Autophagy in Virus Infection: A Race between Host Immune Response and Viral Antagonism
Abstract
:1. Introduction
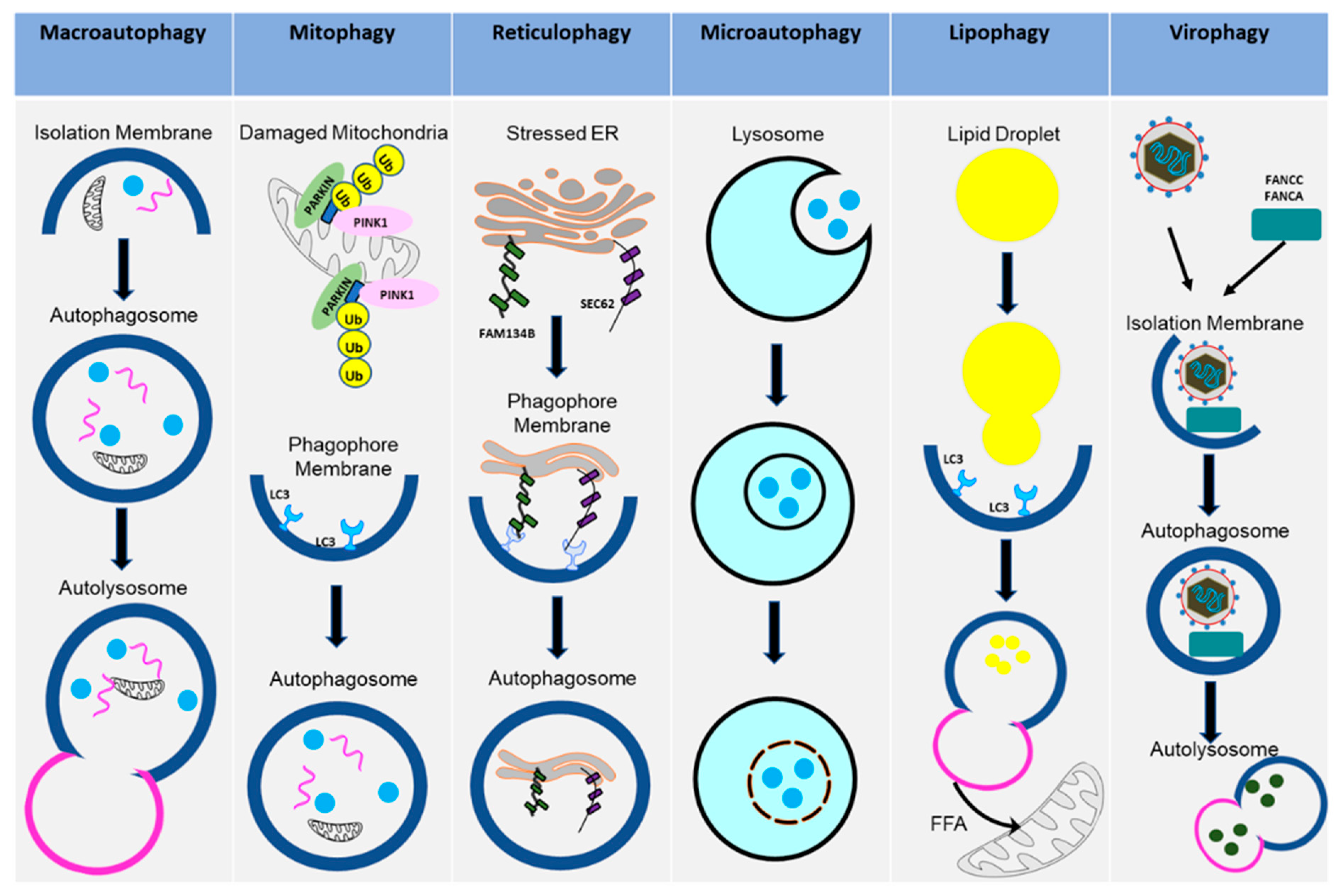
2. Autophagy: Activation and Various Forms
2.1. Cellular Autophagy Pathway
2.2. Mitophagy
2.3. Reticulophagy
2.4. Microautophagy
2.5. Lipophagy
2.6. Virophagy (Viral Xenophagy)
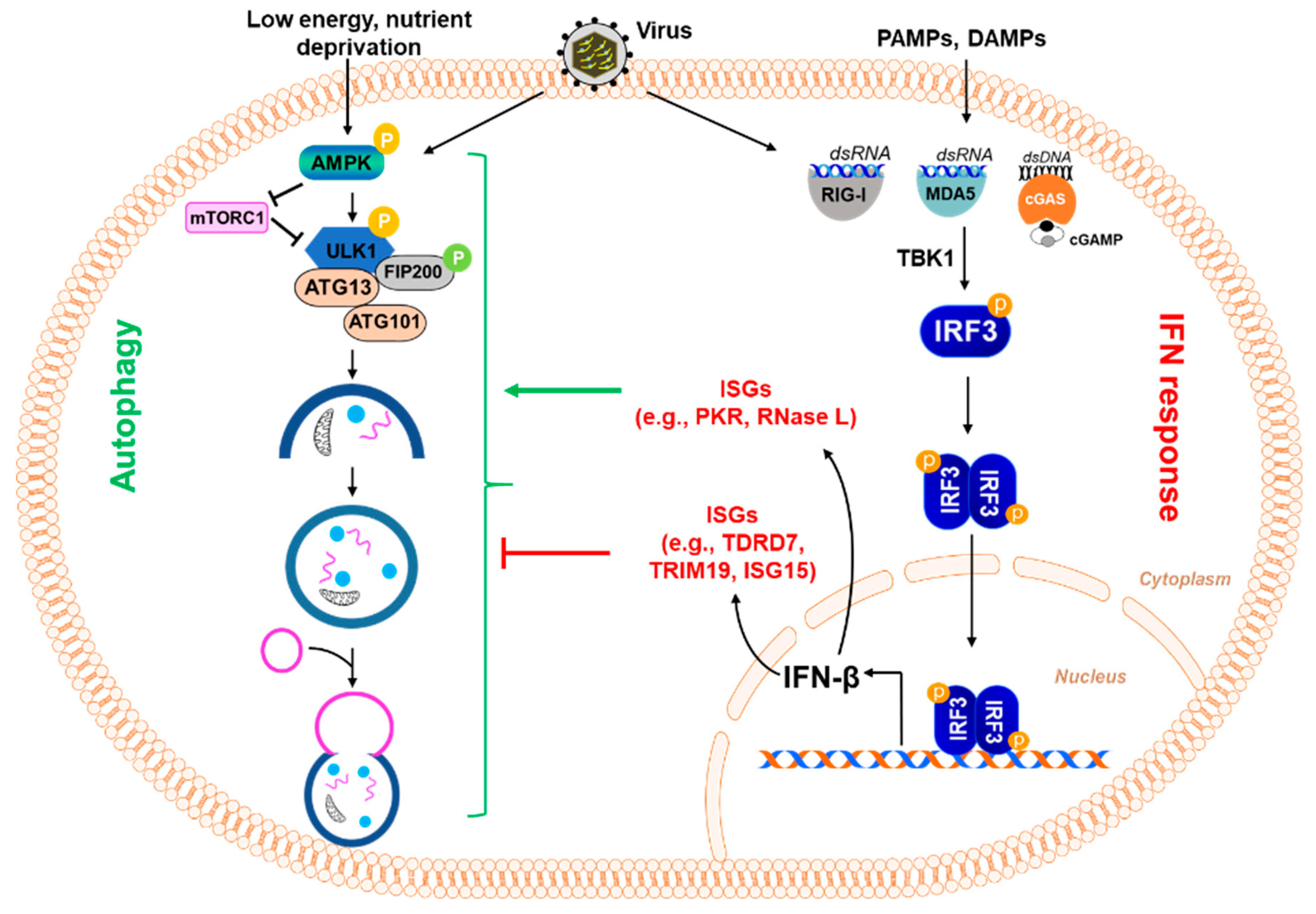
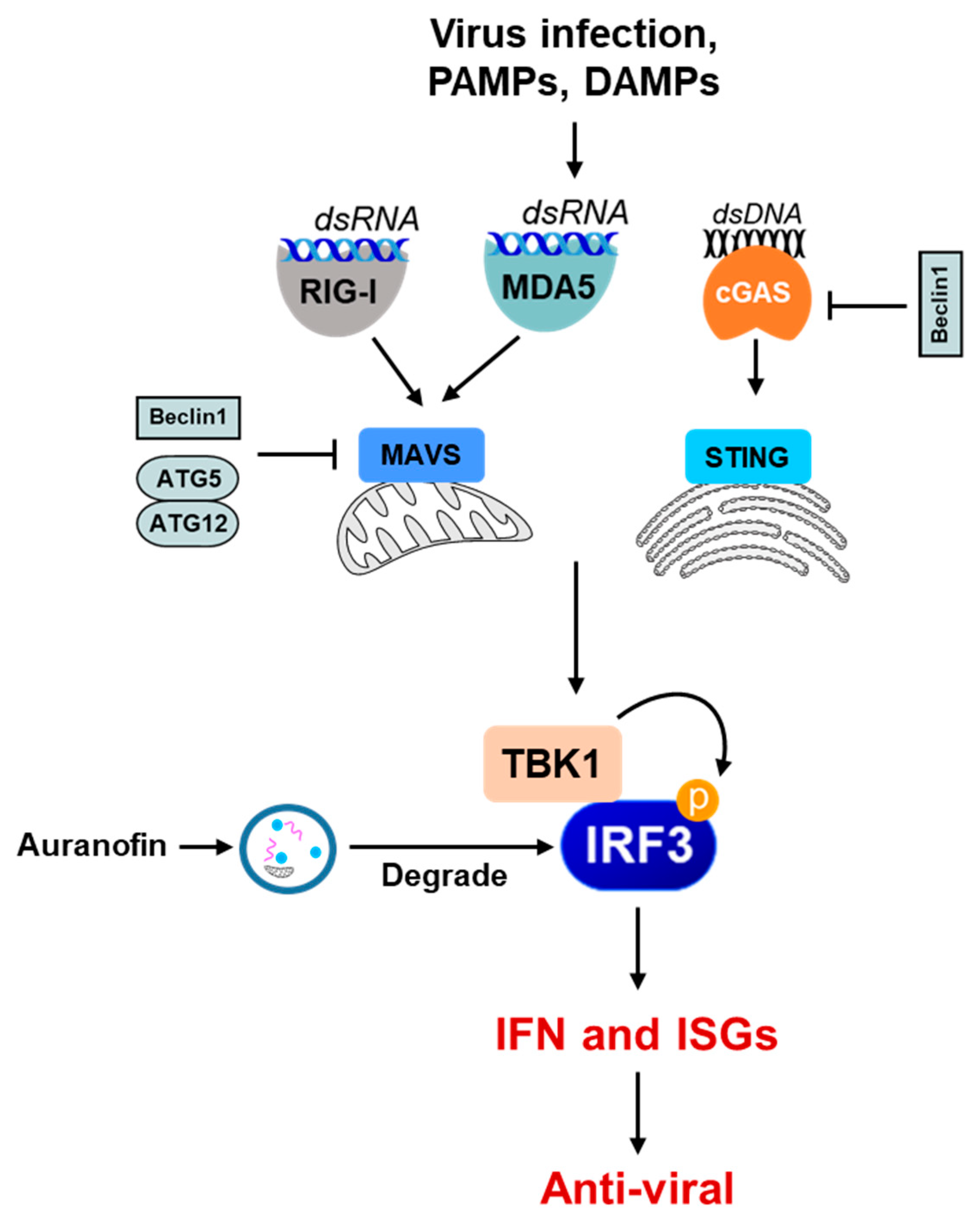
3. Autophagy as an Anti-Viral Immune Response
4. Autophagy and the Interferon System
5. Autophagy as a Pro-Viral Cellular Response
6. Viral Antagonism to Autophagy
Viral Antagonism by SARS-CoV-2
7. Therapeutic Application of Autophagy in Virus Infection
8. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kawai, T.; Akira, S. Innate immune recognition of viral infection. Nat. Immunol. 2006, 7, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, A.; Medzhitov, R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015, 16, 343–353. [Google Scholar] [CrossRef]
- Guidotti, L.G.; Chisari, F.V. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 2001, 19, 65–91. [Google Scholar] [CrossRef]
- Subramanian, G.; Kuzmanovic, T.; Zhang, Y.; Peter, C.B.; Veleeparambil, M.; Chakravarti, R.; Sen, G.C.; Chattopadhyay, S. A new mechanism of interferon’s antiviral action: Induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7. PLoS Pathog. 2018, 14, e1006877. [Google Scholar] [CrossRef] [Green Version]
- Jackson, W.T. Viruses and the autophagy pathway. Virology 2015, 479–480, 450–456. [Google Scholar] [CrossRef]
- Ichimiya, T.; Yamakawa, T.; Hirano, T.; Yokoyama, Y.; Hayashi, Y.; Hirayama, D.; Wagatsuma, K.; Itoi, T.; Nakase, H. Autophagy and Autophagy-Related Diseases: A Review. Int. J. Mol. Sci. 2020, 21, 8974. [Google Scholar] [CrossRef]
- Ahmad, L.; Mostowy, S.; Sancho-Shimizu, V. Autophagy-Virus Interplay: From Cell Biology to Human Disease. Front. Cell Dev. Biol. 2018, 6, 155. [Google Scholar] [CrossRef] [Green Version]
- Mao, J.; Lin, E.; He, L.; Yu, J.; Tan, P.; Zhou, Y. Autophagy and Viral Infection. Adv. Exp. Med. Biol. 2019, 1209, 55–78. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Pereira, G.; Leao, A.; Erustes, A.G.; Morais, I.B.M.; Vrechi, T.A.M.; Zamarioli, L.D.S.; Pereira, C.A.S.; Marchioro, L.O.; Sperandio, L.P.; Lins, I.V.F.; et al. Pharmacological Modulators of Autophagy as a Potential Strategy for the Treatment of COVID-19. Int. J. Mol. Sci. 2021, 22, 4067. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Chiramel, A.I.; Brady, N.R.; Bartenschlager, R. Divergent roles of autophagy in virus infection. Cells 2013, 2, 83–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farre, J.C.; Subramani, S. Mechanistic insights into selective autophagy pathways: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2016, 17, 537–552. [Google Scholar] [CrossRef]
- Quinn, P.M.J.; Moreira, P.I.; Ambrosio, A.F.; Alves, C.H. PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol. Commun. 2020, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Sakakibara, K.; Chen, Q.; Okamoto, K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014, 24, 787–795. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Mochida, K. Reticulophagy and nucleophagy: New findings and unsolved issues. Autophagy 2015, 11, 2377–2378. [Google Scholar] [CrossRef] [Green Version]
