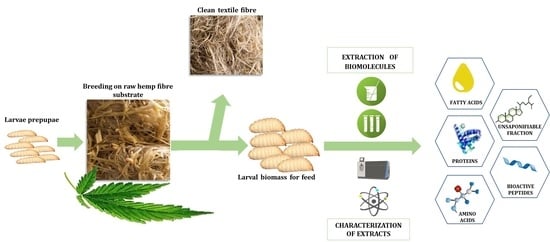
Black Soldier Fly Larvae Grown on Hemp Fiber: Nutritional Composition and Production of Potential Bioactive Peptides
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Black Soldier Fly Sampling and Preliminary Treatments
2.3. Proximate Composition of Larval Biomass
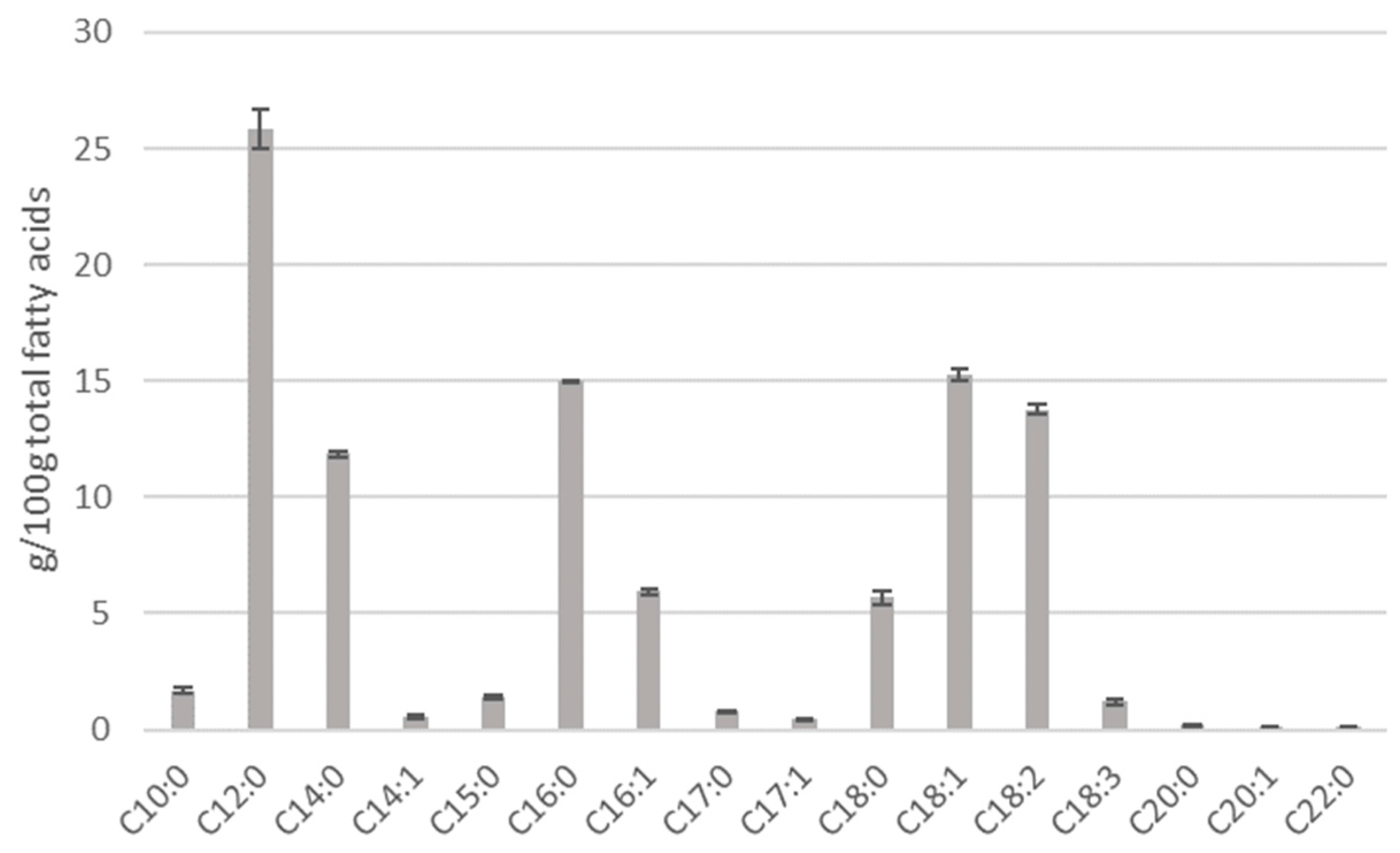
2.4. Determination of Fatty Acid Profile by GC-MS
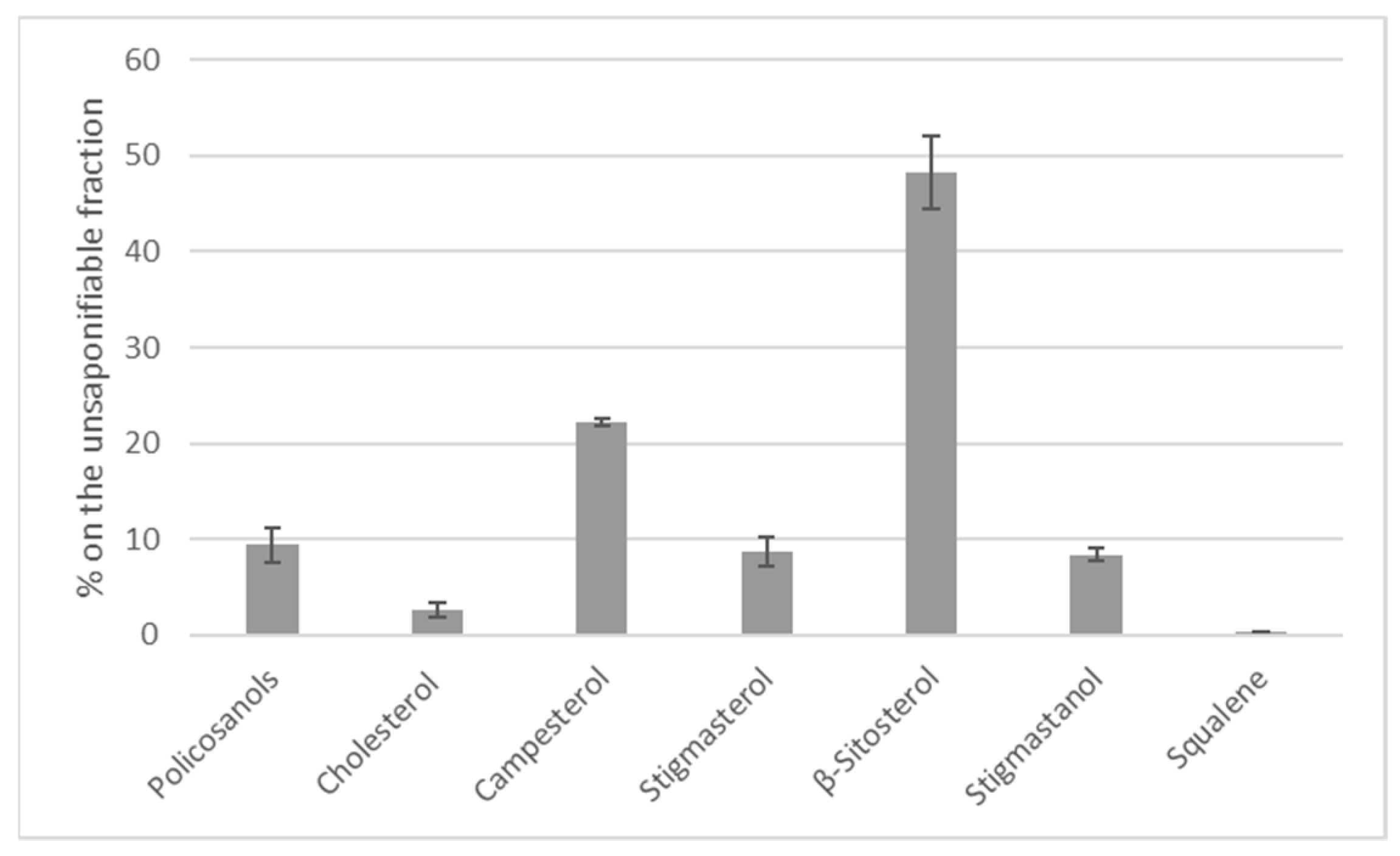
2.5. Determination of Sterols and Other Compounds of Unsaponifiable Fraction
2.6. Amino Acid Analysis
2.7. Protein Hydrolysis and Chemical Characterization of the Hydrolysate
2.7.1. High-Resolution Mass Spectrometry Analysis (μHPLC-LTQ-OrbiTRAP)
2.7.2. In Silico Data Analysis for Putative Bioactive Peptides
2.8. Data Analysis
3. Results
3.1. Black Soldier Fly Larva Composition
3.2. Protein Hydrolysates from BSFL and Peptide Characterization
3.3. Prediction of Potential Bioactive Peptides
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. The Future of Food and Agriculture—Alternative Pathways to 2050; Food and Agriculture Organization of the United Nations Rome: Rome, Italy, 2018; 224p. [Google Scholar]
- European Commission. Communication from the Commission to the European Parliament, The Council, The European Economic and Social Committee and the Committee of the Regions A New Circular Economy Action Plan for a Cleaner and More Competitive Europe COM/2020/98 Final. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=COM:2020:98:FIN&WT.mc_id=Twitter (accessed on 5 January 2022).
