Spermidine Binding to the Acetinobacter baumannii Efflux Protein AceI Observed by Near-UV Synchrotron Radiation Circular Dichroism Spectroscopy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Expression and Purification
2.2. Near-UV SRCD Spectroscopy
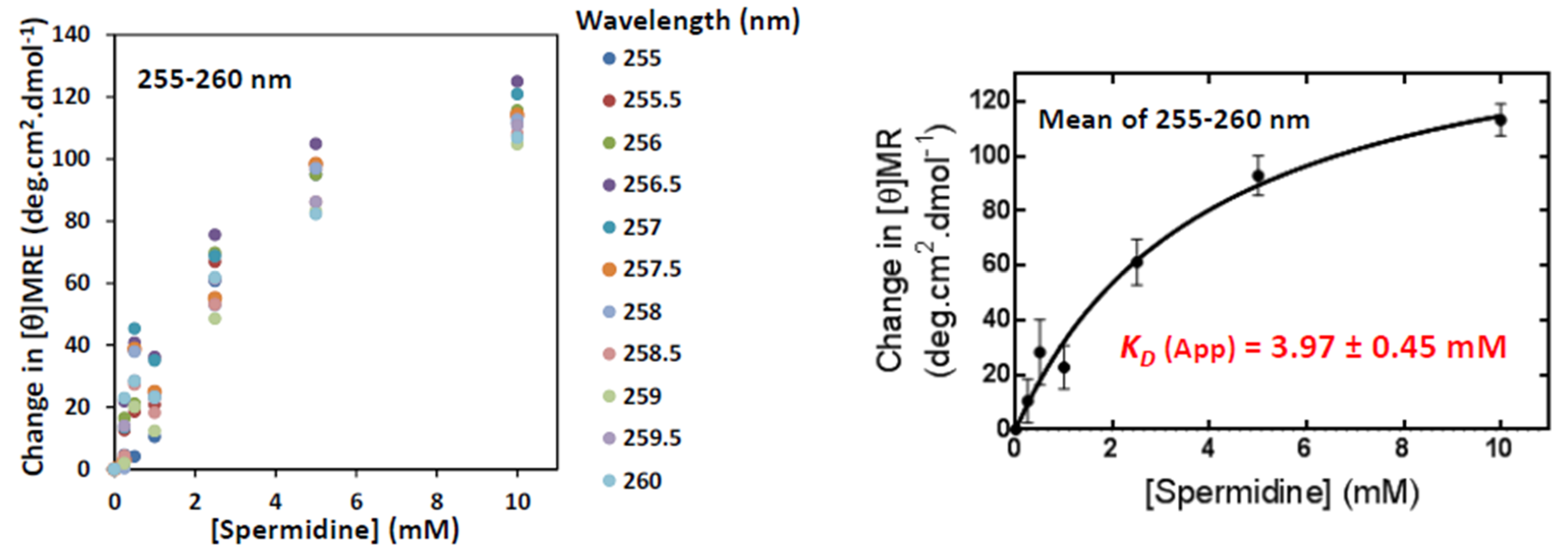
3. Results and Discussion
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Amorim, A.M.; Nascimento, J.D. Acinetobacter: An underrated foodborne pathogen? J. Infect. Dev. Ctries 2017, 11, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Hamidian, M.; Nigro, S.J. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb. Genom. 2019, 5, e000306. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Jackson, S.M.; Penesyan, A.; Patching, S.G.; Tetu, S.G.; Eijkelkamp, B.A.; Brown, M.H.; Henderson, P.J.; Paulsen, I.T. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20254–20259. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Liu, Q.; Henderson, P.J.; Paulsen, I.T. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 2015, 6, e01982-14. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Nawaz, N.; Dermani, F.K.; Kohlan, A.K.; Saidijam, M.; Patching, S.G. Bacterial multidrug efflux proteins: A major mechanism of antimicrobial resistance. Curr. Drug Targets 2018, 19, 1–13. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Elbourne, L.D.H.; Ahmad, I.; Sharples, D.; Naidu, V.; Chan, C.L.; Li, L.; Harborne, S.P.D.; Pokhrel, A.; et al. Pacing across the membrane: The novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 2018, 169, 450–454. [Google Scholar] [CrossRef]
- Bolla, J.R.; Howes, A.C.; Fiorentino, F.; Robinson, C.V. Assembly and regulation of the chlorhexidine-specific efflux pump AceI. Proc. Natl. Acad. Sci. USA 2020, 117, 17011–17018. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 2016, 291, 14896–14903. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Swiatlo, E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008, 68, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Banerji, R.; Kanojiya, P.; Patil, A.; Saroj, S.D. Polyamines in the virulence of bacterial pathogens of respiratory tract. Mol. Oral. Microbiol. 2021, 36, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Patching, S.G.; Edara, S.; Ma, P.; Nakayama, J.; Hussain, R.; Siligardi, G.; Phillips-Jones, M.K. Interactions of the intact FsrC membrane histidine kinase with its pheromone ligand GBAP revealed through synchrotron radiation circular dichroism. Biochim. Biophys. Acta—Biomembranes 2012, 1818, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Jones, M.K.; Patching, S.G.; Edara, S.; Nakayama, J.; Hussain, R.; Siligardi, G. Interactions of the intact FsrC membrane histidine kinase with the tricyclic peptide inhibitor siamycin I revealed through synchrotron radiation circular dichroism. Phys. Chem. Chem. Phys. 2013, 15, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Siligardi, G.; Hussain, R.; Patching, S.G.; Phillips-Jones, M.K. Ligand- and