- Lennemann, N.J.; Coyne, C.B. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy 2017, 13, 322–332. [Google Scholar] [CrossRef] [Green Version]
- Li, W.W.; Li, J.; Bao, J.K. Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 2012, 69, 1125–1136. [Google Scholar] [CrossRef]
- Crotzer, V.L.; Blum, J.S. Autophagy and adaptive immunity. Immunology 2010, 131, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kunz, J.B.; Schwarz, H.; Mayer, A. Determination of four sequential stages during microautophagy in vitro. J. Biol. Chem. 2004, 279, 9987–9996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenberg, A.S.; Coleman, R.A.; Kraemer, F.B.; McManaman, J.L.; Obin, M.S.; Puri, V.; Yan, Q.W.; Miyoshi, H.; Mashek, D.G. The role of lipid droplets in metabolic disease in rodents and humans. J. Clin. Investig. 2011, 121, 2102–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, T.; Parton, R.G. Not just fat: The structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 2011, 3, a004838. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, M.; Waguri, S.; Ueno, T.; Iwata, J.; Murata, S.; Tanida, I.; Ezaki, J.; Mizushima, N.; Ohsumi, Y.; Uchiyama, Y.; et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005, 169, 425–434. [Google Scholar] [CrossRef]
- Ohsaki, Y.; Cheng, J.; Fujita, A.; Tokumoto, T.; Fujimoto, T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol. Biol. Cell 2006, 17, 2674–2683. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Cuervo, A.M. Lipophagy: Connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012, 2012, 282041. [Google Scholar] [CrossRef]
- Koga, H.; Kaushik, S.; Cuervo, A.M. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010, 24, 3052–3065. [Google Scholar] [CrossRef] [Green Version]
- Mei, S.; Ni, H.M.; Manley, S.; Bockus, A.; Kassel, K.M.; Luyendyk, J.P.; Copple, B.L.; Ding, W.X. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J. Pharmacol. Exp. Ther. 2011, 339, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Thoen, L.F.; Guimaraes, E.L.; Dolle, L.; Mannaerts, I.; Najimi, M.; Sokal, E.; van Grunsven, L.A. A role for autophagy during hepatic stellate cell activation. J. Hepatol. 2011, 55, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lan, Y.; Li, M.Y.; Lamers, M.M.; Fusade-Boyer, M.; Klemm, E.; Thiele, C.; Ashour, J.; Sanyal, S. Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host Microbe 2018, 23, 819–831.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.K.; Dang, J.W.; Lin, N.; Qin, Y.; Wang, S.; Rana, T.M. Zika virus depletes neural stem cells and evades selective autophagy by suppressing the Fanconi anemia protein FANCC. EMBO Rep. 2020, 21, e49183. [Google Scholar] [CrossRef] [PubMed]