- The Sustainable Development Goals Report. 2022. Available online: https://unstats.un.org/sdgs/report/2022/ (accessed on 12 December 2022).
- Surendra, K.C.; Tomberlin, J.K.; van Huis, A.; Cammack, J.A.; Heckmann, L.H.L.; Khanal, S.K. Rethinking Organic Wastes Bioconversion: Evaluating the Potential of the Black Soldier Fly (Hermetia Illucens (L.)) (Diptera: Stratiomyidae) (BSF). Waste Manag. 2020, 117, 58–80. [Google Scholar] [CrossRef]
- Gold, M.; Tomberlin, J.K.; Diener, S.; Zurbrügg, C.; Mathys, A. Decomposition of Biowaste Macronutrients, Microbes, and Chemicals in Black Soldier Fly Larval Treatment: A Review. Waste Manag. 2018, 82, 302–318. [Google Scholar] [CrossRef] [PubMed]
- Higa, J.E.; Ruby, M.B.; Rozin, P. Americans’ Acceptance of Black Soldier Fly Larvae as Food for Themselves, Their Dogs, and Farmed Animals. Food Qual. Prefer. 2021, 90, 104119. [Google Scholar] [CrossRef]
- Miron, L.; Montevecchi, G.; Macavei, L.I.; Maistrello, L.; Antonelli, A.; Thomas, M. Effect of Black Soldier Fly Larvae Protein on the Texture of Meat Analogues. LWT 2023, 181, 114745. [Google Scholar] [CrossRef]
- Mishyna, M.; Keppler, J.K.; Chen, J. Techno-Functional Properties of Edible Insect Proteins and Effects of Processing. Curr. Opin. Colloid Interface Sci. 2021, 56, 101508. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Unlocking the Biological Potential of Proteins from Edible Insects through Enzymatic Hydrolysis: A Review. Innov. Food Sci. Emerg. Technol. 2017, 43, 239–252. [Google Scholar] [CrossRef]
- Thiex, N.; Novotny, L.; Crawford, A. Determination of Ash in Animal Feed: AOAC Official Method 942.05 Revisited. J. AOAC Int. 2012, 95, 1392–1397. [Google Scholar] [CrossRef]
- AOAC. Official Method of Analysis, 16th ed.; Association of Official Analytical: Washington, DC, USA, 2002. [Google Scholar]
- Cequier-Sánchez, E.; Rodríguez, C.; Ravelo, Á.G.; Zárate, R. Dichloromethane as a Solvent for Lipid Extraction and Assessment of Lipid Classes and Fatty Acids from Samples of Different Natures. J. Agric. Food Chem. 2008, 56, 4297–4303. [Google Scholar] [CrossRef]
- Luparelli, A.V.; Leni, G.; Fuso, A.; Pedrazzani, C.; Palini, S.; Sforza, S.; Caligiani, A. Development of a Quantitative UPLC-ESI/MS Method for the Simultaneous Determination of the Chitin and Protein Content in Insects. Food Anal. Methods 2023, 16, 252–265. [Google Scholar] [CrossRef]
- Mooney, C.; Haslam, N.J.; Pollastri, G.; Shields, D.C. Towards the Improved Discovery and Design of Functional Peptides: Common Features of Diverse Classes Permit Generalized Prediction of Bioactivity. PLoS ONE 2012, 7, e45012. [Google Scholar] [CrossRef] [PubMed]
- Aguilan, J.T.; Kulej, K.; Sidoli, S. Guide for Protein Fold Change and P-Value Calculation for Non-Experts in Proteomics. Mol. Omics 2020, 16, 573–582. [Google Scholar] [CrossRef] [PubMed]
