Role of KIR Receptor in NK Regulation during Viral Infections
Abstract
1. Introduction
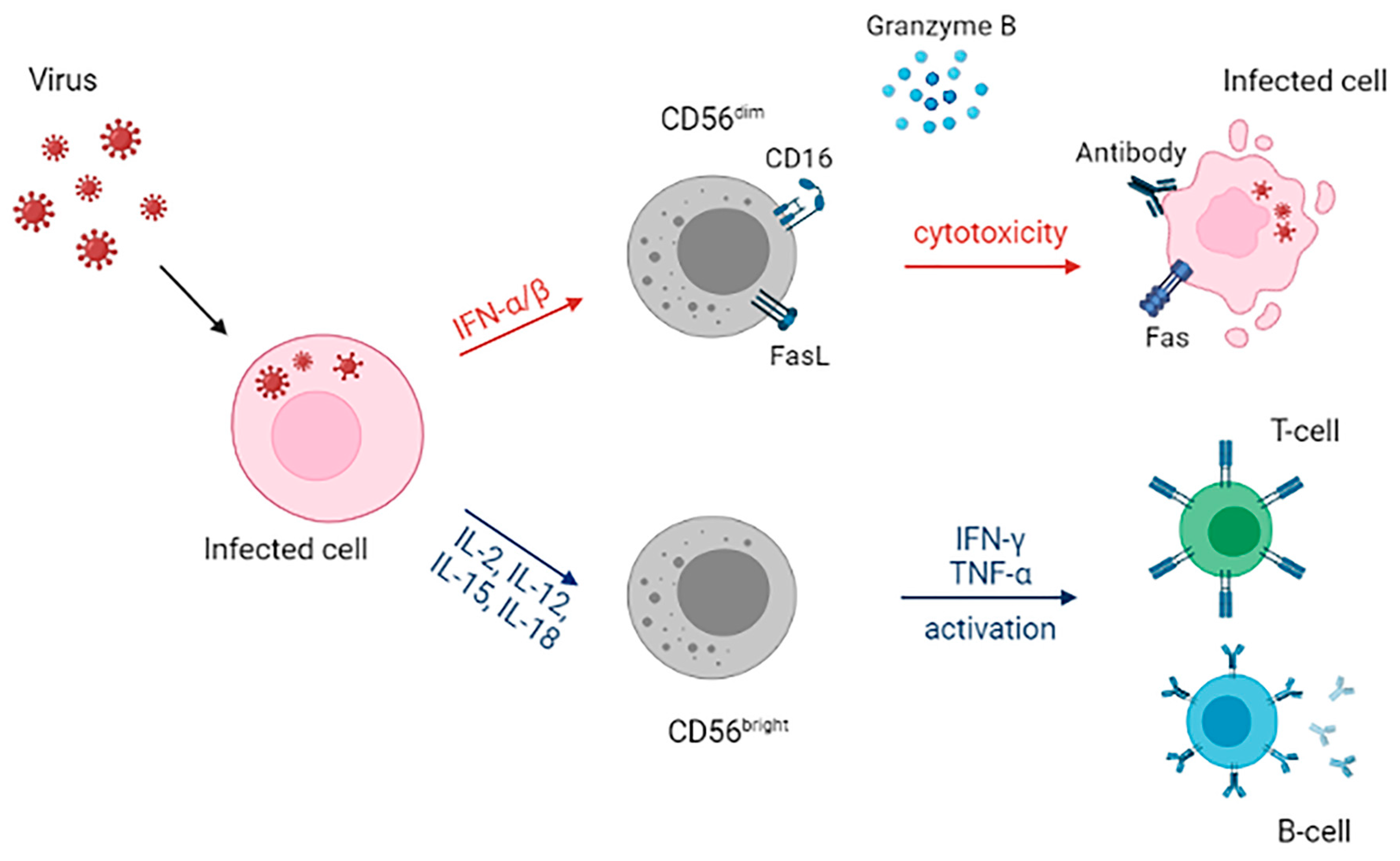
2. NK Receptors
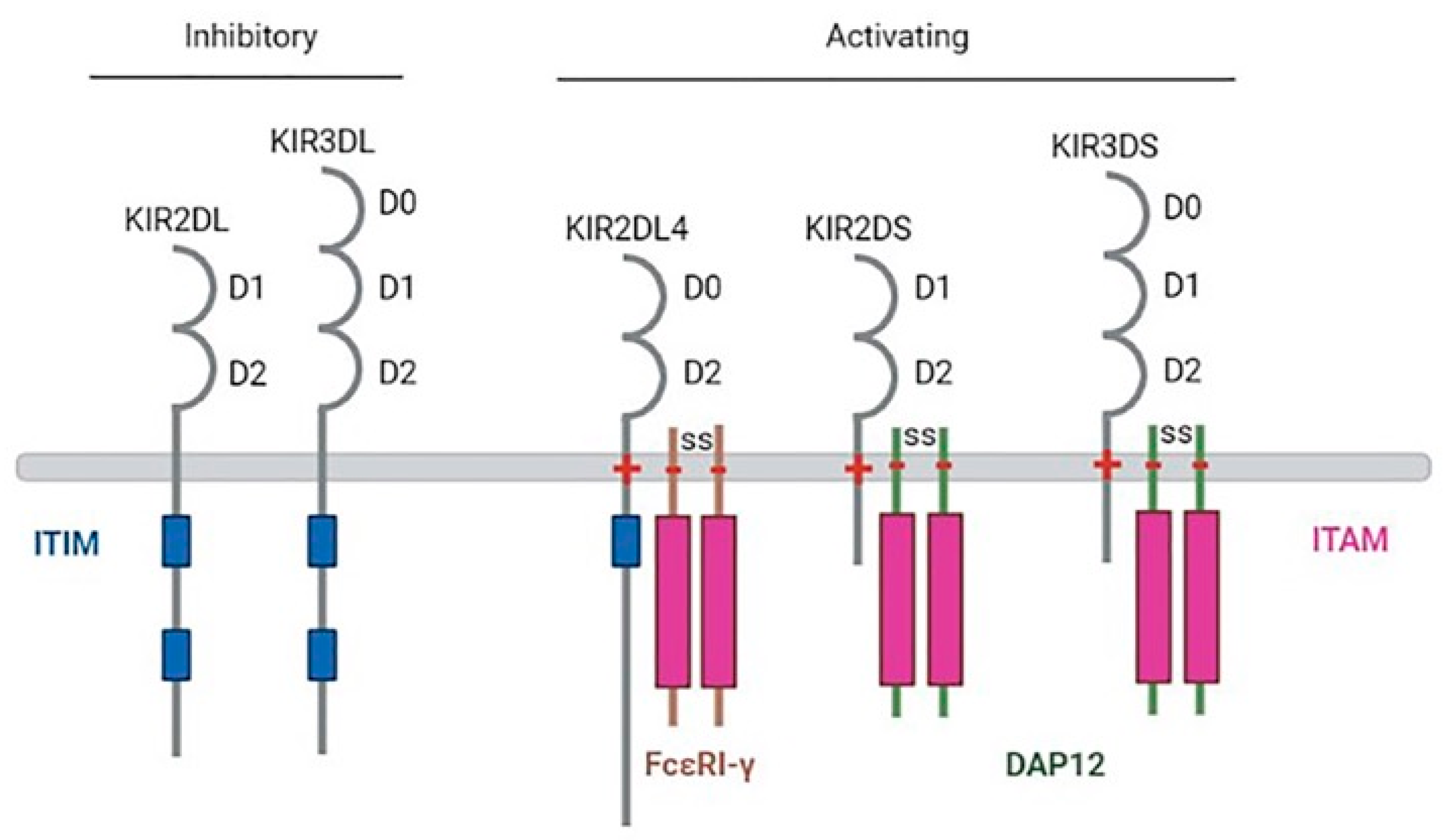
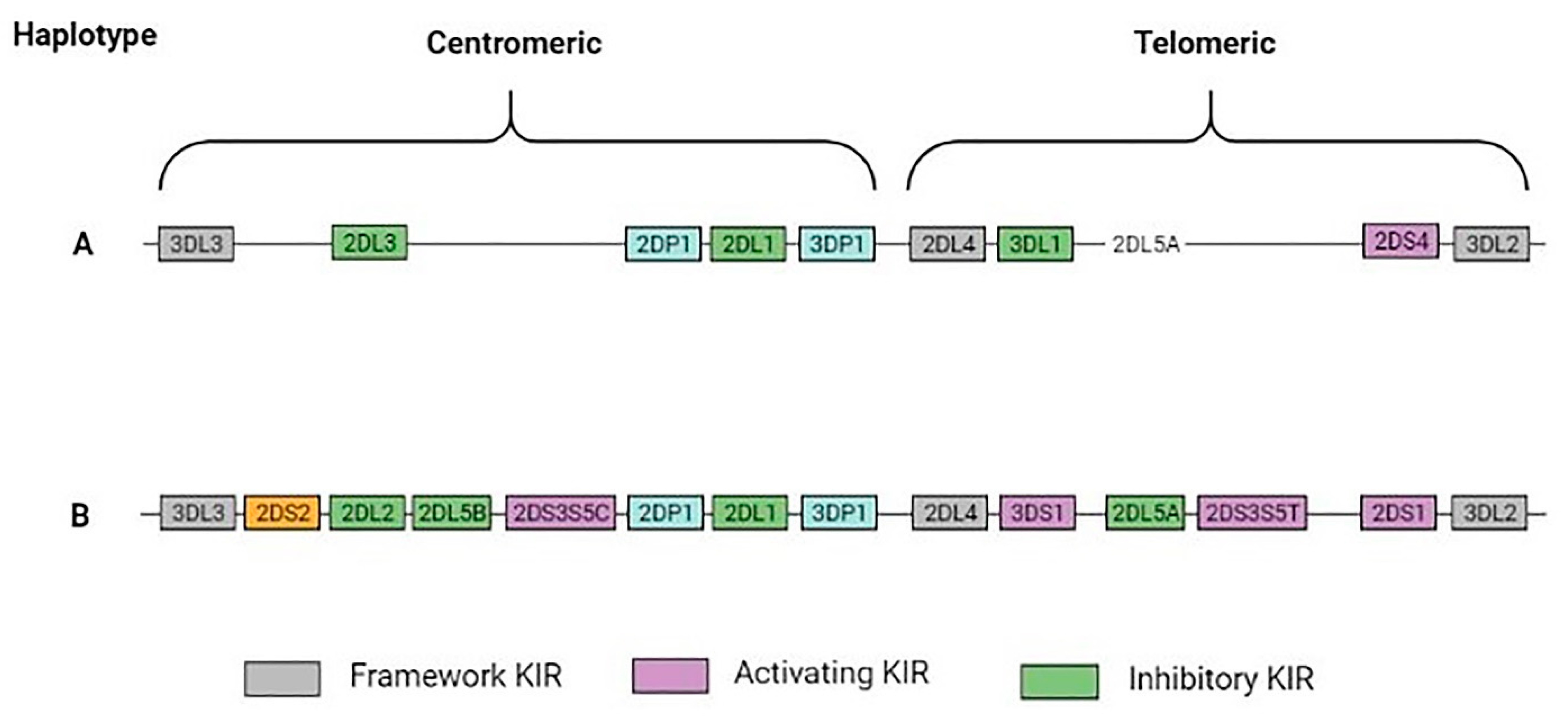
3. Killer Immunoglobulin-like Receptors (KIRs)
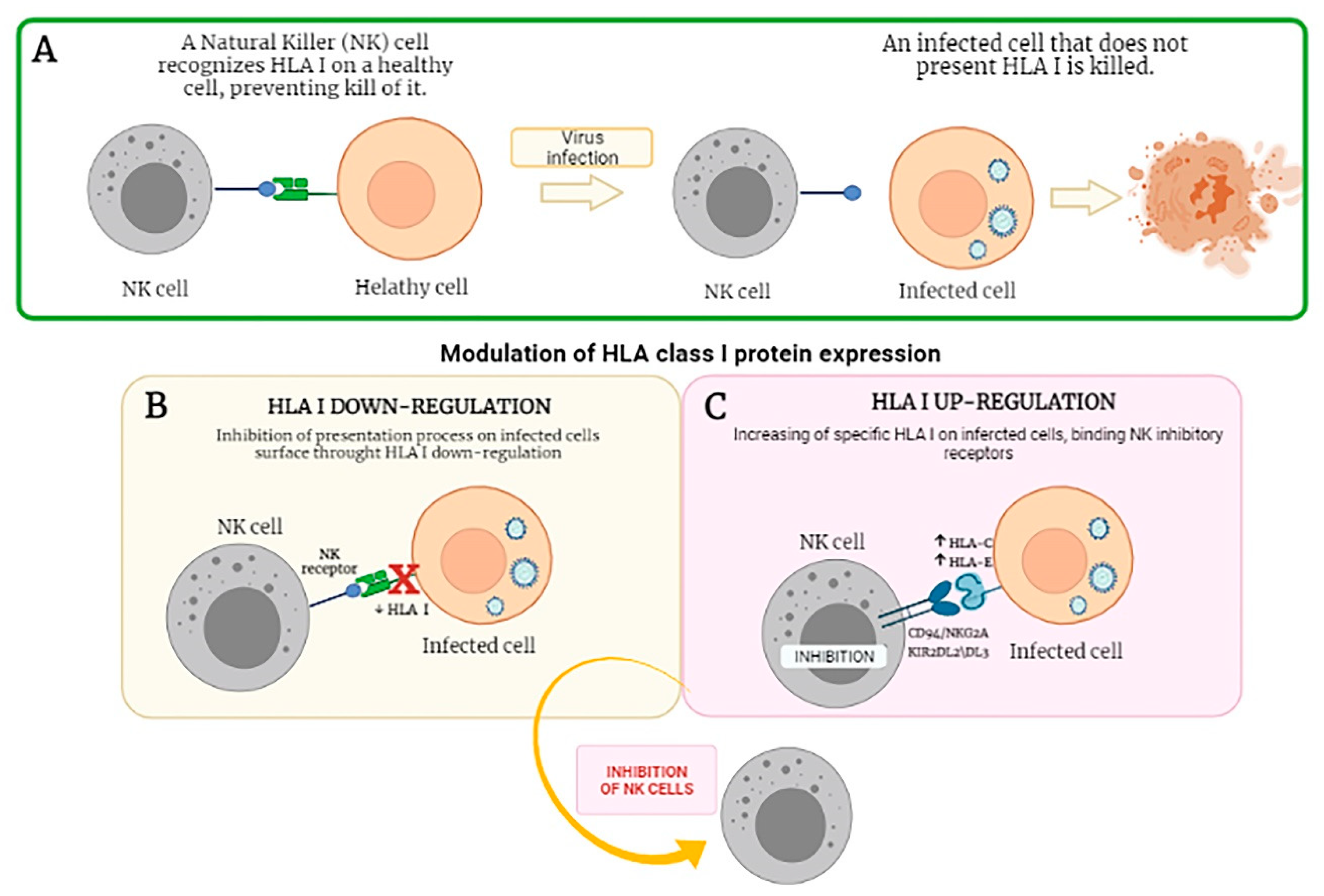
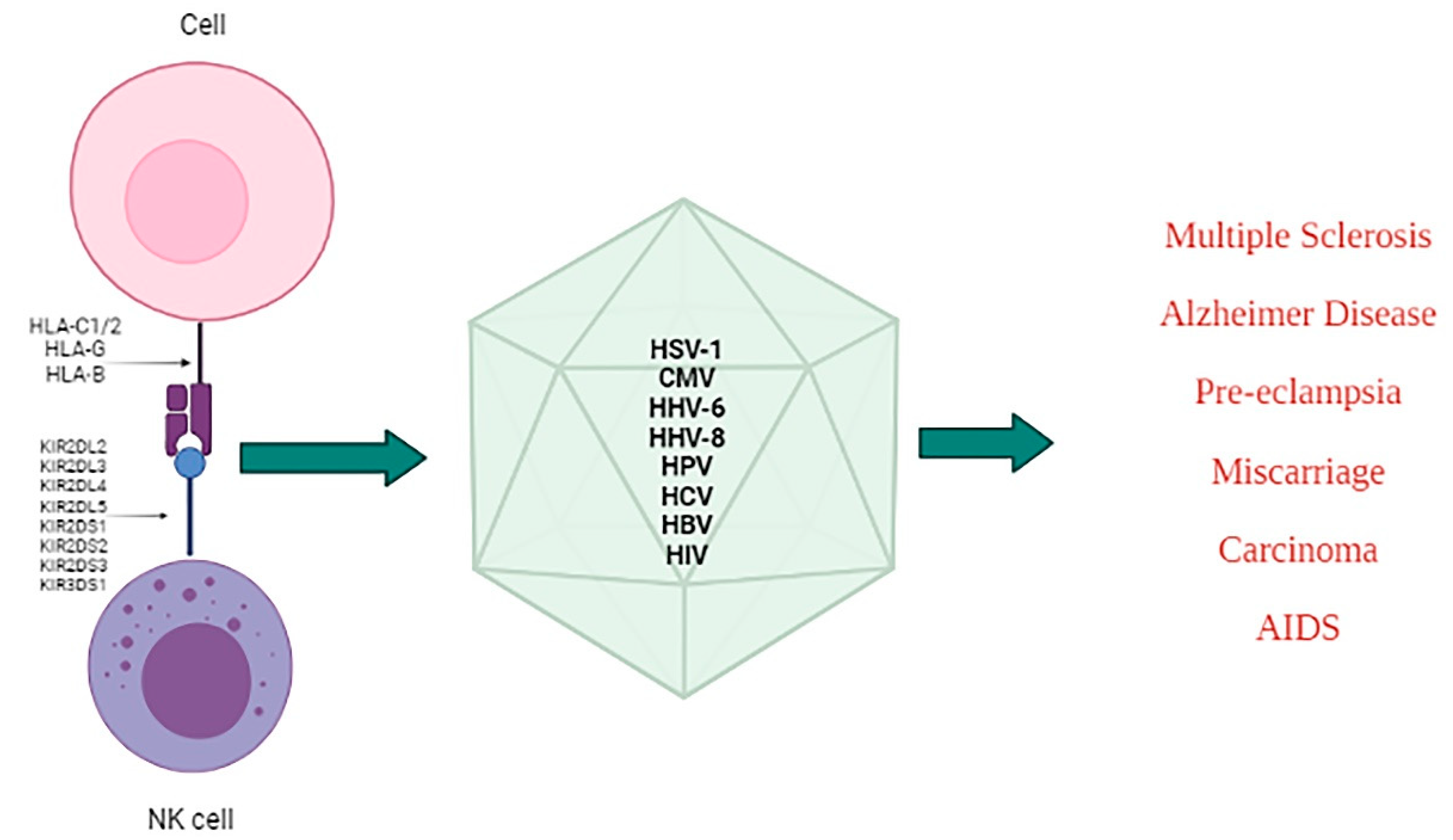
4. Modulation of KIRs/Ligands during Viral Infections and Associated Diseases
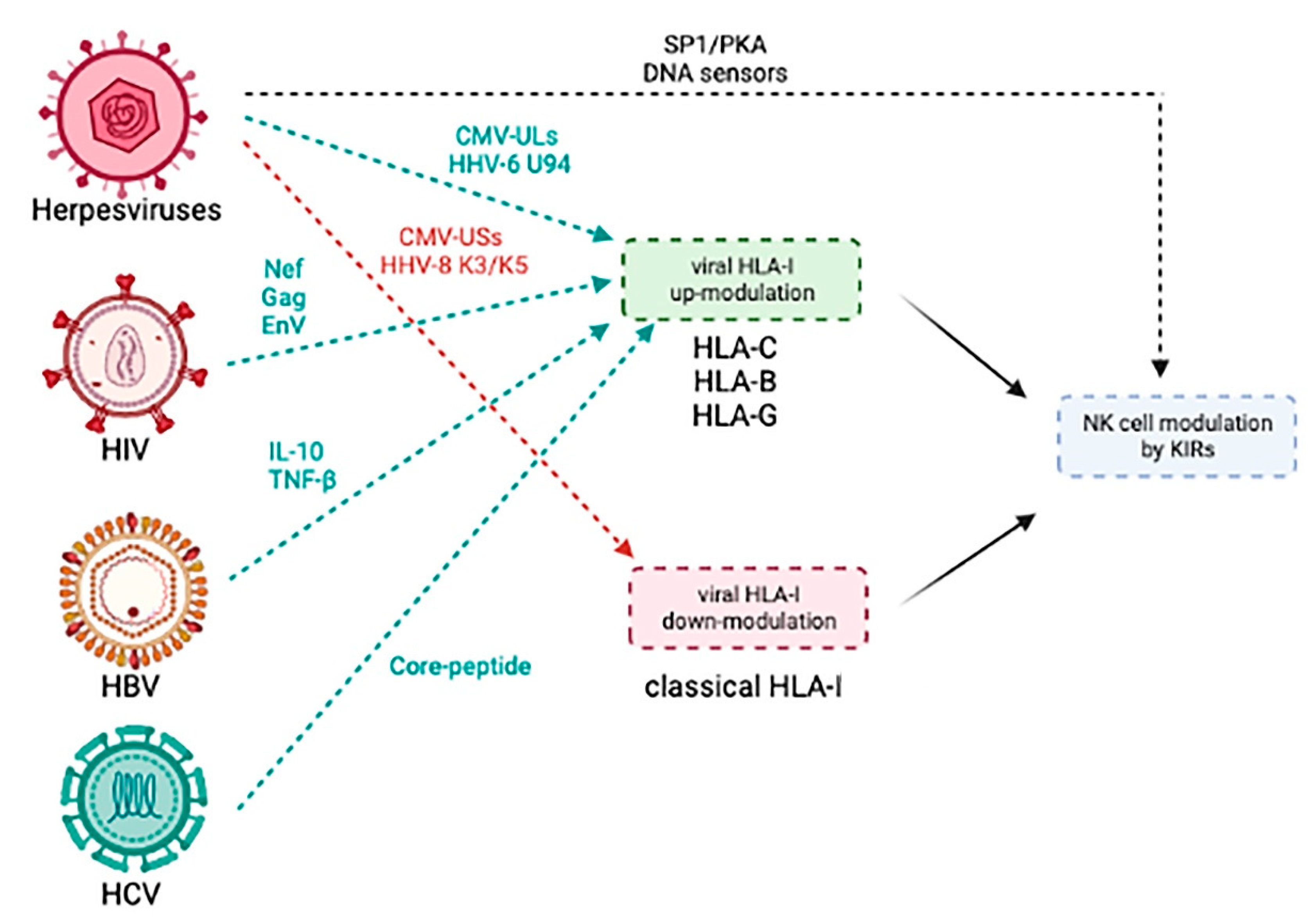
4.1. Herpesviruses
4.2. Human Immunodeficiency Virus (HIV)
4.3. Hepatitis Viruses (HBV and HCV)
4.4. Other Viruses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lanier, L.L. NK cell recognition. Annu. Rev. Immunol. 2005, 23, 225–274. [Google Scholar] [CrossRef] [PubMed]
- Sojka, D.K.; Tian, Z.; Yokoyama, W.M. Tissue-resident natural killer cells and their potential diversity. In Seminars in Immunology; Academic Press: Cambridge, MA, USA, 2014; pp. 127–131. [Google Scholar]
- Yokoyama, W.M.; Sojka, D.K.; Peng, H.; Tian, Z. Tissue-resident natural killer cells. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Long Island, NY, USA, 2013; pp. 149–156. [Google Scholar]
- Poli, A.; Michel, T.; Theresine, M.; Andres, E.; Hentges, F.; Zimmer, J. CD56bright natural killer (NK) cells: An important NK cell subset. Immunology 2009, 126, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Ferlazzo, G.; Thomas, D.; Lin, S.L.; Goodman, K.; Morandi, B.; Muller, W.A.; Moretta, A.; Munz, C. The Abundant Nk Cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J. Immunol. 2004, 172, 1455–1462. [Google Scholar] [CrossRef]
- Perussia, B.; Chen, Y.; Loza, M.J. Peripheral NK cell phenotypes: Multiple changing of faces of an adapting, developing cell. Mol. Immunol. 2005, 42, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Loke, Y.; King, A. Human Implantation: Cell Biology and Immunology; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Moffett-King, A. Natural killer cells and pregnancy. Nat. Rev. Immunol. 2002, 2, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Moffett-King, A.; Entrican, G.; Ellis, S.; Hutchinson, J.; Bainbridge, D. Natural killer cells and reproduction. Trends Immunol. 2002, 23, 332–333. [Google Scholar] [CrossRef]
- Biron, C.A.; Brossay, L. NK cells and NKT cells in innate defense against viral infections. Curr. Opin. Immunol. 2001, 13, 458–464. [Google Scholar] [CrossRef]
- Schmeel, L.C.; Schmeel, F.C.; Coch, C.; Schmidt-Wolf, I.G. Cytokine-induced killer (CIK) cells in cancer immunotherapy: Report of the international registry on CIK cells (IRCC). J. Cancer Res. Clin. Oncol. 2015, 141, 839–849. [Google Scholar] [CrossRef]
- Li, Y.; Schmidt-Wolf, I.G.; Wu, Y.F.; Huang, S.L.; Wei, J.; Fang, J.; Huang, K.; Zhou, D.H. Optimized protocols for generation of cord blood-derived cytokine-induced killer/natural killer cells. Anticancer Res. 2010, 30, 3493–3499. [Google Scholar]
- Cooper, M.A.; Fehniger, T.A.; Caligiuri, M.A. The biology of human natural killer-cell subsets. Trends Immunol. 2001, 22, 633–640. [Google Scholar] [CrossRef]
- Kumar, S. Natural killer cell cytotoxicity and its regulation by inhibitory receptors. Immunology 2018, 154, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Mavilio, D.; Benjamin, J.; Daucher, M.; Lombardo, G.; Kottilil, S.; Planta, M.A.; Marcenaro, E.; Bottino, C.; Moretta, L.; Moretta, A.; et al. Natural killer cells in HIV-1 infection: Dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc. Natl. Acad. Sci. USA 2003, 100, 15011–15016. [Google Scholar] [CrossRef] [PubMed]
- Alter, G.; Teigen, N.; Davis, B.T.; Addo, M.M.; Suscovich, T.J.; Waring, M.T.; Streeck, H.; Johnston, M.N.; Staller, K.D.; Zaman, M.T.; et al. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 2005, 106, 3366–3369. [Google Scholar] [CrossRef]
- Peritt, D.; Robertson, S.; Gri, G.; Showe, L.; Aste-Amezaga, M.; Trinchieri, G. Cutting edge: Differentiation of human NK cells into NK1 and NK2 subsets. J. Immunol. 1998, 161, 5821–5824. [Google Scholar]
- Berahovich, R.D.; Lai, N.L.; Wei, Z.; Lanier, L.L.; Schall, T.J. Evidence for NK cell subsets based on chemokine receptor expression. J. Immunol. 2006, 177, 7833–7840. [Google Scholar] [CrossRef]
- Michel, M.L.; Keller, A.C.; Paget, C.; Fujio, M.; Trottein, F.; Savage, P.B.; Wong, C.H.; Schneider, E.; Dy, M.; Leite-de-Moraes, M.C. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J. Exp. Med. 2007, 204, 995–1001. [Google Scholar] [CrossRef]
- Pandya, A.D.; Al-Jaderi, Z.; Hoglund, R.A.; Holmoy, T.; Harbo, H.F.; Norgauer, J.; Maghazachi, A.A. Identification of human NK17/NK1 cells. PLoS ONE 2011, 6, e26780. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Bortolotti, D.; Fainardi, E.; Gentili, V.; Bolzani, S.; Baldi, E.; Casetta, I.; Granieri, E.; Rotola, A.; Furlan, R.; et al. KIR2DL2 inhibitory pathway enhances Th17 cytokine secretion by NK cells in response to herpesvirus infection in multiple sclerosis patients. J. Neuroimmunol. 2016, 294, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Kang, H.S.; Kim, B.S. Th17 cells enhance viral persistence and inhibit T cell cytotoxicity in a model of chronic virus infection. J. Exp. Med. 2009, 206, 313–328. [Google Scholar] [CrossRef]
- Jost, S.; Altfeld, M. Control of human viral infections by natural killer cells. Annu. Rev. Immunol. 2013, 31, 163–194. [Google Scholar] [CrossRef]
- Sun, J.C.; Lopez-Verges, S.; Kim, C.C.; DeRisi, J.L.; Lanier, L.L. NK cells and immune “memory”. J. Immunol. 2011, 186, 1891–1897. [Google Scholar] [CrossRef]
- Altfeld, M.; Fadda, L.; Frleta, D.; Bhardwaj, N. DCs and NK cells: Critical effectors in the immune response to HIV-1. Nat. Rev. Immunol. 2011, 11, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Tomasello, E.; Baratin, M.; Walzer, T.; Ugolini, S. Functions of natural killer cells. Nat. Immunol. 2008, 9, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Biron, C.A.; Byron, K.S.; Sullivan, J.L. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 1989, 320, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Orange, J.S. Human natural killer cell deficiencies and susceptibility to infection. Microbes Infect. 2002, 4, 1545–1558. [Google Scholar] [CrossRef]
- Biron, C.A.; Dalod, M.; Salazar-Mather, T.P. Innate immunity and viral infections. Immunol. Infect. Dis. 2001, 11, 139–160. [Google Scholar] [CrossRef]