- Sumpter, R., Jr.; Sirasanagandla, S.; Fernandez, A.F.; Wei, Y.; Dong, X.; Franco, L.; Zou, Z.; Marchal, C.; Lee, M.Y.; Clapp, D.W.; et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell 2016, 165, 867–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viret, C.; Rozieres, A.; Faure, M. Novel Insights into NDP52 Autophagy Receptor Functioning. Trends Cell Biol. 2018, 28, 255–257. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, M.J.; Sung, P.S.; Bae, Y.C.; Shin, E.C.; Yoo, J.Y. Interferon-inducible protein SCOTIN interferes with HCV replication through the autolysosomal degradation of NS5A. Nat. Commun. 2016, 7, 10631. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.; Maloney, N.S.; Bruinsma, M.W.; Goel, G.; Duan, E.; Zhang, L.; Shrestha, B.; Diamond, M.S.; Dani, A.; Sosnovtsev, S.V.; et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 2012, 11, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Orvedahl, A.; MacPherson, S.; Sumpter, R., Jr.; Talloczy, Z.; Zou, Z.; Levine, B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 2010, 7, 115–127. [Google Scholar] [CrossRef] [Green Version]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [Green Version]
- Tokarev, A.; Suarez, M.; Kwan, W.; Fitzpatrick, K.; Singh, R.; Guatelli, J. Stimulation of NF-kappaB activity by the HIV restriction factor BST2. J. Virol. 2013, 87, 2046–2057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.K.; Lund, J.M.; Ramanathan, B.; Mizushima, N.; Iwasaki, A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007, 315, 1398–1401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crotzer, V.L.; Blum, J.S. Autophagy and its role in MHC-mediated antigen presentation. J. Immunol. 2009, 182, 3335–3341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glanz, A.; Chakravarty, S.; Varghese, M.; Kottapalli, A.; Fan, S.; Chakravarti, R.; Chattopadhyay, S. Transcriptional and Non-Transcriptional Activation, Posttranslational Modifications, and Antiviral Functions of Interferon Regulatory Factor 3 and Viral Antagonism by the SARS-Coronavirus. Viruses 2021, 13, 575. [Google Scholar] [CrossRef] [PubMed]
- Orvedahl, A.; Alexander, D.; Talloczy, Z.; Sun, Q.; Wei, Y.; Zhang, W.; Burns, D.; Leib, D.A.; Levine, B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007, 1, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, M.A.; Malathi, K. RNase L induces autophagy via c-Jun N-terminal kinase and double-stranded RNA-dependent protein kinase signaling pathways. J. Biol. Chem. 2012, 287, 43651–43664. [Google Scholar] [CrossRef] [Green Version]
- Manivannan, P.; Siddiqui, M.A.; Malathi, K. RNase L Amplifies Interferon Signaling by Inducing Protein Kinase R-Mediated Antiviral Stress Granules. J. Virol. 2020, 94, e00205-20. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.Y.; Yaneva, R.; Cresswell, P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 2011, 10, 534–539. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.Y.; Cresswell, P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013, 9, e1003497. [Google Scholar] [CrossRef]
- Liu, Y.; Gordesky-Gold, B.; Leney-Greene, M.; Weinbren, N.L.; Tudor, M.; Cherry, S. Inflammation-Induced, STING-Dependent Autophagy Restricts Zika Virus Infection in the Drosophila Brain. Cell Host Microbe 2018, 24, 57–68.e3. [Google Scholar] [CrossRef] [Green Version]