drug-binding studies of membrane proteins revealed through circular dichroism spectroscopy. Biochim. Biophys. Acta—Biomembranes 2014, 1838, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Phillips-Jones, M.K.; Channell, G.; Kelsall, C.J.; Hughes, C.S.; Ashcroft, A.E.; Patching, S.G.; Dinu, V.; Gillis, R.B.; Adams, G.G.; Harding, S.E. Hydrodynamics of the VanA-type VanS histidine kinase: An extended solution conformation and first evidence for interactions with vancomycin. Sci. Rep. 2017, 7, 46180. [Google Scholar] [CrossRef] [PubMed]
- Patching, S.G. Recent developments in Nucleobase Cation Symporter-1 (NCS1) family transport proteins from bacteria, archaea, fungi and plants. J. Biosci. 2018, 43, 797–815. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.; Sanderson, N.M.; O’Reilly, J.; Rutherford, N.G.; Poolman, B.; Henderson, P.J.F. The amplified expression, identification, purification, assay, and properties of hexahistidine-tagged bacterial membrane transport proteins. In Membrane Transport: A Practical Approach; Baldwin, S.A., Ed.; Oxford University Press: Oxford, UK, 2000; pp. 141–166. [Google Scholar]
- Hussain, R.; Jávorfi, T.; Siligardi, G. Circular dichroism beamline B23 at the Diamond Light Source. J. Synchrotron Rad. 2012, 19, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Hussain, R.; Siligardi, G. Characterisation of conformational and ligand binding properties of membrane proteins using synchrotron radiation circular dichroism (SRCD). Adv. Exp. Med. Biol. 2016, 922, 43–59. [Google Scholar] [PubMed]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Saidijam, M.; Patching, S.G. Amino acid composition analysis of secondary transport proteins from Escherichia coli with relation to functional classification, ligand specificity and structure. J. Biomol. Struct. Dyn. 2015, 33, 2205–2220. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patching, S.G. Spermidine Binding to the Acetinobacter baumannii Efflux Protein AceI Observed by Near-UV Synchrotron Radiation Circular Dichroism Spectroscopy. Radiation 2022, 2, 228-233. https://doi.org/10.3390/radiation2020016
Patching SG. Spermidine Binding to the Acetinobacter baumannii Efflux Protein AceI Observed by Near-UV Synchrotron Radiation Circular Dichroism Spectroscopy. Radiation. 2022; 2(2):228-233. https://doi.org/10.3390/radiation2020016
Chicago/Turabian StylePatching, Simon G. 2022. "Spermidine Binding to the Acetinobacter baumannii Efflux Protein AceI Observed by Near-UV Synchrotron Radiation Circular Dichroism Spectroscopy" Radiation 2, no. 2: 228-233. https://doi.org/10.3390/radiation2020016
APA StylePatching, S. G. (2022). Spermidine Binding to the Acetinobacter baumannii Efflux Protein AceI Observed by Near-UV Synchrotron Radiation Circular Dichroism Spectroscopy. Radiation, 2(2), 228-233. https://doi.org/10.3390/radiation2020016