- Solerte, S.B.; Cravello, L.; Ferrari, E.; Fioravanti, M. Overproduction of IFN-gamma and TNF-alpha from natural killer (NK) cells is associated with abnormal NK reactivity and cognitive derangement in Alzheimer’s disease. Ann. N. Y. Acad. Sci. 2000, 917, 331–340. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Cooper, M.A.; Nuovo, G.J.; Cella, M.; Facchetti, F.; Colonna, M.; Caligiuri, M.A. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: A potential new link between adaptive and innate immunity. Blood 2003, 101, 3052–3057. [Google Scholar] [CrossRef]
- Walzer, T.; Dalod, M.; Robbins, S.H.; Zitvogel, L.; Vivier, E. Natural-killer cells and dendritic cells: “l’union fait la force”. Blood 2005, 106, 2252–2258. [Google Scholar] [CrossRef]
- Long, E.O. Ready for prime time: NK cell priming by dendritic cells. Immunity 2007, 26, 385–387. [Google Scholar] [CrossRef]
- Sim, G.C.; Radvanyi, L. The IL-2 cytokine family in cancer immunotherapy. Cytokine Growth Factor Rev. 2014, 25, 377–390. [Google Scholar] [CrossRef]
- Giri, J.G.; Kumaki, S.; Ahdieh, M.; Friend, D.J.; Loomis, A.; Shanebeck, K.; DuBose, R.; Cosman, D.; Park, L.S.; Anderson, D.M. Identification and cloning of a novel IL-15 binding protein that is structurally related to the alpha chain of the IL-2 receptor. EMBO J. 1995, 14, 3654–3663. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Shah, M.H.; Turner, M.J.; VanDeusen, J.B.; Whitman, S.P.; Cooper, M.A.; Suzuki, K.; Wechser, M.; Goodsaid, F.; Caligiuri, M.A. Differential cytokine and chemokine gene expression by human NK cells following activation with IL-18 or IL-15 in combination with IL-12: Implications for the innate immune response. J. Immunol. 1999, 162, 4511–4520. [Google Scholar]
- Lucas, M.; Schachterle, W.; Oberle, K.; Aichele, P.; Diefenbach, A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity 2007, 26, 503–517. [Google Scholar] [CrossRef]
- Nandagopal, N.; Ali, A.K.; Komal, A.K.; Lee, S.H. The critical role of IL-15-PI3K-mTOR pathway in natural killer cell effector functions. Front. Immunol. 2014, 5, 187. [Google Scholar] [CrossRef] [PubMed]
- Fehniger, T.A.; Cai, S.F.; Cao, X.; Bredemeyer, A.J.; Presti, R.M.; French, A.R.; Ley, T.J. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity 2007, 26, 798–811. [Google Scholar] [CrossRef]
- Cooper, M.A.; Elliott, J.M.; Keyel, P.A.; Yang, L.; Carrero, J.A.; Yokoyama, W.M. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA 2009, 106, 1915–1919. [Google Scholar] [CrossRef] [PubMed]
- Zamai, L.; Ahmad, M.; Bennett, I.M.; Azzoni, L.; Alnemri, E.S.; Perussia, B. Natural killer (NK) cell-mediated cytotoxicity: Differential use of TRAIL and Fas ligand by immature and mature primary human NK cells. J. Exp. Med. 1998, 188, 2375–2380. [Google Scholar] [CrossRef]
- Bryceson, Y.T.; Chiang, S.C.; Darmanin, S.; Fauriat, C.; Schlums, H.; Theorell, J.; Wood, S.M. Molecular mechanisms of natural killer cell activation. J. Innate Immun. 2011, 3, 216–226. [Google Scholar] [CrossRef]
- Lanier, L.L. Up on the tightrope: Natural killer cell activation and inhibition. Nat. Immunol. 2008, 9, 495–502. [Google Scholar] [CrossRef]
- Veillette, A. SLAM-family receptors: Immune regulators with or without SAP-family adaptors. Cold Spring Harb. Perspect. Biol. 2010, 2, a002469. [Google Scholar] [CrossRef]
- Wu, N.; Veillette, A. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 2016, 38, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Cerwenka, A.; Lanier, L.L. Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 2016, 16, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Afonina, I.S.; Cullen, S.P.; Martin, S.J. Cytotoxic and non-cytotoxic roles of the CTL/NK protease granzyme B. Immunol. Rev. 2010, 235, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Arnon, T.I.; Markel, G.; Mandelboim, O. Tumor and viral recognition by natural killer cells receptors. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2006; pp. 348–358. [Google Scholar]
- Jost, S.; Altfeld, M. Evasion from NK cell-mediated immune responses by HIV-1. Microbes Infect. 2012, 14, 904–915. [Google Scholar] [CrossRef]
- Arnon, T.I.; Lev, M.; Katz, G.; Chernobrov, Y.; Porgador, A.; Mandelboim, O. Recognition of viral hemagglutinins by NKp44 but not by NKp30. Eur. J. Immunol. 2001, 31, 2680–2689. [Google Scholar] [CrossRef]
- Mandelboim, O.; Lieberman, N.; Lev, M.; Paul, L.; Arnon, T.I.; Bushkin, Y.; Davis, D.M.; Strominger, J.L.; Yewdell, J.W.; Porgador, A. Recognition of haemagglutinins on virus-infected cells by NKp46 activates lysis by human NK cells. Nature 2001, 409, 1055–1060. [Google Scholar] [CrossRef]
- Baugh, R.; Khalique, H.; Seymour, L.W. Convergent evolution by cancer and viruses in evading the NKG2D immune response. Cancers 2020, 12, 3827. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Sun, C. The rise of NK cell checkpoints as promising therapeutic targets in cancer immunotherapy. Front. Immunol. 2019, 10, 2354. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.S.; Purdy, A.K. Structure/function of human killer cell immunoglobulin-like receptors: Lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 2011, 132, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Pende, D.; Falco, M.; Vitale, M.; Cantoni, C.; Vitale, C.; Munari, E.; Bertaina, A.; Moretta, F.; Del Zotto, G.; Pietra, G.; et al. Killer Ig-like receptors (KIRs): Their role in NK cell modulation and developments leading to their clinical exploitation. Front. Immunol. 2019, 10, 1179. [Google Scholar] [CrossRef] [PubMed]
- Colonna, M.; Samaridis, J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science 1995, 268, 405–408. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, A.; Chang, C.; Franz-Bacon, K.; McClanahan, T.; Phillips, J.H.; Lanier, L.L. Molecular cloning of NKB1. A natural killer cell receptor for HLA-B allotypes. J. Immunol. 1995, 155, 2306–2310. [Google Scholar] [PubMed]
- Scharenberg, A.M.; Kinet, J.P. The emerging field of receptor-mediated inhibitory signaling: SHP or SHIP? Cell 1996, 87, 961–964. [Google Scholar] [CrossRef][Green Version]
- Rajagopalan, S.; Fu, J.; Long, E.O. Cutting edge: Induction of IFN-gamma production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J. Immunol. 2001, 167, 1877–1881. [Google Scholar] [CrossRef]
- Kikuchi-Maki, A.; Catina, T.L.; Campbell, K.S. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the fc receptor gamma protein. J. Immunol. 2005, 174, 3859–3863. [Google Scholar] [CrossRef] [PubMed]
- Binstadt, B.A.; Brumbaugh, K.M.; Dick, C.J.; Scharenberg, A.M.; Williams, B.L.; Colonna, M.; Lanier, L.L.; Kinet, J.P.; Abraham, R.T.; Leibson, P.J. Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity 1996, 5, 629–638. [Google Scholar] [CrossRef]