- van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, G.; Popli, S.; Chakravarty, S.; Taylor, R.T.; Chakravarti, R.; Chattopadhyay, S. The interferon-inducible protein TDRD7 inhibits AMP-activated protein kinase and thereby restricts autophagy-independent virus replication. J. Biol. Chem. 2020, 295, 6811–6822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.; Feng, C.; Tian, X.; Zheng, N.; Wu, Z. Promyelocytic Leukemia Restricts Enterovirus 71 Replication by Inhibiting Autophagy. Front. Immunol. 2018, 9, 1268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glanz, A.; Chawla, K.; Fabry, S.; Subramanian, G.; Garcia, J.; Jay, B.; Ciricillo, J.; Chakravarti, R.; Taylor, R.T.; Chattopadhyay, S. High Throughput Screening of FDA-Approved Drug Library Reveals the Compounds that Promote IRF3-Mediated Pro-Apoptotic Pathway Inhibit Virus Replication. Viruses 2020, 12, 442. [Google Scholar] [CrossRef] [Green Version]
- Glanz, A.; Chakravarty, S.; Fan, S.; Chawla, K.; Subramanian, G.; Rahman, T.; Walters, D.; Chakravarti, R.; Chattopadhyay, S. Autophagic degradation of IRF3 induced by the small-molecule auranofin inhibits its transcriptional and proapoptotic activities. J. Biol. Chem. 2021, 297, 101274. [Google Scholar] [CrossRef]
- Xie, W.; Jin, S.; Zhang, C.; Yang, S.; Wu, Y.; Zhao, Y.; Songyang, Z.; Cui, J. Selective autophagy controls the stability of TBK1 via NEDD4 to balance host defense. Cell Death Differ. 2021, 29, 40–53. [Google Scholar] [CrossRef]
- Liu, J.; Wu, X.; Wang, H.; Wei, J.; Wu, Q.; Wang, X.; Yan, Y.; Cui, J.; Min, J.; Wang, F.; et al. HFE inhibits type I IFNs signaling by targeting the SQSTM1-mediated MAVS autophagic degradation. Autophagy 2021, 17, 1962–1977. [Google Scholar] [CrossRef]
- Wu, Y.; Jin, S.; Liu, Q.; Zhang, Y.; Ma, L.; Zhao, Z.; Yang, S.; Li, Y.P.; Cui, J. Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy 2021, 17, 1379–1392. [Google Scholar] [CrossRef]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Schroder, K.; Johansen, T.; Deretic, V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 2015, 210, 973–989. [Google Scholar] [CrossRef] [Green Version]
- Gui, X.; Yang, H.; Li, T.; Tan, X.; Shi, P.; Li, M.; Du, F.; Chen, Z.J. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 2019, 567, 262–266. [Google Scholar] [CrossRef]
- Prantner, D.; Perkins, D.J.; Vogel, S.N. AMP-activated Kinase (AMPK) Promotes Innate Immunity and Antiviral Defense through Modulation of Stimulator of Interferon Genes (STING) Signaling. J. Biol. Chem. 2017, 292, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Seo, G.J.; Choi, Y.J.; Ge, J.; Rodgers, M.A.; Shi, M.; Jung, J.U. Autophagy side of MB21D1/cGAS DNA sensor. Autophagy 2014, 10, 1146–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herhaus, L.; Bhaskara, R.M.; Lystad, A.H.; Gestal-Mato, U.; Covarrubias-Pinto, A.; Bonn, F.; Simonsen, A.; Hummer, G.; Dikic, I. TBK1-mediated phosphorylation of LC3C and GABARAP-L2 controls autophagosome shedding by ATG4 protease. EMBO Rep. 2020, 21, e48317. [Google Scholar] [CrossRef] [PubMed]
- Weidberg, H.; Elazar, Z. TBK1 mediates crosstalk between the innate immune response and autophagy. Sci. Signal. 2011, 4, pe39. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Gastaminza, P.; Wieland, S.F.; Chisari, F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 2009, 106, 14046–14051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Qin, Y.; Chen, M. Viral strategies for triggering and manipulating mitophagy. Autophagy 2018, 14, 1665–1673. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Zhang, L.; Li, Z.; Zhong, Y.; Tang, Q.; Qin, Y.; Chen, M. The Matrix Protein of Human Parainfluenza Virus Type 3 Induces Mitophagy that Suppresses Interferon Responses. Cell Host Microbe 2017, 21, 538–547.e4. [Google Scholar] [CrossRef] [Green Version]