- De Marco, M.; Martínez, S.; Hernandez, F.; Madrid, J.; Gai, F.; Rotolo, L.; Belforti, M.; Bergero, D.; Katz, H.; Dabbou, S.; et al. Nutritional Value of Two Insect Larval Meals (Tenebrio Molitor and Hermetia Illucens) for Broiler Chickens: Apparent Nutrient Digestibility, Apparent Ileal Amino Acid Digestibility and Apparent Metabolizable Energy. Anim. Feed Sci. Technol. 2015, 209, 211–218. [Google Scholar] [CrossRef]
- Kroeckel, S.; Harjes, A.G.E.; Roth, I.; Katz, H.; Wuertz, S.; Susenbeth, A.; Schulz, C. When a Turbot Catches a Fly: Evaluation of a Pre-Pupae Meal of the Black Soldier Fly (Hermetia Illucens) as Fish Meal Substitute—Growth Performance and Chitin Degradation in Juvenile Turbot (Psetta Maxima). Aquaculture 2012, 364–365, 345–352. [Google Scholar] [CrossRef]
- Yakti, W.; Förster, N.; Müller, M.; Mewis, I.; Ulrichs, C. Hemp Waste as a Substrate for Hermetia illucens (L.) (Diptera: Stratiomyidae) and Tenebrio molitor L. (Coleoptera: Tenebrionidae) Rearing. Insects 2023, 14, 183. [Google Scholar] [CrossRef] [PubMed]
- Fuso, A.; Barbi, S.; Macavei, L.I.; Luparelli, A.V.; Maistrello, L.; Montorsi, M.; Sforza, S.; Caligiani, A. Effect of the Rearing Substrate on Total Protein and Amino Acid Composition in Black Soldier Fly. Foods 2021, 10, 1773. [Google Scholar] [CrossRef]
- Boukid, F.; Riudavets, J.; Del Arco, L.; Castellari, M.; Savoldelli, S.; Spranghers, T.; Francis, F. Impact of Diets Including Agro-Industrial By-Products on the Fatty Acid and Sterol Profiles of Larvae Biomass from Ephestia Kuehniella, Tenebrio Molitor and Hermetia Illucens. Insects 2021, 12, 672. [Google Scholar] [CrossRef]
- Gao, Z.; Wang, W.; Lu, X.; Zhu, F.; Liu, W.; Wang, X.; Lei, C. Bioconversion Performance and Life Table of Black Soldier Fly (Hermetia Illucens) on Fermented Maize Straw. J. Clean. Prod. 2019, 230, 974–980. [Google Scholar] [CrossRef]
- Caligiani, A.; Marseglia, A.; Sorci, A.; Bonzanini, F.; Lolli, V.; Maistrello, L.; Sforza, S. Influence of the Killing Method of the Black Soldier Fly on Its Lipid Composition. Food Res. Int. 2019, 116, 276–282. [Google Scholar] [CrossRef]
- Ras, R.T.; Geleijnse, J.M.; Trautwein, E.A. LDL-Cholesterol-Lowering Effect of Plant Sterols and Stanols across Different Ranges: A Meta-Analysis of Randomised Controlled Studies. Br. J. Nutr. 2014, 112, 214. [Google Scholar] [CrossRef]
- Mannarino, M.R.; Ministrini, S.; Pirro, M. Nutraceuticals for the Treatment of Hypercholesterolemia. Eur. J. Intern. Med. 2014, 25, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Dietary Protein Quality Evaluation in Human Nutrition Report of an FAO Expert Consultation. FAO FOOD AND NUTRITION PAPER, 92, ISSN 0254-4725. Available online: https://www.fao.org/ag/humannutrition/35978-02317b979a686a57aa4593304ffc17f06.pdf (accessed on 12 January 2024).