- Valiante, N.M.; Phillips, J.H.; Lanier, L.L.; Parham, P. Killer cell inhibitory receptor recognition of human leukocyte antigen (HLA) class I blocks formation of a pp36/PLC-gamma signaling complex in human natural killer (NK) cells. J. Exp. Med. 1996, 184, 2243–2250. [Google Scholar] [CrossRef]
- Pegram, H.J.; Andrews, D.M.; Smyth, M.J.; Darcy, P.K.; Kershaw, M.H. Activating and inhibitory receptors of natural killer cells. Immunol. Cell Biol. 2011, 89, 216–224. [Google Scholar] [CrossRef]
- Jonsson, A.H.; Yokoyama, W.M. Natural killer cell tolerance licensing and other mechanisms. Adv. Immunol. 2009, 101, 27–79. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.S.; Poulton, K.V.; Poles, A. Factors affecting HLA expression: A review. Int. J. Immunogenet. 2019, 46, 307–320. [Google Scholar] [CrossRef]
- Van den Elsen, P.J. Expression regulation of major histocompatibility complex class I and class II encoding genes. Front. Immunol. 2011, 2, 48. [Google Scholar] [CrossRef]
- Van den Elsen, P.J.; Holling, T.M.; Kuipers, H.F.; van der Stoep, N. Transcriptional regulation of antigen presentation. Curr. Opin. Immunol. 2004, 16, 67–75. [Google Scholar] [CrossRef]
- Stewart, C.A.; Laugier-Anfossi, F.; Vely, F.; Saulquin, X.; Riedmuller, J.; Tisserant, A.; Gauthier, L.; Romagne, F.; Ferracci, G.; Arosa, F.A.; et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 13224–13229. [Google Scholar] [CrossRef] [PubMed]
- Dohring, C.; Colonna, M. Human natural killer cell inhibitory receptors bind to HLA class I molecules. Eur. J. Immunol. 1996, 26, 365–369. [Google Scholar] [CrossRef]
- Pende, D.; Biassoni, R.; Cantoni, C.; Verdiani, S.; Falco, M.; di Donato, C.; Accame, L.; Bottino, C.; Moretta, A.; Moretta, L. The natural killer cell receptor specific for HLA-A allotypes: A novel member of the p58/p70 family of inhibitory receptors that is characterized by three immunoglobulin-like domains and is expressed as a 140-kD disulphide-linked dimer. J. Exp. Med. 1996, 184, 505–518. [Google Scholar] [CrossRef]
- Tajik, N.; Shahsavar, F.; Mousavi, T.; Radjabzadeh, M.F. Distribution of KIR genes in the Iranian population. Tissue Antigens 2009, 74, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Kulski, J.K.; Gaudieri, S.; Witt, C.S.; Freitas, E.M.; Trowsdale, J.; Christiansen, F.T. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene 2004, 335, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Lozano, N.; Estefania, E.; Williams, F.; Halfpenny, I.; Middleton, D.; Solis, R.; Vilches, C. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur. J. Immunol. 2005, 35, 16–24. [Google Scholar] [CrossRef]
- Gomez-Lozano, N.; Gardiner, C.M.; Parham, P.; Vilches, C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics 2002, 54, 314–319. [Google Scholar] [CrossRef]
- Uhrberg, M. The KIR gene family: Life in the fast lane of evolution. Eur. J. Immunol. 2005, 35, 10–15. [Google Scholar] [CrossRef]
- Uhrberg, M.; Valiante, N.M.; Shum, B.P.; Shilling, H.G.; Lienert-Weidenbach, K.; Corliss, B.; Tyan, D.; Lanier, L.L.; Parham, P. Human diversity in killer cell inhibitory receptor genes. Immunity 1997, 7, 753–763. [Google Scholar] [CrossRef]
- Shilling, H.G.; Guethlein, L.A.; Cheng, N.W.; Gardiner, C.M.; Rodriguez, R.; Tyan, D.; Parham, P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J. Immunol. 2002, 168, 2307–2315. [Google Scholar] [CrossRef]
- Yawata, M.; Yawata, N.; Abi-Rached, L.; Parham, P. Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit. Rev. Immunol. 2002, 22, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.C.; Liu, X.R.; Selvakumar, A.; Mickelson, E.; O’Reilly, R.J.; Dupont, B. Killer Ig-like receptor haplotype analysis by gene content: Evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J. Immunol. 2002, 169, 5118–5129. [Google Scholar] [CrossRef] [PubMed]
- Orange, J.S.; Fassett, M.S.; Koopman, L.A.; Boyson, J.E.; Strominger, J.L. Viral evasion of natural killer cells. Nat. Immunol. 2002, 3, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Machold, R.P.; Wiertz, E.J.; Jones, T.R.; Ploegh, H. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J. Exp. Med. 1997, 185, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Früh, K.; Gruhler, A.; Krishna, R.M.; Schoenhals, G.J. A comparison of viral immune escape strategies targeting the MHC class I assembly pathway. Immunol. Rev. 1999, 168, 157–166. [Google Scholar] [CrossRef]
- Chan, H.-W.; Kurago, Z.B.; Stewart, C.A.; Wilson, M.J.; Martin, M.P.; Mace, B.E.; Carrington, M.; Trowsdale, J.; Lutz, C.T. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J. Exp. Med. 2003, 197, 245–255. [Google Scholar] [CrossRef]
- Milavetz, B.I.; Balakrishnan, L. Viral epigenetics. Methods Mol. Biol. 2015, 1238, 569–596. [Google Scholar] [CrossRef]
- Blazkova, J.; Trejbalova, K.; Gondois-Rey, F.; Halfon, P.; Philibert, P.; Guiguen, A.; Verdin, E.; Olive, D.; Van Lint, C.; Hejnar, J. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009, 5, e1000554. [Google Scholar] [CrossRef]
- Chan, H.W.; Miller, J.S.; Moore, M.B.; Lutz, C.T. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J. Immunol. 2005, 175, 5966–5974. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, S.; Long, E.O. Understanding how combinations of HLA and KIR genes influence disease. J. Exp. Med. 2005, 201, 1025–1029. [Google Scholar] [CrossRef]
- Snyder, M.R.; Weyand, C.M.; Goronzy, J.J. The double life of NK receptors: Stimulation or co-stimulation? Trends Immunol. 2004, 25, 25–32. [Google Scholar] [CrossRef]
- López-Botet, M.; Llano, M.; Ortega, M. Human cytomegalovirus and natural killer-mediated surveillance of HLA class I expression: A paradigm of host–pathogen adaptation. Immunol. Rev. 2001, 181, 193–202. [Google Scholar] [CrossRef]
- Miller, J.S. Biology of Natural Killer Cells in Cancer and Infection: Miniseries/Special Article. Cancer Investig. 2002, 20, 405–419. [Google Scholar] [CrossRef]
- Hopfensperger, K.; Richard, J.; Stürzel, C.M.; Bibollet-Ruche, F.; Apps, R.; Leoz, M.; Plantier, J.-C.; Hahn, B.H.; Finzi, A.; Kirchhoff, F. Convergent Evolution of HLA-C Downmodulation in HIV-1 and HIV-2. mBio 2020, 11, e00782-20. [Google Scholar] [CrossRef] [PubMed]
- Jugovic, P.; Hill, A.M.; Tomazin, R.; Ploegh, H.; Johnson, D.C. Inhibition of major histocompatibility complex class I antigen presentation in pig and primate cells by herpes simplex virus type 1 and 2 ICP47. J. Virol. 1998, 72, 5076–5084. [Google Scholar] [CrossRef] [PubMed]
- Tortorella, D.; Gewurz, B.E.; Furman, M.H.; Schust, D.J.; Ploegh, H.L. Viral subversion of the immune system. Annu. Rev. Immunol. 2000, 18, 861–926. [Google Scholar] [CrossRef]