- Lennemann, N.J.; Coyne, C.B. Catch me if you can: The link between autophagy and viruses. PLoS Pathog. 2015, 11, e1004685. [Google Scholar] [CrossRef]
- Richetta, C.; Gregoire, I.P.; Verlhac, P.; Azocar, O.; Baguet, J.; Flacher, M.; Tangy, F.; Rabourdin-Combe, C.; Faure, M. Sustained autophagy contributes to measles virus infectivity. PLoS Pathog. 2013, 9, e1003599. [Google Scholar] [CrossRef]
- Hou, L.; Ge, X.; Xin, L.; Zhou, L.; Guo, X.; Yang, H. Nonstructural proteins 2C and 3D are involved in autophagy as induced by the encephalomyocarditis virus. Virol. J. 2014, 11, 156. [Google Scholar] [CrossRef] [Green Version]
- Green, A.M.; Beatty, P.R.; Hadjilaou, A.; Harris, E. Innate immunity to dengue virus infection and subversion of antiviral responses. J. Mol. Biol. 2014, 426, 1148–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, V.; Lai, C.K.; Chao, T.C.; Jeng, K.S.; Lai, M.M. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J. Virol. 2012, 86, 4139–4150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alirezaei, M.; Flynn, C.T.; Wood, M.R.; Whitton, J.L. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe 2012, 11, 298–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beale, R.; Wise, H.; Stuart, A.; Ravenhill, B.J.; Digard, P.; Randow, F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 2014, 15, 239–247. [Google Scholar] [CrossRef] [Green Version]
- Yakoub, A.M.; Shukla, D. Basal Autophagy Is Required for Herpes simplex Virus-2 Infection. Sci. Rep. 2015, 5, 12985. [Google Scholar] [CrossRef]
- Buckingham, E.M.; Carpenter, J.E.; Jackson, W.; Grose, C. Autophagy and the effects of its inhibition on varicella-zoster virus glycoprotein biosynthesis and infectivity. J. Virol. 2014, 88, 890–902. [Google Scholar] [CrossRef] [Green Version]
- Lukhele, S.; Cohen, E.A. Conserved residues within the HIV-1 Vpu transmembrane-proximal hinge region modulate BST2 binding and antagonism. Retrovirology 2017, 14, 18. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, J.; Zeng, R.; Yang, J.; Liu, J.; Zhang, Z.; Song, X.; Yao, Z.; Ma, C.; Li, W.; et al. Respiratory Syncytial Virus Replication Is Promoted by Autophagy-Mediated Inhibition of Apoptosis. J. Virol. 2018, 92, e02193-17. [Google Scholar] [CrossRef] [Green Version]
- Jordan, T.X.; Randall, G. Dengue Virus Activates the AMP Kinase-mTOR Axis To Stimulate a Proviral Lipophagy. J. Virol. 2017, 91, e02020-16. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Buck, M.D.; Desai, C.; Zhang, X.; Loginicheva, E.; Martinez, J.; Freeman, M.L.; Saitoh, T.; Akira, S.; Guan, J.L.; et al. Autophagy Genes Enhance Murine Gammaherpesvirus 68 Reactivation from Latency by Preventing Virus-Induced Systemic Inflammation. Cell Host Microbe 2016, 19, 91–101. [Google Scholar] [CrossRef] [Green Version]
