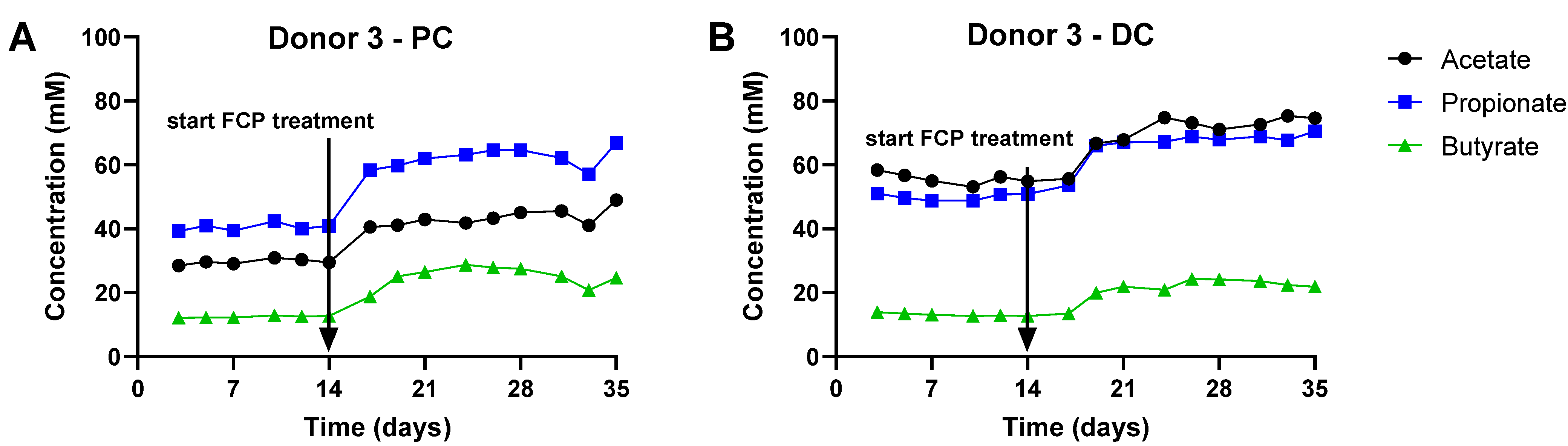
3.2. Repeated Adminsitration of FCP Consistently Boosted SCFA and Lowered bCFA Levels in the Simulated Proximal and Distal Colon of Three Human Adults Tested in the Triple-M-SHIME (Test 2)
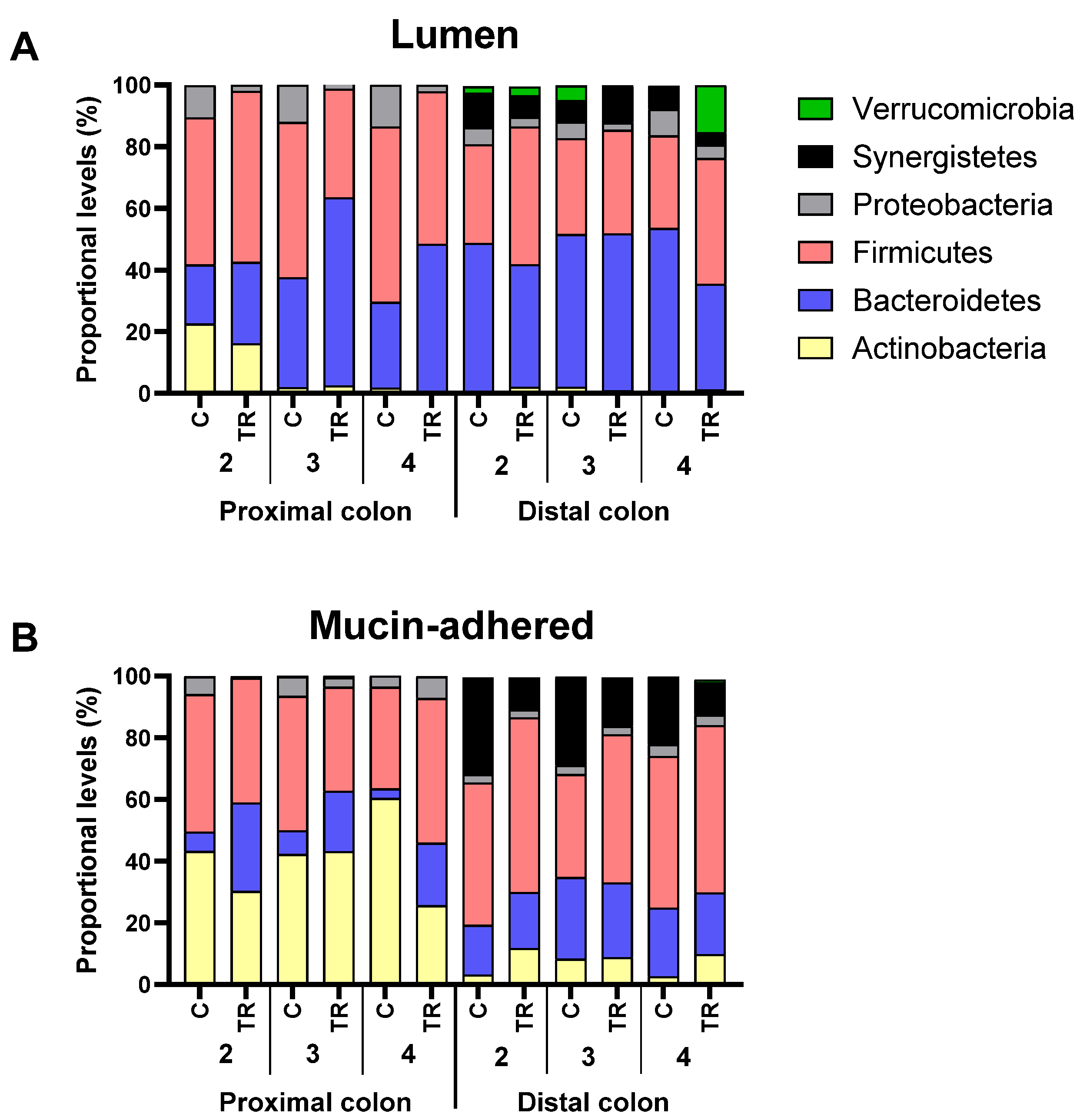
3.3. Repeated Adminsitration of FCP Consistently Altered Microbial Composition in the Simulated Proximal and Distal Colon with Some Interpersonal Differences across the Three Human Adults Tested in the Triple-M-SHIME (Test 2)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment Dominates over Host Genetics in Shaping Human Gut Microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filippo, C.D.; Cavalieri, D.; Paola, M.D.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of Diet in Shaping Gut Microbiota Revealed by a Comparative Study in Children from Europe and Rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, D.; Hyde, E.; Debelius, J.W.; Morton, J.T.; Gonzalez, A.; Ackermann, G.; Aksenov, A.A.; Behsaz, B.; Brennan, C.; Chen, Y.; et al. American Gut: An Open Platform for Citizen Science Microbiome Research. mSystems 2018, 3, e00031-18. [Google Scholar] [CrossRef] [Green Version]
- Almeida, A.; Mitchell, A.L.; Boland, M.; Forster, S.C.; Gloor, G.B.; Tarkowska, A.; Lawley, T.D.; Finn, R.D. A New Genomic Blueprint of the Human Gut Microbiota. Nature 2019, 568, 499–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosca, A.; Leclerc, M.; Hugot, J.P. Gut Microbiota Diversity and Human Diseases: Should We Reintroduce Key Predators in Our Ecosystem? Front. Microbiol. 2016, 7, 455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.A.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The Gut Microbiota and Host Health: A New Clinical Frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [Green Version]
- Blumberg, J.B.; Camesano, T.A.; Cassidy, A.; Kris-Etherton, P.; Howell, A.; Manach, C.; Ostertag, L.M.; Sies, H.; Skulas-Ray, A.; Vita, J.A. Cranberries and Their Bioactive Constituents in Human Health. Adv. Nutr. 2013, 4, 618–632. [Google Scholar] [CrossRef] [Green Version]
- Neto, C.C. Cranberry and Its Phytochemicals: A Review of in Vitro Anticancer Studies. J. Nutr. 2007, 137, 186S–193S. [Google Scholar] [CrossRef] [Green Version]
- Clifford, M.N. Diet-Derived Phenols in Plasma and Tissues and Their Implications for Health. Planta Med. 2004, 70, 1103–1114. [Google Scholar] [CrossRef] [Green Version]
- Le Roy, C.; Wells, P.; Si, J.; Raes, J.; Bell, J.; Spector, T. Red Wine Consumption Associated With Increased Gut Microbiota α-Diversity in 3 Independent Cohorts. Gastroenterology 2019, 158, 270–272.e2. [Google Scholar] [CrossRef] [PubMed]