- Collins, K.L.; Chen, B.K.; Kalams, S.A.; Walker, B.D.; Baltimore, D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 1998, 391, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.; Martin, M.P.; Carrington, M. The Yin and Yang of HLA and KIR in human disease. In Seminars in Immunology; Academic Press: Cambridge, MA, USA, 2008; pp. 343–352. [Google Scholar]
- Romero, V.; Azocar, J.; Zúñiga, J.; Clavijo, O.P.; Terreros, D.; Gu, X.; Husain, Z.; Chung, R.T.; Amos, C.; Yunis, E.J. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol. Immunol. 2008, 45, 2429–2436. [Google Scholar] [CrossRef]
- Rizzo, R.; Gentili, V.; Casetta, I.; Caselli, E.; De Gennaro, R.; Granieri, E.; Cassai, E.; Di Luca, D.; Rotola, A. Altered natural killer cells’ response to herpes virus infection in multiple sclerosis involves KIR2DL2 expression. J. Neuroimmunol. 2012, 251, 55–64. [Google Scholar] [CrossRef]
- Gewurz, B.E.; Wang, E.W.; Tortorella, D.; Schust, D.J.; Ploegh, H.L. Human cytomegalovirus US2 endoplasmic reticulum-lumenal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J. Virol. 2001, 75, 5197–5204. [Google Scholar] [CrossRef] [PubMed]
- Guma, M.; Angulo, A.; Lopez-Botet, M. NK cell receptors involved in the response to human cytomegalovirus infection. Curr. Top Microbiol. Immunol. 2006, 298, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Lanteri, M.; Giordanengo, V.; Hiraoka, N.; Fuzibet, J.G.; Auberger, P.; Fukuda, M.; Baum, L.G.; Lefebvre, J.C. Altered T cell surface glycosylation in HIV-1 infection results in increased susceptibility to galectin-1-induced cell death. Glycobiology 2003, 13, 909–918. [Google Scholar] [CrossRef]
- Kielczewska, A.; Pyzik, M.; Sun, T.; Krmpotic, A.; Lodoen, M.B.; Munks, M.W.; Babic, M.; Hill, A.B.; Koszinowski, U.H.; Jonjic, S.; et al. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J. Exp. Med. 2009, 206, 515–523. [Google Scholar] [CrossRef]
- Crespo, A.C.; Strominger, J.L.; Tilburgs, T. Expression of KIR2DS1 by decidual natural killer cells increases their ability to control placental HCMV infection. Proc. Natl. Acad. Sci. USA 2016, 113, 15072–15077. [Google Scholar] [CrossRef] [PubMed]
- Kenneson, A.; Cannon, M.J. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev. Med. Virol. 2007, 17, 253–276. [Google Scholar] [CrossRef]
- Schust, D.J.; Tortorella, D.; Seebach, J.; Phan, C.; Ploegh, H.L. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J. Exp. Med. 1998, 188, 497–503. [Google Scholar] [CrossRef]
- Gazit, R.; Garty, B.Z.; Monselise, Y.; Hoffer, V.; Finkelstein, Y.; Markel, G.; Katz, G.; Hanna, J.; Achdout, H.; Gruda, R.; et al. Expression of KIR2DL1 on the entire NK cell population: A possible novel immunodeficiency syndrome. Blood 2004, 103, 1965–1966. [Google Scholar] [CrossRef]
- Van Duin, D.; Avery, R.K.; Hemachandra, S.; Yen-Lieberman, B.; Zhang, A.; Jain, A.; Butler, R.S.; Barnard, J.; Schold, J.D.; Fung, J.; et al. KIR and HLA interactions are associated with control of primary CMV infection in solid organ transplant recipients. Am. J. Transplant. 2014, 14, 156–162. [Google Scholar] [CrossRef]
- Hou, Y.F.; Zhang, Y.C.; Jiao, Y.L.; Wang, L.C.; Li, J.F.; Pan, Z.L.; Yang, Q.R.; Sun, H.S.; Zhao, Y.R. Disparate distribution of activating and inhibitory killer cell immunoglobulin-like receptor genes in patients with systemic lupus erythematosus. Lupus 2010, 19, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Pellett, F.; Siannis, F.; Vukin, I.; Lee, P.; Urowitz, M.; Gladman, D. KIRs and autoimmune disease: Studies in systemic lupus erythematosus and scleroderma. In Tissue Antigens; Wiley Online Library: Hoboken, NJ, USA, 2007. [Google Scholar]
- Majorczyk, E.; Pawlik, A.; Luszczek, W.; Nowak, I.; Wisniewski, A.; Jasek, M.; Kusnierczyk, P. Associations of killer cell immunoglobulin-like receptor genes with complications of rheumatoid arthritis. Genes Immun. 2007, 8, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Hoteit, R.; Bazarbachi, A.; Antar, A.; Salem, Z.; Shammaa, D.; Mahfouz, R. KIR genotype distribution among patients with multiple myeloma: Higher prevalence of KIR 2DS4 and KIR 2DS5 genes. Meta Gene 2014, 2, 730–736. [Google Scholar] [CrossRef] [PubMed]
- Coscoy, L.; Ganem, D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 2000, 97, 8051–8056. [Google Scholar] [CrossRef] [PubMed]
- Ishido, S.; Wang, C.; Lee, B.S.; Cohen, G.B.; Jung, J.U. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 2000, 74, 5300–5309. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lybarger, L.; Connors, R.; Harris, M.R.; Hansen, T.H. Model for the interaction of gammaherpesvirus 68 RING-CH finger protein mK3 with major histocompatibility complex class I and the peptide-loading complex. J. Virol. 2004, 78, 8673–8686. [Google Scholar] [CrossRef]
- Caselli, E.; Rizzo, R.; Ingianni, A.; Contini, P.; Pompei, R.; Di Luca, D. High prevalence of HHV8 infection and specific killer cell immunoglobulin-like receptors allotypes in Sardinian patients with type 2 diabetes mellitus. J. Med. Virol. 2014, 86, 1745–1751. [Google Scholar] [CrossRef]
- Borghi, A.; D’Accolti, M.; Rizzo, R.; Virgili, A.; Di Luca, D.; Corazza, M.; Caselli, E. High prevalence of specific KIR types in patients with HHV-8 positive cutaneous vascular lesions: A possible predisposing factor? Arch. Dermatol. Res. 2016, 308, 373–377. [Google Scholar] [CrossRef]
- Guerini, F.R.; Mancuso, R.; Agostini, S.; Agliardi, C.; Zanzottera, M.; Hernis, A.; Tourlaki, A.; Calvo, M.G.; Bellinvia, M.; Brambilla, L.; et al. Activating KIR/HLA complexes in classic Kaposi’s Sarcoma. Infect. Agent. Cancer 2012, 7, 9. [Google Scholar] [CrossRef]
- Eimer, W.A.; Vijaya Kumar, D.K.; Navalpur Shanmugam, N.K.; Rodriguez, A.S.; Mitchell, T.; Washicosky, K.J.; Gyorgy, B.; Breakefield, X.O.; Tanzi, R.E.; Moir, R.D. Alzheimer’s disease-associated beta-amyloid is rapidly seeded by herpesviridae to protect against brain infection. Neuron 2018, 99, 56–63. [Google Scholar] [CrossRef]
- Rizzo, R.; Bortolotti, D.; Gentili, V.; Rotola, A.; Bolzani, S.; Caselli, E.; Tola, M.R.; Di Luca, D. KIR2DS2/KIR2DL2/HLA-C1 Haplotype is associated with Alzheimer’s disease: Implication for the role of herpesvirus infections. J. Alzheimers Dis. 2019, 67, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Machado-Sulbaran, A.C.; Ramirez-Duenas, M.G.; Navarro-Zarza, J.E.; Munoz-Valle, J.F.; Mendoza-Carrera, F.; Banos-Hernandez, C.J.; Parra-Rojas, I.; Montoya-Buelna, M.; Sanchez-Hernandez, P.E. KIR/HLA gene profile implication in systemic sclerosis patients from Mexico. J. Immunol. Res. 2019, 2019, 6808061. [Google Scholar] [CrossRef] [PubMed]