- Yordy, B.; Iijima, N.; Huttner, A.; Leib, D.; Iwasaki, A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 2012, 12, 334–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaumorcel, M.; Lussignol, M.; Mouna, L.; Cavignac, Y.; Fahie, K.; Cotte-Laffitte, J.; Geballe, A.; Brune, W.; Beau, I.; Codogno, P.; et al. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with Beclin 1. J. Virol. 2012, 86, 2571–2584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouna, L.; Hernandez, E.; Bonte, D.; Brost, R.; Amazit, L.; Delgui, L.R.; Brune, W.; Geballe, A.P.; Beau, I.; Esclatine, A. Analysis of the role of autophagy inhibition by two complementary human cytomegalovirus BECN1/Beclin 1-binding proteins. Autophagy 2016, 12, 327–342. [Google Scholar] [CrossRef] [Green Version]
- Liang, Q.; Wei, D.; Chung, B.; Brulois, K.F.; Guo, C.; Dong, S.; Gao, S.J.; Feng, P.; Liang, C.; Jung, J.U. Novel Role of vBcl2 in the Virion Assembly of Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2018, 92, e00914-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, W.T.; Giddings, T.H., Jr.; Taylor, M.P.; Mulinyawe, S.; Rabinovitch, M.; Kopito, R.R.; Kirkegaard, K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005, 3, e156. [Google Scholar] [CrossRef] [Green Version]
- Staring, J.; von Castelmur, E.; Blomen, V.A.; van den Hengel, L.G.; Brockmann, M.; Baggen, J.; Thibaut, H.J.; Nieuwenhuis, J.; Janssen, H.; van Kuppeveld, F.J.; et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 2017, 541, 412–416. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Pacheco, J.M.; LaRocco, M.; Burrage, T.; Jackson, W.; Rodriguez, L.L.; Borca, M.V.; Baxt, B. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology 2011, 410, 142–150. [Google Scholar] [CrossRef]
- Liang, Q.; Luo, Z.; Zeng, J.; Chen, W.; Foo, S.S.; Lee, S.A.; Ge, J.; Wang, S.; Goldman, S.A.; Zlokovic, B.V.; et al. Zika Virus NS4A and NS4B Proteins Deregulate Akt-mTOR Signaling in Human Fetal Neural Stem Cells to Inhibit Neurogenesis and Induce Autophagy. Cell Stem Cell 2016, 19, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Sun, H.; Pei, R.; Mao, B.; Zhao, Z.; Li, H.; Lin, Y.; Lu, K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021, 7, 31. [Google Scholar] [CrossRef]
- Gassen, N.C.; Papies, J.; Bajaj, T.; Emanuel, J.; Dethloff, F.; Chua, R.L.; Trimpert, J.; Heinemann, N.; Niemeyer, C.; Weege, F.; et al. SARS-CoV-2-mediated dysregulation of metabolism and autophagy uncovers host-targeting antivirals. Nat. Commun. 2021, 12, 3818. [Google Scholar] [CrossRef]
- Kyei, G.B.; Dinkins, C.; Davis, A.S.; Roberts, E.; Singh, S.B.; Dong, C.; Wu, L.; Kominami, E.; Ueno, T.; Yamamoto, A.; et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009, 186, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maity, S.; Saha, A. Therapeutic Potential of Exploiting Autophagy Cascade Against Coronavirus Infection. Front. Microbiol. 2021, 12, 675419. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Stone, S.; Natekar, J.; Kumari, P.; Arora, K.; Kumar, M. The FDA-approved gold drug auranofin inhibits novel coronavirus (SARS-CoV-2) replication and attenuates inflammation in human cells. Virology 2020, 547, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Jiang, K.; Ding, C.; Meng, S. Targeting Autophagy for Oncolytic Immunotherapy. Biomedicines 2017, 5, 5. [Google Scholar] [CrossRef]