- Young, V.R.; Pellett, P.L. Protein Evaluation, Amino Acid Scoring and the Food and Drug Administration’s Proposed Food Labeling Regulations. J. Nutr. 1991, 121, 145–150. [Google Scholar] [CrossRef]
- Yandi, I.; Öztürk, R.Ç.; Kocabas, M.; Kurtoglu, I.Z.; Altinok, I. Nutritional Composition of Black Soldier Fly (Hermetia illucens) Reared on Chicken Waste Meal, Fruit & Vegetable Waste, and Their Mixture. J. Insects Food Feed. 2022, 9, 557–567. [Google Scholar] [CrossRef]
- Van Huis, A.; Rumpold, B.; Maya, C.; Roos, N. Annual Review of Nutrition Nutritional Qualities and Enhancement of Edible Insects. Annu. Rev. Nutr. 2021, 41, 551–576. [Google Scholar] [CrossRef]
- Leni, G.; Tedeschi, T.; Faccini, A.; Pratesi, F.; Folli, C.; Puxeddu, I.; Migliorini, P.; Gianotten, N.; Jacobs, J.; Depraetere, S.; et al. Shotgun Proteomics, in-Silico Evaluation and Immunoblotting Assays for Allergenicity Assessment of Lesser Mealworm, Black Soldier Fly and Their Protein Hydrolysates. Sci. Rep. 2020, 10, 1228. [Google Scholar] [CrossRef] [PubMed]
- Correnig, M. Bioactive Milk Protein and Peptide Functionality. In Dairy-Derived Ingredients: Food and Nutraceutical Uses; Woodhead Publishing: Cambridge, UK, 2009; pp. 238–268. [Google Scholar] [CrossRef]
- Capriotti, A.L.; Caruso, G.; Cavaliere, C.; Samperi, R.; Ventura, S.; Zenezini Chiozzi, R.; Laganà, A. Identification of Potential Bioactive Peptides Generated by Simulated Gastrointestinal Digestion of Soybean Seeds and Soy Milk Proteins. J. Food Compos. Anal. 2015, 44, 205–213. [Google Scholar] [CrossRef]
- Vercruysse, L.; Van Camp, J.; Smagghe, G. ACE Inhibitory Peptides Derived from Enzymatic Hydrolysates of Animal Muscle Protein: A Review. J. Agric. Food Chem. 2005, 53, 8106–8115. [Google Scholar] [CrossRef]
- Nongonierma, A.B.; FitzGerald, R.J. Features of Dipeptidyl Peptidase IV (DPP-IV) Inhibitory Peptides from Dietary Proteins. J. Food Biochem. 2019, 43, e12451. [Google Scholar] [CrossRef]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A Useful Resource for Research on Antimicrobial Peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef]


| Hydrolysis Time | ||
|---|---|---|
| Composition% | 3 h | 15 h |
| Proteins | 63.96 ± 1.80 a | 64.23 ± 2.27 a |
| Lipid | 1.15 ± 0.27 a | 0.41 ± 0.1 a |
| Ash | 22.31 ± 1.23 a | 20.33 ± 0.62 a |
| Accession | Protein | Function | Average Normalized Peptide Area |
|---|---|---|---|
| 3 h of hydrolysis | |||
| A0A7R8V1D0 | Hypothetical protein | Structural constituent of cuticle | 1.369 |
| A0A6J1S563 | Larval cuticle protein 65Ag1-like | Structural constituent of cuticle | 1.172 |
| A0A7R8V0P4 | Hypothetical protein | Structural constituent of cuticle | 1.172 |
| A0A7R8UKB6 | Hypothetical protein | Structural constituent of cuticle | 0.760 |
| A0A7R8YSD0 | Hypothetical protein | Structural constituent of cuticle | 0.592 |
| A0A0L7LRD1 | Putative cuticle protein | Structural constituent of cuticle | 0.339 |