- Ben Fredj, N.; Rizzo, R.; Bortolotti, D.; Nefzi, F.; Chebel, S.; Rotola, A.; Frih-Ayed, M.; Di Luca, D.; Aouni, M. Evaluation of the implication of KIR2DL2 receptor in multiple sclerosis and herpesvirus susceptibility. J. Neuroimmunol. 2014, 271, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Bortolotti, D.; Gentili, V.; Santi, E.; Taliento, C.; Vitagliano, A.; Schiuma, G.; Beltrami, S.; Rizzo, S.; Lanza, G.; Rizzo, R.; et al. Late-onset intrauterine growth restriction and HHV-6 infection: A pilot study. J. Med. Virol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; D’Accolti, M.; Bortolotti, D.; Caccuri, F.; Caruso, A.; Di Luca, D.; Caselli, E. Human herpesvirus 6A and 6B inhibit in vitro angiogenesis by induction of human leukocyte antigen G. Sci. Rep. 2018, 8, 17683. [Google Scholar] [CrossRef] [PubMed]
- Komaroff, A.L.; Rizzo, R.; Ecker, J.L. Human herpesviruses 6A and 6B in reproductive diseases. Front. Immunol. 2021, 12, 648945. [Google Scholar] [CrossRef]
- Caselli, E.; Bortolotti, D.; Marci, R.; Rotola, A.; Gentili, V.; Soffritti, I.; D’Accolti, M.; Lo Monte, G.; Sicolo, M.; Barao, I.; et al. HHV-6A infection of endometrial epithelial cells induces increased endometrial NK cell-mediated cytotoxicity. Front. Microbiol. 2017, 8, 2525. [Google Scholar] [CrossRef]
- Marci, R.; Gentili, V.; Bortolotti, D.; Lo Monte, G.; Caselli, E.; Bolzani, S.; Rotola, A.; Di Luca, D.; Rizzo, R. Presence of HHV-6A in endometrial epithelial cells from women with primary unexplained infertility. PLoS ONE 2016, 11, e0158304. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, A.; Bortolotti, D.; Marci, R.; Caselli, E.; Falzoni, S.; De Marchi, E.; Di Virgilio, F.; Rizzo, R.; Adinolfi, E. The P2X7 receptor 489C>T gain of function polymorphism favors HHV-6A infection and associates with female idiopathic infertility. Front. Pharmacol. 2020, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Lo Monte, G.; Bortolotti, D.; Graziano, A.; Gentili, V.; Di Luca, D.; Marci, R. Impact of soluble HLA-G levels and endometrial NK cells in uterine flushing samples from primary and secondary unexplained infertile women. Int. J. Mol. Sci. 2015, 16, 5510–5516. [Google Scholar] [CrossRef]
- Djulejic, E.; Petlickovski, A.; Trajkov, D.; Dimitrov, G.; Alabakovska, S. KIR Gene Frequencies in Women with Infertility Problems. South East Eur. J. Immunol. 2015, 2015, 1–5. [Google Scholar] [CrossRef]
- El Borai, N.; Inoue, M.; Lefevre, C.; Naumova, E.N.; Sato, B.; Yamamura, M. Detection of herpes simplex DNA in semen and menstrual blood of individuals attending an infertility clinic. J. Obstet. Gynaecol. Res. 1997, 23, 17–24. [Google Scholar] [CrossRef]
- Gimenes, F.; Souza, R.P.; Bento, J.C.; Teixeira, J.J.; Maria-Engler, S.S.; Bonini, M.G.; Consolaro, M.E. Male infertility: A public health issue caused by sexually transmitted pathogens. Nat. Rev. Urol. 2014, 11, 672–687. [Google Scholar] [CrossRef]
- Apps, R.; Qi, Y.; Carlson, J.M.; Chen, H.; Gao, X.; Thomas, R.; Yuki, Y.; Del Prete, G.Q.; Goulder, P.; Brumme, Z.L.; et al. Influence of HLA-C expression level on HIV control. Science 2013, 340, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, S.; Erdtmann, L.; Benichou, S.; Berlioz-Torrent, C.; Liu, L.; Benarous, R.; Heard, J.M.; Schwartz, O. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 1998, 8, 483–495. [Google Scholar] [CrossRef]
- Cohen, G.B.; Gandhi, R.T.; Davis, D.M.; Mandelboim, O.; Chen, B.K.; Strominger, J.L.; Baltimore, D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999, 10, 661–671. [Google Scholar] [CrossRef]
- Fadda, L.; Korner, C.; Kumar, S.; van Teijlingen, N.H.; Piechocka-Trocha, A.; Carrington, M.; Altfeld, M. HLA-Cw*0102-restricted HIV-1 p24 epitope variants can modulate the binding of the inhibitory KIR2DL2 receptor and primary NK cell function. PLoS Pathog. 2012, 8, e1002805. [Google Scholar] [CrossRef] [PubMed]
- Parolini, F.; Biswas, P.; Serena, M.; Sironi, F.; Muraro, V.; Guizzardi, E.; Cazzoletti, L.; Scupoli, M.T.; Gibellini, D.; Ugolotti, E.; et al. Stability and expression levels of HLA-C on the cell membrane modulate HIV-1 infectivity. J. Virol. 2018, 92, e01711–e01717. [Google Scholar] [CrossRef] [PubMed]
- Serena, M.; Parolini, F.; Biswas, P.; Sironi, F.; Blanco Miranda, A.; Zoratti, E.; Scupoli, M.T.; Ziglio, S.; Valenzuela-Fernandez, A.; Gibellini, D.; et al. HIV-1 Env associates with HLA-C free-chains at the cell membrane modulating viral infectivity. Sci. Rep. 2017, 7, 40037. [Google Scholar] [CrossRef]
- Long, B.R.; Ndhlovu, L.C.; Oksenberg, J.R.; Lanier, L.L.; Hecht, F.M.; Nixon, D.F.; Barbour, J.D. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J. Virol. 2008, 82, 4785–4792. [Google Scholar] [CrossRef]
- Omosun, Y.O.; Blackstock, A.J.; Williamson, J.; van Eijk, A.M.; Ayisi, J.; Otieno, J.; Lal, R.B.; Ter Kuile, F.O.; Slutsker, L.; Shi, Y.P. Association of maternal KIR gene content polymorphisms with reduction in perinatal transmission of HIV-1. PLoS ONE 2018, 13, e0191733. [Google Scholar] [CrossRef]
- Sorgho, P.A.; Djigma, F.W.; Martinson, J.J.; Yonli, A.T.; Nagalo, B.M.; Compaore, T.R.; Diarra, B.; Sombie, H.K.; Simpore, A.; Zongo, A.W.; et al. Role of killer cell immunoglobulin-like receptors (KIR) genes in stages of HIV-1 infection among patients from Burkina Faso. Biomol. Concepts 2019, 10, 226–236. [Google Scholar] [CrossRef]
- Paximadis, M.; Minevich, G.; Winchester, R.; Schramm, D.B.; Gray, G.E.; Sherman, G.G.; Coovadia, A.H.; Kuhn, L.; Tiemessen, C.T. KIR-HLA and maternal-infant HIV-1 transmission in sub-Saharan Africa. PLoS ONE 2011, 6, e16541. [Google Scholar] [CrossRef]
- Martin, M.P.; Gao, X.; Lee, J.-H.; Nelson, G.W.; Detels, R.; Goedert, J.J.; Buchbinder, S.; Hoots, K.; Vlahov, D.; Trowsdale, J. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002, 31, 429–434. [Google Scholar] [CrossRef]
- Khakoo, S.I.; Carrington, M. KIR and disease: A model system or system of models? Immunol. Rev. 2006, 214, 186–201. [Google Scholar] [CrossRef] [PubMed]
- Ravet, S.; Scott-Algara, D.; Bonnet, E.; Tran, H.K.; Tran, T.; Nguyen, N.; Truong, L.X.; Theodorou, I.; Barre-Sinoussi, F.; Pancino, G.; et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood 2007, 109, 4296–4305. [Google Scholar] [CrossRef] [PubMed]
- Boulet, S.; Sharafi, S.; Simic, N.; Bruneau, J.; Routy, J.P.; Tsoukas, C.M.; Bernard, N.F. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS 2008, 22, 595–599. [Google Scholar] [CrossRef]
- Shah-Hosseini, A.; Jafari, M.; Mohammadi, A.; Sanaei, R.; Alavian, S.M.; Doosti-Irani, A.; Nooradeh Keykavousi, M.; Tajik, N. The impact of KIR-HLA genotype on hepatitis B virus clearance in Iranian infected individuals. Med. Microbiol. Immunol. 2017, 206, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Jamil, K.M.; Khakoo, S.I. KIR/HLA interactions and pathogen immunity. J. Biomed. Biotechnol. 2011, 2011, 298348. [Google Scholar] [CrossRef] [PubMed]