- Cheney, L.; Barbaro, J.M.; Berman, J.W. Antiretroviral Drugs Impact Autophagy with Toxic Outcomes. Cells 2021, 10, 909. [Google Scholar] [CrossRef]
- Cloherty, A.P.M.; van Teijlingen, N.H.; Eisden, T.; van Hamme, J.L.; Rader, A.G.; Geijtenbeek, T.B.H.; Schreurs, R.; Ribeiro, C.M.S. Autophagy-enhancing drugs limit mucosal HIV-1 acquisition and suppress viral replication ex vivo. Sci. Rep. 2021, 11, 4767. [Google Scholar] [CrossRef]
- Jounai, N.; Takeshita, F.; Kobiyama, K.; Sawano, A.; Miyawaki, A.; Xin, K.Q.; Ishii, K.J.; Kawai, T.; Akira, S.; Suzuki, K.; et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14050–14055. [Google Scholar] [CrossRef] [Green Version]
- Viret, C.; Rozieres, A.; Faure, M. Autophagy during Early Virus-Host Cell Interactions. J. Mol. Biol. 2018, 430, 1696–1713. [Google Scholar] [CrossRef]
- Gregoire, I.P.; Rabourdin-Combe, C.; Faure, M. Autophagy and RNA virus interactomes reveal IRGM as a common target. Autophagy 2012, 8, 1136–1137. [Google Scholar] [CrossRef] [Green Version]

| Virus Inhibited | Autophagy Step | Mechanism | References |
|---|---|---|---|
| Hepatitis C Virus (HCV) | Elongation | SCOTIN associates with viral NS5A, leading to its degradation | [37] |
| Poliovirus | Cargo Selection | Galectin 8 marks permeated endosomes for autophagic destruction of the viral genome | [10] |
| Norovirus (NoV) | Nucleation | IFN-ɣ and GTPases recruit Atg5-Atg12-Atg16L1 complex to restrict the virus | [38] |
| Sindbis Virus (SINV) | Cargo Selection | p62 binds to viral capsid protein to target the virus to autophagosome | [39] |
| Vesicular Stomatitis Virus (VSV) | Initiation and Nucleation | VSV-G surface glycoprotein initiates the anti-viral autophagic pathway controlled by PI3K/Akt | [40] |
| Herpes Simplex Virus (HSV) | Cargo Selection | An LC3 like protein is derived from the viral nucleus to bind to the autophagosome | [36] |
| Human Immunodeficiency Virus (HIV-1) | Initiation | BST2 ectodomain anchors the HIV genome to cell membranes, to restrict virion release | [41] |
| Virus | Viral Protein | Mechanism | References |
|---|---|---|---|
| Human Parainfluenza Virus 3 (HPIV3) | Induces autophagy through AMPK for replication | [6,66,67] | |
| Measles Virus (MeV) | MeV-C protein | MeV binds CD46 to induce initial autophagy C protein binds host IRGM to induce a second autophagy wave after viral replication, to prevent cell death | [68,69] |
| Sendai Virus (SeV) | Induces autophagy through AMPK for replication | [6] | |
| Encephalomyocarditis Virus (EMCV) | Non-structural proteins 2C and 3D (NS2C/3D) | Induces autophagy through the ER stress pathway | [70] |
| Dengue Virus (DENV) | NS2B/3 | Induces ER stress through XBP1 and lipophagy to use the resulting ATP | [36,71] |
| West Nile Virus (WNV) | NS2B/3 | Induces lipophagy to use the resulting ATP | [36] |
| Hepatitis C Virus (HCV) | NS3/4A | Interacts with host annexin-A2 to use autophagosomal lipid rafts during viral RNA translation | [36,72] |
| Coxsackievirus B | Induces autophagy, to use the membrane for viral RNA replication | [73] | |