| A0A7R8YZ13 | Hypothetical protein | Structural constituent of cuticle | 0.307 |
| A0A7R8URU7 | Hypothetical protein | Unknown | 0.274 |
| A0A7R8URQ8 | Hypothetical protein | Enzyme | 0.211 |
| A0A7R8V147 | Hypothetical protein | Structural constituent of cuticle | 0.173 |
| B0WR94 | Hypothetical protein | Unknown | 0.001 |
| A0A087ZMI4 | Putative actin-related protein | Muscular | −0.011 |
| A0A7R8UJC0 | Hypothetical protein | Hemocyanin | −0.055 |
| A0A7R8V0P3 | Hypothetical protein | Structural constituent of cuticle | −0.088 |
| B6ZCL2 | Elongation factor 1 alpha | RNA binding | −0.132 |
| A0A7R8UKG1 | Catalase | Enzyme | −0.144 |
| A0A7R8URW5 | Hypothetical protein | Muscular | −0.249 |
| A0A7R8YKR5 | Hypothetical protein | Enzyme | −0.335 |
| A0A7R8V4G4 | Fructose-bisphosphate aldolase | Enzyme | −0.364 |
| A0A310SDR1 | ATP synthase subunit beta | Enzyme | −0.380 |
| A0A7R8UM91 | Hypothetical protein | Structural constituent of cuticle | −0.453 |
| A0A7R8YNH0 | Hypothetical protein | Structural constituent of cuticle | −0.462 |
| A0A7R8YSW8 | Hypothetical protein | Hemolymphatic | −0.521 |
| A0A7R8YVB4 | Hypothetical protein | Ca2+ binding | −0.811 |
| A0A7R8UHR0 | Hypothetical protein | Hemolymphatic | −0.833 |
| A0A7R8UR83 | Hypothetical protein | Unknown | −0.970 |
| A0A7R8Z362 | Hypothetical protein | ATP binding | −1.118 |
| A0A2Z5REH8 | Pyruvate kinase | Enzyme | −1.141 |
| A0A7R8YT79 | Hypothetical protein | Enzyme | −1.183 |
| A0A023EQM6 | Fructose-bisphosphate aldolase | Enzyme | −1.476 |
| B4QW45 | Fructose-bisphosphate aldolase | Enzyme | −1.476 |
| A0A1J1IKW3 | CLUMA_CG013677 | Enzyme | −1.768 |
| 15 h of hydrolysis | |||
| A0A7R8V1D0 | Hypothetical protein | Structural constituent of cuticle | 1.120 |
| A0A6J1S563 | larval cuticle protein 65Ag1-like | Structural constituent of cuticle | 0.802 |
| A0A7R8V0Q1 | Hypothetical protein | Structural constituent of cuticle | 0.802 |
| A0A7R8URU7 | Hypothetical protein | Unknown | 0.279 |
| A0A7R8URQ8 | Hypothetical protein | Enzyme | 0.154 |
| A0A7R8V147 | Hypothetical protein | Structural constituent of cuticle | 0.021 |
| A0A7R8UHR0 | Hypothetical protein | Hemolymphatic | −0.099 |
| A0A087ZMI4 | Putative actin-related protein | Muscular | −0.103 |
| A0A7R8YSD0 | Hypothetical protein | Structural constituent of cuticle | −0.105 |
| A0A7R8V0P3 | Hypothetical protein | Structural constituent of cuticle | −0.383 |
| A0A7R8UM91 | Hypothetical protein | Structural constituent of cuticle | −0.996 |
| A0A067RLP1 | Catalase | Enzyme | −1.043 |
| A0A7R8V1D0 | Hypothetical protein | Structural constituent of cuticle | 1.120 |
| Peptide | UNIPROT Accession | Protein Function | Peptide Ranker | Antimicrobial Activity | Encrypted Bioactive Properties | ||||
|---|---|---|---|---|---|---|---|---|---|
| SVM | RFC | ANN | ACE Inhibitor | DPP IV Inhibitor | Antioxidative | ||||
| 3 h hydrolysis | |||||||||