- Zhernakova, A.; Kurilshikov, A.; Bonder, M.J.; Tigchelaar, E.F.; Schirmer, M.; Vatanen, T.; Mujagic, Z.; Vila, A.V.; Falony, G.; Vieira-Silva, S.; et al. Population-Based Metagenomics Analysis Reveals Markers for Gut Microbiome Composition and Diversity. Science 2016, 352, 565–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anhê, F.F.; Roy, D.; Pilon, G.; Dudonné, S.; Matamoros, S.; Varin, T.V.; Garofalo, C.; Moine, Q.; Desjardins, Y.; Levy, E.; et al. A Polyphenol-Rich Cranberry Extract Protects from Diet-Induced Obesity, Insulin Resistance and Intestinal Inflammation in Association with Increased Akkermansia Spp. Population in the Gut Microbiota of Mice. Gut 2015, 64, 872–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Verstrepen, L.; De Medts, J.; Duysburgh, C.; Van den Abbeele, P.; Marzorati, M.; Khoo, C. A Cranberry Concentrate Decreases Adhesion and Invasion of Escherichia coli (AIEC) LF82 In Vitro. Pathogens 2021, 10, 1217. [Google Scholar] [CrossRef]
- Straub, T.J.; Chou, W.-C.; Manson, A.L.; Schreiber, H.L.; Walker, B.J.; Desjardins, C.A.; Chapman, S.B.; Kaspar, K.L.; Kahsai, O.J.; Traylor, E.; et al. Limited Effects of Long-Term Daily Cranberry Consumption on the Gut Microbiome in a Placebo-Controlled Study of Women with Recurrent Urinary Tract Infections. BMC Microbiol. 2021, 21, 53. [Google Scholar] [CrossRef]
- O’Connor, K.; Morrissette, M.; Strandwitz, P.; Ghiglieri, M.; Caboni, M.; Liu, H.; Khoo, C.; D’Onofrio, A.; Lewis, K. Cranberry Extracts Promote Growth of Bacteroidaceae and Decrease Abundance of Enterobacteriaceae in a Human Gut Simulator Model. PLoS ONE 2019, 14, e0224836. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Marzorati, M.; Derde, M.; De Weirdt, R.; Joan, V.; Possemiers, S.; Van de Wiele, T. Arabinoxylans, Inulin and Lactobacillus Reuteri 1063 Repress the Adherent-Invasive Escherichia coli from Mucus in a Mucosa-Comprising Gut Model. NPJ Biofilms Microbiomes 2016, 2, 16016. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Belzer, C.; Goossens, M.; Kleerebezem, M.; De Vos, W.M.; Thas, O.; De Weirdt, R.; Kerckhof, F.-M.; Van de Wiele, T. Butyrate-Producing Clostridium Cluster XIVa Species Specifically Colonize Mucins in an in Vitro Gut Model. ISME J. 2013, 7, 949–961. [Google Scholar] [CrossRef] [Green Version]
- Van den Abbeele, P.; Taminiau, B.; Pinheiro, I.; Duysburgh, C.; Jacobs, H.; Pijls, L.; Marzorati, M. Arabinoxylo-Oligosaccharides and Inulin Impact Inter-Individual Variation on Microbial Metabolism and Composition, Which Immunomodulates Human Cells. J. Agric. Food Chem. 2018, 66, 1121–1130. [Google Scholar] [CrossRef]