- Di Bona, D.; Aiello, A.; Colomba, C.; Bilancia, M.; Accardi, G.; Rubino, R.; Giannitrapani, L.; Tuttolomondo, A.; Cascio, A.; Caiaffa, M.F.; et al. KIR2DL3 and the KIR ligand groups HLA-A-Bw4 and HLA-C2 predict the outcome of hepatitis B virus infection. J. Viral Hepat. 2017, 24, 768–775. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, B.; Chen, S.; Gai, Z.; Feng, Z.; Liu, X.; Liu, Y.; Wen, X.; Li, L.; Jiao, Y.; et al. Association of KIR genotypes and haplotypes with susceptibility to chronic hepatitis B virus infection in Chinese Han population. Cell. Mol. Immunol. 2008, 5, 457–463. [Google Scholar] [CrossRef]
- Kibar, F.; Goruroglu Ozturk, O.; Ulu, A.; Erken, E.; Inal, S.; Dinkci, S.; Kurtaran, B.; Tasova, Y.; Aksu, H.S.; Yaman, A. Role of KIR genes and genotypes in susceptibility to or protection against hepatitis B virus infection in a Turkish cohort. Med. Sci. Monit. 2014, 20, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Pan, N.; Jiang, W.; Sun, H.; Miao, F.; Qiu, J.; Jin, H.; Xu, J.; Shi, Q.; Xie, W.; Zhang, J. KIR and HLA loci are associated with hepatocellular carcinoma development in patients with hepatitis B virus infection: A case-control study. PLoS ONE 2011, 6, e25682. [Google Scholar] [CrossRef] [PubMed]
- Suppiah, V.; Gaudieri, S.; Armstrong, N.J.; O’Connor, K.S.; Berg, T.; Weltman, M.; Abate, M.L.; Spengler, U.; Bassendine, M.; Dore, G.J.; et al. IL28B, HLA-C, and KIR variants additively predict response to therapy in chronic hepatitis C virus infection in a European Cohort: A cross-sectional study. PLoS Med. 2011, 8, e1001092. [Google Scholar] [CrossRef] [PubMed]
- Golden-Mason, L.; Bambha, K.M.; Cheng, L.; Howell, C.D.; Taylor, M.W.; Clark, P.J.; Afdhal, N.; Rosen, H.R.; Virahep, C.S.G. Natural killer inhibitory receptor expression associated with treatment failure and interleukin-28B genotype in patients with chronic hepatitis C. Hepatology 2011, 54, 1559–1569. [Google Scholar] [CrossRef]
- Lunemann, S.; Martrus, G.; Holzemer, A.; Chapel, A.; Ziegler, M.; Korner, C.; Garcia Beltran, W.; Carrington, M.; Wedemeyer, H.; Altfeld, M. Sequence variations in HCV core-derived epitopes alter binding of KIR2DL3 to HLA-C *03:04 and modulate NK cell function. J. Hepatol. 2016, 65, 252–258. [Google Scholar] [CrossRef]
- Schneidewind, A.; Brockman, M.A.; Yang, R.; Adam, R.I.; Li, B.; Le Gall, S.; Rinaldo, C.R.; Craggs, S.L.; Allgaier, R.L.; Power, K.A.; et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 2007, 81, 12382–12393. [Google Scholar] [CrossRef]
- Marcilla, M.; Lopez de Castro, J.A. Peptides: The cornerstone of HLA-B27 biology and pathogenetic role in spondyloarthritis. Tissue Antigens 2008, 71, 495–506. [Google Scholar] [CrossRef]
- Stewart-Jones, G.B.; di Gleria, K.; Kollnberger, S.; McMichael, A.J.; Jones, E.Y.; Bowness, P. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur. J. Immunol. 2005, 35, 341–351. [Google Scholar] [CrossRef]
- Oliviero, B.; Varchetta, S.; Paudice, E.; Michelone, G.; Zaramella, M.; Mavilio, D.; De Filippi, F.; Bruno, S.; Mondelli, M.U. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology 2009, 137, 1151–1160. [Google Scholar] [CrossRef]
- Njiomegnie, G.F.; Read, S.A.; Fewings, N.; George, J.; McKay, F.; Ahlenstiel, G. Immunomodulation of the natural killer cell phenotype and response during HCV infection. J. Clin. Med. 2020, 9, 1030. [Google Scholar] [CrossRef] [PubMed]
- De Re, V.; Caggiari, L.; De Zorzi, M.; Repetto, O.; Zignego, A.L.; Izzo, F.; Tornesello, M.L.; Buonaguro, F.M.; Mangia, A.; Sansonno, D.; et al. Genetic diversity of the KIR/HLA system and susceptibility to hepatitis C virus-related diseases. PLoS ONE 2015, 10, e0117420. [Google Scholar] [CrossRef]
- Knapp, S.; Warshow, U.; Hegazy, D.; Brackenbury, L.; Guha, I.N.; Fowell, A.; Little, A.M.; Alexander, G.J.; Rosenberg, W.M.; Cramp, M.E.; et al. Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology 2010, 51, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Ahlenstiel, G.; Martin, M.P.; Gao, X.; Carrington, M.; Rehermann, B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J. Clin. Investig. 2008, 118, 1017–1026. [Google Scholar] [CrossRef]
- Qin, H.; Wang, Z.; Du, W.; Lee, W.H.; Wu, X.; Riggs, A.D.; Liu, C.P. Killer cell Ig-like receptor (KIR) 3DL1 down-regulation enhances inhibition of type 1 diabetes by autoantigen-specific regulatory T cells. Proc. Natl. Acad. Sci. USA 2011, 108, 2016–2021. [Google Scholar] [CrossRef] [PubMed]
- Weider, T.; Richardson, S.J.; Morgan, N.G.; Paulsen, T.H.; Dahl-Jorgensen, K.; Hammerstad, S.S. Upregulation of HLA class I and antiviral tissue responses in Hashimoto’s thyroiditis. Thyroid 2020, 30, 432–442. [Google Scholar] [CrossRef]
- Aslanidis, S.; Pyrpasopoulou, A.; Kontotasios, K.; Doumas, S.; Zamboulis, C. Parvovirus B19 infection and systemic lupus erythematosus: Activation of an aberrant pathway? Eur. J. Intern. Med. 2008, 19, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Gasser, S.; Raulet, D. The DNA damage response, immunity and cancer. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2006; pp. 344–347. [Google Scholar]
- Rizzo, R.; Gentili, V.; Rotola, A.; Bortolotti, D.; Cassai, E.; Di Luca, D. Implication of HLA-C and KIR alleles in human papillomavirus infection and associated cervical lesions. Viral Immunol. 2014, 27, 468–470. [Google Scholar] [CrossRef]
- Kollnberger, S.; Bird, L.; Sun, M.Y.; Retiere, C.; Braud, V.M.; McMichael, A.; Bowness, P. Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheum. 2002, 46, 2972–2982. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Holzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.E.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef]
- Burian, A.; Wang, K.L.; Finton, K.A.; Lee, N.; Ishitani, A.; Strong, R.K.; Geraghty, D.E. HLA-F and MHC-I open conformers bind natural killer cell Ig-like receptor KIR3DS1. PLoS ONE 2016, 11, e0163297. [Google Scholar] [CrossRef]
- Cook, M.; Briggs, D.; Craddock, C.; Mahendra, P.; Milligan, D.; Fegan, C.; Darbyshire, P.; Lawson, S.; Boxall, E.; Moss, P. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood 2006, 107, 1230–1232. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Prakash, S. Significance of KIR like natural killer cell receptors in autoimmune disorders. Clin. Immunol. 2020, 216, 108449. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhang, C.; Xu, D.; Sun, H. Association of killer cell immunoglobulin-like receptor and human leucocyte antigen-C w gene combinations with systemic lupus erythematosus. Clin. Exp. Immunol. 2015, 180, 250–254. [Google Scholar] [CrossRef]