| GPPGPPGPPGPPGPPGKFPL | A0A7R8UM91 | Cuticular | 0.982 | N | Y | Y | X | X | X |
| GPGGPGGPKGPGGPGGPF | A0A7R8UM91 | Cuticular | 0.971 | Y | Y | Y | x | x | |
| GPPGPPGPPGLPGKFPGST | A0A7R8UM91 | Cuticular | 0.969 | N | Y | Y | x | x | X |
| NIYPVPDPRFPFL | A0A1J1IKW3 | Enzymatic | 0.917 | Y | N | Y | X | X | X |
| ISPPPPLVSIPVGGIL | A0A7R8YVB4 | Ca2+ binding | 0.916 | Y | Y | Y | X | X | |
| GNIYPVPDPRFPFL | A0A1J1IKW3 | Enzymatic | 0.908 | Y | N | Y | X | X | |
| IISPPPPLVSIPVGGIL | A0A7R8YVB4 | Ca2+ binding | 0.906 | Y | Y | Y | X | X | |
| GKPGPAGPPGPPGPL | A0A7R8UM91 | Cuticular | 0.892 | N | Y | Y | X | X | X |
| GFPFDRPIDL | A0A7R8YSW8 | Hemolymphatic | 0.885 | N | N | N | X | X | |
| SRPSIGLPGSPGLPGL | A0A7R8UM91 | Cuticular | 0.870 | N | N | N | X | X | |
| GAHLPIPPPIPDYILR | A0A7R8YNH0 | Cuticular | 0.841 | N | N | Y | X | X | X |
| SRPSLGLPGSPGSPGLPGL | A0A7R8UM91 | Cuticular | 0.840 | N | N | N | X | X | |
| PPPPQPIPDYIVR | A0A7R8V147 | Cuticular | 0.834 | Y | N | N | X | X | |
| GAHLPIPPPIPDYIL | A0A7R8YNH0 | Cuticular | 0.799 | N | N | Y | X | X | X |
| KNPFDEPGKPGNL | A0A7R8Z362 | ATP binding | 0.798 | N | N | N | X | X | X |
| GSRFFDDLPL | A0A7R8UJC0 | Hemolymphatic | 0.789 | N | N | N | X | X | X |
| 15 h hydrolysis | |||||||||
| GPPGPPGPPGPPGPPGKFPL | A0A7R8UM91 | Cuticular | 0.982 | N | Y | Y | X | X | X |
| GPGGPGGPKGPGGPGGPF | A0A7R8UM91 | Cuticular | 0.971 | Y | Y | Y | X | X | |
| GPPGPPGPPGLPGKFPGST | A0A7R8UM91 | Cuticular | 0.969 | N | Y | Y | X | X | X |
| GPGGPGGPGGKGPGGPL | A0A7R8UM91 | Cuticular | 0.963 | Y | Y | Y | X | X | |
| SRPSIGLPGSPGLPGL | A0A7R8UM91 | Cuticular | 0.870 | N | N | N | X | X | |
| GAPGAPGSPGRPGSPIRPS | A0A7R8UM91 | Cuticular | 0.804 | N | Y | Y | X | X | X |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leni, G.; Del Vecchio, L.; Dellapina, C.; Moliterni, V.M.C.; Caligiani, A.; Cirlini, M. Black Soldier Fly Larvae Grown on Hemp Fiber: Nutritional Composition and Production of Potential Bioactive Peptides. Macromol 2024, 4, 135-149. https://doi.org/10.3390/macromol4010007
Leni G, Del Vecchio L, Dellapina C, Moliterni VMC, Caligiani A, Cirlini M. Black Soldier Fly Larvae Grown on Hemp Fiber: Nutritional Composition and Production of Potential Bioactive Peptides. Macromol. 2024; 4(1):135-149. https://doi.org/10.3390/macromol4010007
Chicago/Turabian StyleLeni, Giulia, Lorenzo Del Vecchio, Claudia Dellapina, Vita Maria Cristiana Moliterni, Augusta Caligiani, and Martina Cirlini. 2024. "Black Soldier Fly Larvae Grown on Hemp Fiber: Nutritional Composition and Production of Potential Bioactive Peptides" Macromol 4, no. 1: 135-149. https://doi.org/10.3390/macromol4010007
APA StyleLeni, G., Del Vecchio, L., Dellapina, C., Moliterni, V. M. C., Caligiani, A., & Cirlini, M. (2024). Black Soldier Fly Larvae Grown on Hemp Fiber: Nutritional Composition and Production of Potential Bioactive Peptides. Macromol, 4(1), 135-149. https://doi.org/10.3390/macromol4010007