- De Weirdt, R.; Possemiers, S.; Vermeulen, G.; Moerdijk-Poortvliet, T.C.W.; Boschker, H.T.S.; Verstraete, W.; Van de Wiele, T. Human Faecal Microbiota Display Variable Patterns of Glycerol Metabolism. FEMS Microbiol. Ecol. 2010, 74, 601–611. [Google Scholar] [CrossRef] [Green Version]
- Boon, N.; Top, E.M.; Verstraete, W.; Siciliano, S.D. Bioaugmentation as a Tool To Protect the Structure and Function of an Activated-Sludge Microbial Community against a 3-Chloroaniline Shock Load. Appl. Environ. Microbiol. 2003, 69, 1511–1520. [Google Scholar] [CrossRef] [Green Version]
- Duysburgh, C.; Van den Abbeele, P.; Krishnan, K.; Bayne, T.F.; Marzorati, M. A Synbiotic Concept Containing Spore-Forming Bacillus Strains and a Prebiotic Fiber Blend Consistently Enhanced Metabolic Activity by Modulation of the Gut Microbiome in Vitro. Int. J. Pharm. X 2019, 1, 100021. [Google Scholar] [CrossRef]
- Rinttilä, T.; Kassinen, A.; Malinen, E.; Krogius, L.; Palva, A. Development of an Extensive Set of 16S RDNA-Targeted Primers for Quantification of Pathogenic and Indigenous Bacteria in Faecal Samples by Real-Time PCR. J. Appl. Microbiol. 2004, 97, 1166–1177. [Google Scholar] [CrossRef]
- Van den Abbeele, P.; Verstrepen, L.; Ghyselinck, J.; Albers, R.; Marzorati, M.; Mercenier, A. A Novel Non-Digestible, Carrot-Derived Polysaccharide (CRG-I) Selectively Modulates the Human Gut Microbiota While Promoting Gut Barrier Integrity: An Integrated in Vitro Approach. Nutrients 2020, 12, 1917. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L. Assessing and Improving Methods Used in Operational Taxonomic Unit-Based Approaches for 16S RRNA Gene Sequence Analysis. Appl. Environ. Microbiol. 2011, 77, 3219–3226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved Alignments and New Tools for RRNA Analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Lee, S.; Lee, D.K. What Is the Proper Way to Apply the Multiple Comparison Test? Korean J. Anesthesiol. 2018, 71, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Davila, A.-M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.-H.; Sanz, Y.; Tomé, D. Intestinal Luminal Nitrogen Metabolism: Role of the Gut Microbiota and Consequences for the Host. Pharmacol. Res. 2013, 68, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of Short Chain Fatty Acids by the Gut Microbiota and Their Impact on Human Metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Flint, H.J. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Guo, X.; Zhang, J.; Zhang, M.; Ou, Z.; Peng, Y. Phascolarctobacterium Faecium Abundant Colonization in Human Gastrointestinal Tract. Exp. Ther. Med. 2017, 14, 3122–3126. [Google Scholar] [CrossRef] [Green Version]