| Influenza A Virus (IAV) | M2 protein LC3-interacting region | M2 protein interacts with LC3 to relocate the virus to the plasma membrane for budding | [74] |
| Herpes Simplex Virus-1 (HSV-1) | TAP-blocking protein | Promotes use of autophagosome for protection during antigen presentation | [36] |
| Herpes Simplex Virus-2 (HSV-2) | Maintains basal level of autophagy for infection | [75] | |
| Varicella Zoster Virus (VZV) | Induction of autophagy for viral glycoprotein synthesis | [76] | |
| Human Immunodeficiency Virus (HIV-1) | Viral protein u (Vpu) | Removes BST2 from the viral budding sites to allow the spread of new virions | [77] |
| Virus | Viral Protein | Mechanism | References |
|---|---|---|---|
| Poliovirus | 2BC, 3A | Induces LC3 lipidation and double-membraned vesicle formation for replication | [36,85] |
| HRAS-like suppressor 3 (PLA2G16) | Escapes autophagic degradation by evading detection of its genome-containing endosomes | [36,86] | |
| Hepatitis C Virus (HCV) | RdRP NS5B | Binds Atg5 | [36] |
| NS4B | Induces UVRAG and Rubicon to enhance autophagic flux temporarily | ||
| Unknown | Targets host IRGM to fragment the Golgi apparatus | ||
| Foot-and-Mouth Disease Virus (FMDV) | VP1 capsid protein | Associates with p62 to use autophagosomes after the initial induction of autophagy | [36,87] |
| Zika Virus (ZIKV) | NS4A/4B | Inhibit AKT phosphorylation and mTOR activation | [36,88] |
| Coronavirus (SARS-CoV-2) | ORF3a | Binds to VPS39 and inhibits recruitment of Rab7 and the subsequent assembly of the SNARE complex, preventing autophagosome fusion with the lysosome | [89,90] |
| Human Simplex Virus 1 (HSV-1) | ICP34.5 | Inhibits Beclin 1 | [36] |
| Human Cytomegalovirus (HCMV) | TRS1, IRS1 | Inhibits Beclin 1 | [36,82] |
| Kaposi’s Sarcoma-associated Herpesvirus (KSHV) | vBcl-2 | Inhibits Beclin 1 | [84] |
| vFLIP | Blocks Atg3 E2 enzyme and the lipidation of LC3 | ||
| Human Immuno-deficiency Virus (HIV-1) | Nef | Inhibits Beclin 1 | [36,91] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chawla, K.; Subramanian, G.; Rahman, T.; Fan, S.; Chakravarty, S.; Gujja, S.; Demchak, H.; Chakravarti, R.; Chattopadhyay, S. Autophagy in Virus Infection: A Race between Host Immune Response and Viral Antagonism. Immuno 2022, 2, 153-169. https://doi.org/10.3390/immuno2010012
Chawla K, Subramanian G, Rahman T, Fan S, Chakravarty S, Gujja S, Demchak H, Chakravarti R, Chattopadhyay S. Autophagy in Virus Infection: A Race between Host Immune Response and Viral Antagonism. Immuno. 2022; 2(1):153-169. https://doi.org/10.3390/immuno2010012
Chicago/Turabian StyleChawla, Karan, Gayatri Subramanian, Tia Rahman, Shumin Fan, Sukanya Chakravarty, Shreyas Gujja, Hayley Demchak, Ritu Chakravarti, and Saurabh Chattopadhyay. 2022. "Autophagy in Virus Infection: A Race between Host Immune Response and Viral Antagonism" Immuno 2, no. 1: 153-169. https://doi.org/10.3390/immuno2010012
APA StyleChawla, K., Subramanian, G., Rahman, T., Fan, S., Chakravarty, S., Gujja, S., Demchak, H., Chakravarti, R., & Chattopadhyay, S. (2022). Autophagy in Virus Infection: A Race between Host Immune Response and Viral Antagonism. Immuno, 2(1), 153-169. https://doi.org/10.3390/immuno2010012