- Beck, S.; Barrell, B.G. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature 1988, 331, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R. Controversial role of herpesviruses in Alzheimer’s disease. PLoS Pathog. 2020, 16, e1008575. [Google Scholar] [CrossRef]
- Itzhaki, R.F. Herpes simplex virus type 1 and Alzheimer’s disease: Increasing evidence for a major role of the virus. Front. Aging Neurosci. 2014, 6, 202. [Google Scholar] [CrossRef]
- Readhead, B.; Haure-Mirande, J.V.; Funk, C.C.; Richards, M.A.; Shannon, P.; Haroutunian, V.; Sano, M.; Liang, W.S.; Beckmann, N.D.; Price, N.D.; et al. Multiscale analysis of independent Alzheimer’s cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron 2018, 99, 64–82.e67. [Google Scholar] [CrossRef]
- Challoner, P.B.; Smith, K.T.; Parker, J.D.; MacLeod, D.L.; Coulter, S.N.; Rose, T.M.; Schultz, E.R.; Bennett, J.L.; Garber, R.L.; Chang, M.; et al. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc. Natl. Acad. Sci. USA 1995, 92, 7440–7444. [Google Scholar] [CrossRef]
- Alvarez-Lafuente, R.; De las Heras, V.; Bartolome, M.; Picazo, J.J.; Arroyo, R. Relapsing-remitting multiple sclerosis and human herpesvirus 6 active infection. Arch. Neurol. 2004, 61, 1523–1527. [Google Scholar] [CrossRef]
- Gómez-Lozano, N.; de Pablo, R.; Puente, S.; Vilches, C. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur. J. Immunol. 2003, 33, 639–644. [Google Scholar] [CrossRef]
- Boulet, S.; Kleyman, M.; Kim, J.Y.; Kamya, P.; Sharafi, S.; Simic, N.; Bruneau, J.; Routy, J.P.; Tsoukas, C.M.; Bernard, N.F. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 2008, 22, 1487–1491. [Google Scholar] [CrossRef] [PubMed]
- Jennes, W.; Verheyden, S.; Demanet, C.; Menten, J.; Vuylsteke, B.; Nkengasong, J.N.; Kestens, L. Low CD4+ T cell counts among African HIV-1 infected subjects with group B KIR haplotypes in the absence of specific inhibitory KIR ligands. PLoS ONE 2011, 6, e17043. [Google Scholar] [CrossRef] [PubMed]
- Roeth, J.F.; Williams, M.; Kasper, M.R.; Filzen, T.M.; Collins, K.L. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol. 2004, 167, 903–913. [Google Scholar] [CrossRef]
- Wonderlich, E.R.; Williams, M.; Collins, K.L. The tyrosine binding pocket in the adaptor protein 1 (AP-1) mu1 subunit is necessary for Nef to recruit AP-1 to the major histocompatibility complex class I cytoplasmic tail. J. Biol. Chem. 2008, 283, 3011–3022. [Google Scholar] [CrossRef]
- Vidricaire, G.; Imbeault, M.; Tremblay, M.J. Endocytic host cell machinery plays a dominant role in intracellular trafficking of incoming human immunodeficiency virus type 1 in human placental trophoblasts. J. Virol. 2004, 78, 11904–11915. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martin, M.P.; Qi, Y.; Gao, X.; Yamada, E.; Martin, J.N.; Pereyra, F.; Colombo, S.; Brown, E.E.; Shupert, W.L.; Phair, J.; et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007, 39, 733–740. [Google Scholar] [CrossRef]
- Qi, Y.; Martin, M.P.; Gao, X.; Jacobson, L.; Goedert, J.J.; Buchbinder, S.; Kirk, G.D.; O’Brien, S.J.; Trowsdale, J.; Carrington, M. KIR/HLA pleiotropism: Protection against both HIV and opportunistic infections. PLoS Pathog. 2006, 2, 741–745. [Google Scholar] [CrossRef]
- Singh, R.; Kaul, R.; Kaul, A.; Khan, K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J. Gastroenterol. 2007, 13, 1770–1787. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, J.M.; Jamieson, S.E.; Burgner, D. HLA and infectious diseases. Clin. Microbiol. Rev. 2009, 22, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, H.; Gao, B.; Hu, Z.; Zheng, S.; Tian, Z. Activation and function of hepatic NK cells in hepatitis B infection: An underinvestigated innate immune response. J. Viral Hepat. 2005, 12, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Burt, B.M.; Plitas, G.; Zhao, Z.; Bamboat, Z.M.; Nguyen, H.M.; Dupont, B.; DeMatteo, R.P. The lytic potential of human liver NK cells is restricted by their limited expression of inhibitory killer Ig-like receptors. J. Immunol. 2009, 183, 1789–1796. [Google Scholar] [CrossRef]
- Crispe, I.N. The liver as a lymphoid organ. Annu. Rev. Immunol. 2009, 27, 147–163. [Google Scholar] [CrossRef]
- Protzer, U.; Maini, M.K.; Knolle, P.A. Living in the liver: Hepatic infections. Nat. Rev. Immunol. 2012, 12, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Tu, Z.; Bozorgzadeh, A.; Pierce, R.H.; Kurtis, J.; Crispe, I.N.; Orloff, M.S. TLR-dependent cross talk between human Kupffer cells and NK cells. J. Exp. Med. 2008, 205, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wei, H.; Sun, R.; Dong, Z.; Zhang, J.; Tian, Z. Increased susceptibility to liver injury in hepatitis B virus transgenic mice involves NKG2D-ligand interaction and natural killer cells. Hepatology 2007, 46, 706–715. [Google Scholar] [CrossRef]
- Parham, P.; Abi-Rached, L.; Matevosyan, L.; Moesta, A.K.; Norman, P.J.; Older Aguilar, A.M.; Guethlein, L.A. Primate-specific regulation of natural killer cells. J. Med. Primatol. 2010, 39, 194–212. [Google Scholar] [CrossRef] [PubMed]
- Ursu, L.D.; Calenic, B.; Diculescu, M.; Dima, A.; Stoian, I.T.; Constantinescu, I. Clinical and histopathological changes in different KIR gene profiles in chronic HCV Romanian patients. Int. J. Immunogenet. 2021, 48, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Bost, A.; Glass, J.I.; Tan, S.L. Cytokine-activated natural killer cells exert direct killing of hepatoma cells harboring hepatitis C virus replicons. J. Interferon Cytokine Res. 2006, 26, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, K.A.; Bjorkstrom, N.K.; Veber, H.; Ciesek, S.; Riese, P.; Wiegand, J.; Hadem, J.; Suneetha, P.V.; Jaroszewicz, J.; Wang, C.; et al. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology 2010, 138, 1885–1897. [Google Scholar] [CrossRef] [PubMed]
- Nitschke, K.; Barriga, A.; Schmidt, J.; Timm, J.; Viazov, S.; Kuntzen, T.; Kim, A.Y.; Lauer, G.M.; Allen, T.M.; Gaudieri, S.; et al. HLA-B*27 subtype specificity determines targeting and viral evolution of a hepatitis C virus-specific CD8+ T cell epitope. J. Hepatol. 2014, 60, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Hausen, H.z. Papillomavirus infections–a major cause of human cancers. Biochim. Biophys. Acta Rev. Cancer 1996, 1288, F55–F78. [Google Scholar] [CrossRef]

| Receptor Family | Receptor | Known Ligand | Function |
|---|---|---|---|
| NCR | NKp46 | Viral HA, HSPG, PfEMP-1 | Activating |