- Dot, T.; Osawa, R.; Stackebrandt, E. Phascolarctobacterium Faecium Gen. Nov, Spec. Nov., a Novel Taxon of the Sporomusa Group of Bacteria. Syst. Appl. Microbiol. 1993, 16, 380–384. [Google Scholar] [CrossRef]
- van der Beek, C.M.; Bloemen, J.G.; van den Broek, M.A.; Lenaerts, K.; Venema, K.; Buurman, W.A.; Dejong, C.H. Hepatic Uptake of Rectally Administered Butyrate Prevents an Increase in Systemic Butyrate Concentrations in Humans. J. Nutr. 2015, 145, 2019–2024. [Google Scholar] [CrossRef] [PubMed]
- Clausen, M.R.; Mortensen, P.B. Kinetic Studies on Colonocyte Metabolism of Short Chain Fatty Acids and Glucose in Ulcerative Colitis. Gut 1995, 37, 684–689. [Google Scholar] [CrossRef] [Green Version]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of Human Colonic Butyrate-Producing Bacteria Revealed by Analysis of the Butyryl-CoA:Acetate CoA-Transferase Gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef]
- Lacombe, A.; McGivney, C.; Tadepalli, S.; Sun, X.; Wu, V.C.H. The Effect of American Cranberry (Vaccinium macrocarpon) Constituents on the Growth Inhibition, Membrane Integrity, and Injury of Escherichia coli O157:H7 and Listeria Monocytogenes in Comparison to Lactobacillus Rhamnosus. Food Microbiol. 2013, 34, 352–359. [Google Scholar] [CrossRef]
- Feldman, M.; Grenier, D. Cranberry Proanthocyanidins Act in Synergy with Licochalcone A to Reduce Porphyromonas Gingivalis Growth and Virulence Properties, and to Suppress Cytokine Secretion by Macrophages. J. Appl. Microbiol. 2012, 113, 438–447. [Google Scholar] [CrossRef]
- González de Llano, D.; Liu, H.; Khoo, C.; Moreno-Arribas, M.V.; Bartolomé, B. Some New Findings Regarding the Antiadhesive Activity of Cranberry Phenolic Compounds and Their Microbial-Derived Metabolites against Uropathogenic Bacteria. J. Agric. Food Chem. 2019, 67, 2166–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maisuria, V.B.; Los Santos, Y.L.; Tufenkji, N.; Déziel, E. Cranberry-Derived Proanthocyanidins Impair Virulence and Inhibit Quorum Sensing of Pseudomonas Aeruginosa. Sci. Rep. 2016, 6, 30169. [Google Scholar] [CrossRef] [Green Version]
- Jumas-Bilak, E.; Marchandin, H. The Phylum Synergistetes. In The Prokaryotes: Other Major Lineages of Bacteria and The Archaea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 931–954. ISBN 978-3-642-38954-2. [Google Scholar]
- de la Fuente-Nunez, C.; Meneguetti, B.T.; Franco, O.L.; Lu, T.K. Neuromicrobiology: How Microbes Influence the Brain. ACS Chem. Neurosci. 2018, 9, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Cultivation of a Synergistetes Strain Representing a Previously Uncultivated Lineage. Environ. Microbiol. 2010, 12, 916–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.-F.; Yu, T.-C.; Hong, J.; Fang, J.-Y. Emerging Roles of Hydrogen Sulfide in Inflammatory and Neoplastic Colonic Diseases. Front. Physiol. 2016, 7, 156. [Google Scholar] [CrossRef] [Green Version]
- Markou, P.; Apidianakis, Y. Pathogenesis of Intestinal Pseudomonas Aeruginosa Infection in Patients with Cancer. Front. Cell. Infect. Microbiol. 2014, 3, 115. [Google Scholar] [CrossRef]
- Crossman, L.C.; Gould, V.C.; Dow, J.M.; Vernikos, G.S.; Okazaki, A.; Sebaihia, M.; Saunders, D.; Arrowsmith, C.; Carver, T.; Peters, N.; et al. The Complete Genome, Comparative and Functional Analysis of Stenotrophomonas Maltophilia Reveals an Organism Heavily Shielded by Drug Resistance Determinants. Genome Biol. 2008, 9, R74. [Google Scholar] [CrossRef] [Green Version]
- Gallo, S.W.; Figueiredo, T.P.; Bessa, M.C.; Pagnussatti, V.E.; Ferreira, C.A.S.; Oliveira, S.D. Isolation and Characterization of Stenotrophomonas Maltophilia Isolates from a Brazilian Hospital. Microb. Drug Resist. 2016, 22, 688–695. [Google Scholar] [CrossRef]
- Wu, G.D.; Compher, C.; Chen, E.Z.; Smith, S.A.; Shah, R.D.; Bittinger, K.; Chehoud, C.; Albenberg, L.G.; Nessel, L.; Gilroy, E.; et al. Comparative Metabolomics in Vegans and Omnivores Reveal Constraints on Diet-Dependent Gut Microbiota Metabolite Production. Gut 2016, 65, 63–72. [Google Scholar] [CrossRef]
- Rodríguez-Daza, M.-C.; Roquim, M.; Dudonné, S.; Pilon, G.; Levy, E.; Marette, A.; Roy, D.; Desjardins, Y. Berry Polyphenols and Fibers Modulate Distinct Microbial Metabolic Functions and Gut Microbiota Enterotype-Like Clustering in Obese Mice. Front. Microbiol. 2020, 11, 2032. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You Are What You Eat: Diet, Health and the Gut Microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Healey, G.R.; Murphy, R.; Brough, L.; Butts, C.A.; Coad, J. Interindividual Variability in Gut Microbiota and Host Response to Dietary Interventions. Nutr. Rev. 2017, 75, 1059–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tap, J.; Furet, J.-P.; Bensaada, M.; Philippe, C.; Roth, H.; Rabot, S.; Lakhdari, O.; Lombard, V.; Henrissat, B.; Corthier, G.; et al. Gut Microbiota Richness Promotes Its Stability upon Increased Dietary Fibre Intake in Healthy Adults. Environ. Microbiol. 2015, 17, 4954–4964. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Marais, J.P.J.; Khoo, C.; LaPlante, K.; Vejborg, R.M.; Givskov, M.; Tolker-Nielsen, T.; Seeram, N.P.; Rowley, D.C. Cranberry (Vaccinium macrocarpon) Oligosaccharides Decrease Biofilm Formation by Uropathogenic Escherichia coli. J. Funct. Foods 2015, 17, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hotchkiss, A.T.; Nuñez, A.; Strahan, G.D.; Chau, H.K.; White, A.K.; Marais, J.P.J.; Hom, K.; Vakkalanka, M.S.; Di, R.; Yam, K.L.; et al. Cranberry Xyloglucan Structure and Inhibition of Escherichia coli Adhesion to Epithelial Cells. J. Agric. Food Chem. 2015, 63, 5622–5633. [Google Scholar] [CrossRef]
- Cervenka, I.; Agudelo, L.Z.; Ruas, J.L. Kynurenines: Tryptophan’s Metabolites in Exercise, Inflammation, and Mental Health. Science 2017, 357, eaaf9794. [Google Scholar] [CrossRef] [Green Version]
- Pierre, J.; Heneghan, A.; Feliciano, R.; Shanmuganayagam, D.; Roenneburg, D.; Krueger, C.; Reed, J.; Kudsk, K. Cranberry Proanthocyanidins Improve the Gut Mucous Layer Morphology and Function in Mice Receiving Elemental Enteral Nutrition. J. Parenter. Enteral Nutr. 2012, 37, 401–409. [Google Scholar] [CrossRef] [Green Version]
- Van den Abbeele, P.; Roos, S.; Eeckhaut, V.; MacKenzie, D.A.; Derde, M.; Verstraete, W.; Marzorati, M.; Possemiers, S.; Vanhoecke, B.; Van Immerseel, F.; et al. Incorporating a Mucosal Environment in a Dynamic Gut Model Results in a More Representative Colonization by Lactobacilli. Microb. Biotechnol. 2012, 5, 106–115. [Google Scholar] [CrossRef] [Green Version]
- Vandeputte, D.; Kathagen, G.; D’hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative Microbiome Profiling Links Gut Community Variation to Microbial Load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [Green Version]



| Components | Freeze-Dried Whole Cranberry Powder (FCP) |
|---|---|
| total anthocyanins (mg/g) | 5.98 |
| organic acids (%) | 22.68 |
| flavonols (mg/g) | 9.01 |
| phenolic acids (mg/g) | 1.81 |
| PAC—BL DMAC (mg/g) | 8.77 |
| PAC—OSC DMAC (mg/g) | 31.20 |
| total phenolics (mg/g)—measured by Folin-Ciocalteu method | 28.35 |
| Phylum | Family | Proximal Colon | Distal Colon | ||||
|---|---|---|---|---|---|---|---|
| Donor 2 | Donor 3 | Donor 4 | Donor 2 | Donor 3 | Donor 4 | ||
| Actinobacteria | Bifidobacteriaceae | −6.4% | 0.5% | −1.3% | 1.2% | −1.1% | −0.1% |
| Coriobacteriaceae | 0.0% | 0.1% | 0.0% | 0.3% | 0.1% | 0.7% | |
| Bacteroidetes | Bacteroidaceae | 7.2% | 0.6% | 0.0% | −8.3% | −17.6% | −29.4% |
| Bacteroidales_S24-7_group | 0.0% | 0.0% | 0.0% | 0.0% | 0.5% | 0.3% | |
| Porphyromonadaceae | 0.0% | 0.0% | 0.0% | -1.8% | −0.5% | −2.3% | |
| Prevotellaceae | 0.0% | 24.7% | 20.1% | 0.0% | 19.0% | 12.6% | |
| Rikenellaceae | 0.0% | 0.0% | 0.0% | 1.7% | 0.0% | 0.0% | |
| Firmicutes | Acidaminococcaceae | 19.6% | 2.5% | 7.4% | 2.8% | 3.6% | 7.2% |
| Christensenellaceae | 0.0% | 0.0% | 0.0% | 0.1% | 0.0% | 0.0% | |
| Clostridiaceae_1 | 0.0% | −0.7% | −1.3% | 0.0% | 0.0% | 0.0% | |
| Erysipelotrichaceae | 0.0% | 0.0% | 0.0% | 0.1% | 0.0% | 0.2% | |
| Eubacteriaceae | 0.0% | 0.0% | 0.0% | 0.5% | 0.1% | 0.3% | |
| Lachnospiraceae | −3.5% | 1.6% | 2.1% | 11.9% | 0.1% | 2.2% | |
| Ruminococcaceae | 0.0% | 0.0% | 0.0% | 2.8% | 1.5% | 4.2% | |
| Veillonellaceae | −8.3% | −18.6% | −15.5% | −5.7% | −2.7% | −3.2% | |
| Lentisphaerae | Victivallaceae | 0.0% | 0.0% | 0.0% | -0.2% | −0.2% | −0.2% |
| Proteobacteria | Alcaligenaceae | 0.0% | 0.6% | 0.2% | 0.0% | 0.2% | 0.2% |
| Campylobacteraceae | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | |
| Desulfovibrionaceae | 0.0% | 0.1% | 0.0% | 0.4% | −0.5% | −2.0% | |
| Enterobacteriaceae | −4.9% | −2.5% | −3.3% | 0.0% | −0.1% | -0.1% | |
| Pseudomonadaceae | −1.8% | −8.1% | −4.1% | −2.7% | −2.9% | −4.3% | |
| Rhodospirillaceae | 0.0% | 0.0% | 0.0% | 0.2% | 0.3% | 2.4% | |
| Xanthomonadaceae | −1.9% | −0.8% | −4.2% | −0.1% | −0.1% | −0.3% | |
| Synergistetes | Synergistaceae | 0.0% | 0.0% | 0.0% | −4.2% | 4.6% | −3.4% |
| Verrucomicrobia | Verrucomicrobiaceae | 0.0% | 0.0% | 0.0% | 0.7% | −4.2% | 15.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khoo, C.; Duysburgh, C.; Marzorati, M.; Van den Abbeele, P.; Zhang, D. A Freeze-Dried Cranberry Powder Consistently Enhances SCFA Production and Lowers Abundance of Opportunistic Pathogens In Vitro. BioTech 2022, 11, 14. https://doi.org/10.3390/biotech11020014
Khoo C, Duysburgh C, Marzorati M, Van den Abbeele P, Zhang D. A Freeze-Dried Cranberry Powder Consistently Enhances SCFA Production and Lowers Abundance of Opportunistic Pathogens In Vitro. BioTech. 2022; 11(2):14. https://doi.org/10.3390/biotech11020014
Chicago/Turabian StyleKhoo, Christina, Cindy Duysburgh, Massimo Marzorati, Pieter Van den Abbeele, and Derek Zhang. 2022. "A Freeze-Dried Cranberry Powder Consistently Enhances SCFA Production and Lowers Abundance of Opportunistic Pathogens In Vitro" BioTech 11, no. 2: 14. https://doi.org/10.3390/biotech11020014
APA StyleKhoo, C., Duysburgh, C., Marzorati, M., Van den Abbeele, P., & Zhang, D. (2022). A Freeze-Dried Cranberry Powder Consistently Enhances SCFA Production and Lowers Abundance of Opportunistic Pathogens In Vitro. BioTech, 11(2), 14. https://doi.org/10.3390/biotech11020014