| NKp30 | BAT-3, HCMV pp65, B7-H6, | Activating | |
| NKp44 | HSPG, PfEMP-1, Viral HA, HSPG | Activating | |
| C-type lectin | CD94/NKG2A | HLA-E | Inhibitory |
| CD94/NKG2C | HLA-E | Activating | |
| CD94/NKG2E | HLA-E | Activating | |
| NKG2D | MICA/B, ULBPs | Activating | |
| NKR-P1A | LLT1 | Inhibitory | |
| KIR | KIR2DL1 | HLA-C2 (Lys 80) | Inhibitory |
| KIR2DL2/3 | HLA-C1 (Asn 80) | Inhibitory | |
| KIR2DL4 | HLA-G | Activating | |
| KIR2DL5 | Unknown | Inhibitory | |
| KIR2DS1 | HLA-C2 (Lys 80) | Activating | |
| KIR2DS2 | HLA-C1 (Asn 80) | Activating | |
| KIR2DS3 | Unknown | Activating | |
| KIR2DS4 | HLA-Cw4 | Activating | |
| KIR2DS5 | Unknown | Activating | |
| KIR3DL1 | HLA-Bw4 | Inhibitory | |
| KIR3DS1 | HLA-Bw4 (possible) | Activating | |
| KIR3DL2 | HLA-A3/A11 | Inhibitory | |
| LILR | LILRB1/ILT2/LIR-1 | HLA class I, UL18 | Inhibitory |
| CD2 | 2B4 | CD48 | Activating/Inhibitory |
| CD2 | CD58 | Activating | |
| NTB-A | NTB-A | Activating | |
| CRACC | CRACC | Activating | |
| DNAM-1 | PVR, CD122 | Activating | |
| CEACAM | CEACAM1 | NKG2D receptor–ligand system | Inhibitory |
| TIGIT | CD96 | CD155 | Inhibitory |
| TIGIT | CD112, CD113, CD155 | Inhibitory | |
| TIM | Tim-3 | Ceacam-1, PtdSer, Galectin-9 | Inhibitory |
| DC-SIGN | Lag-3 | HLA class II, IL-2R, IL-15R | Inhibitory |
| Type I cytokine receptor | IL-2Rα | IL-2 | Activating |
| IL-15Rα | IL-15 | Activating |
| Virus | KIRs/HLA | KIRs/HLA Frequency | System/Disease or Effect on Infection Outcome | References |
|---|---|---|---|---|
| CMV | KIR2DL2 | increased | Increased susceptibility to herpetic infections | [97] |
| HLA class I | decreased | Viral immune-escape | [81,98] | |
| HLA-C | Increased | Viral immune-escape | [89,99,100,101] | |
| KIR2DS1 | increased | Control of placental CMV infection | [102] | |
| KIR2DS1, KIR2DS5 | decreased | Lower CMV infection control; abortion, pre-term delivery | [102,103] | |
| HLA-G/KIR2DL4 | increased | Trophoblast cells; viral immune-escape | [104] | |
| KIR2DL1/HLA-C2 | increased | immunodeficiency syndrome, recurrent CMV reactivation | [105] | |
| KIR2DS2, KIR2DL3, HLA-C1 | increased | organ transplant; protection from CMV reactivation | [106] | |
| KIR2DL2, KIR2DS1 | increased | SLE | [107] | |
| KIR2DS2 | decreased | SLE | [108] | |
| KIR2DS2, KIR2DL3 | increased | Rheumatoid Arthritis; worst prognosis | [109] | |
| EBV | HLA class I | decreased | Reduced NK cell killing | [93] |
| KIR2DS5, KIR2DS4 | increased | Increased risk of lymphoproliferative diseases | [110] | |
| KIR2DS2, KIR2DL3 | increased | Rheumatoid Arthritis; worst prognosis | [109] | |
| KIR2DL2 | increased | Multiple Sclerosis, increased susceptibility to herpetic infections | [97] | |
| HHV-8 | HLA class I | decreased | Viral immune-escape | [111,112,113] |
| KIR2DL2/HLA-C1 | increased | type-2 diabetes | [114] | |
| KIR2DL2, KIR2DS2 | increased | Cutaneous vascular lesions | [115] | |
| KIR2DS1, KIR3DS1 | increased | Kaposi Sarcoma | [116] | |
| HHV-6 | KIR2DL2/HLA-C1 | increased | Alzheimer’s Disease, NK cell inhibition | [117,118] |
| KIR2DL2/HLA-C1 | increased | Multiple Sclerosis, worst disease outcome, NK cell inhibition, herpetic reactivation | [97,119,120] | |
| HLA-G | increased | IUGR, alterated KIR2DL4+ dNK cell function | [121,122] | |
| HLA-G/KIR2DL4 | decreased | Infertility; KIR2DL4+ dNK cell function | [123,124,125,126,127] | |
| KIR2DL2 | increased | Infertility, Multiple Sclerosis, increased susceptibility to herpetic infections | [97,128] | |
| KIR2DL5 | decreased | infertility | [128] | |
| HSV-1 | KIR2DL2/HLA-C | increased | infertility | [129,130] |
| KIR2DL2/HLA-C | increased | Multiple Sclerosis | [119,120] | |
| HIV | KIR2DL5, KIR2DS5 | increased | Protection from HIV transmission | [95] |
| HLA-C | increased | Protection from HIV transmission | [131] | |
| HLA-C | increased | Viral immune-escape (via Nef protein) | [132] | |
| HLA-A, HLA-B | decreased | Viral immune-escape (via Nef protein) | [133] | |
| KIR2DL2, HLA-Cw*0102 | increased | Viral immune-escape (via Gag protein) | [134] | |
| HLA-C | increased | Viral immune-escape (via Env protein) | [135,136] | |
| KIR3DS1 | decreased | Higher NK cell function, reduced perinatal transmission | [137,138] | |
| KIR3DL1, KIR2DL2, KIR2DL3, KIR2DL5, KIR2DS5, HLA-C | increased | Reduced perinatal transmission | [139,140] | |
| KIR2DL2, KIR2DS2, KIR2DS3, KIR2DS4, KIR3DS1 | increased | Higher susceptibility in the newborn | [138] | |
| KIR3DS1/HLA-Bw4-80I | increased | Decreased AIDS progression; protective effect | [141,142,143,144] | |
| HBV | KIR2DS1, KIR2DS2 | increased | HBV recovered subjects, lower NK cell activation | [145] |
| KIR2DL3/HLA-C1 | increased | Protective toward HBV susceptibility | [146] | |
| KIR2DL3 | decreased | CHB | [147] | |
| KIR2DL1/HLA-C2 | increased | Higher HBV infection rate | [146] | |
| KIR2DS2, KIR2DS3 | increased | CHB, increased HBV susceptibility | [148] | |
| KIR2DS1, KIR3DS1, KIR2DL5, KIR2DL3 | increased | CHB, protective effect, HBV clearance | [147,148,149] | |
| HLA-C1 | increased | Increased risk of HCC progression in HBV-infected patients | [150] | |
| HLA-C2, HLA-A-Bw4 | increased | Increased risk for CHB development | [147] | |
| HCV | KIR3DL2, KIR2DL1, KIR2DL2, KIR2DL3, HLA-C2 | increased | Poor HCV clearance after treatment | [151,152] |
| HLA-C∗03:04, KIR2DL3 | increased | Viral immune-escape | [153] | |
| HLA-B*27/KIR3DL1 | decreased | Viral immune-escape | [154,155,156] | |
| KIR3DL1 KIR2DS3 | decreased increased | Increased risk for chronic HCV infection | [157] [158] | |
| HLA-Bw4/KIR3DS1 | increased | Protective against liver disease in HCV patients | [159] | |
| HLA-C1/KIR2DL3 | increased | Spontaneous infection resolution and sustained response to HCV antiviral therapy | [160,161] | |
| Enterovirus | KIR3DL1 | decreased | Type-1 diabetes | [162] |
| HLA class I | increased | Induction of antiviral response that triggers Hashimoto’s Thyroiditis condition | [163] | |
| Parvovirus | HLA-I | increased | SLE, persistent viral infection | [164,165] |
| HPV | KIR2DL2/HLA-C1, KIR2DL3/HLA-C1 | increased | Cervix cancer, risk factor for HPV high-risk infection and neoplastic transformation | [166] |
| HLA-C2 | increased | Cervix cancer, no increased risk for HPV high-risk infection and neoplastic transformation | [166] | |
| KIR2DL1 | increased | Cervix cancer | [166] | |
| KIR2DL2/HLA-C2 | decreased | Cervix cancer | [166] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rizzo, S.; Schiuma, G.; Beltrami, S.; Gentili, V.; Rizzo, R.; Bortolotti, D. Role of KIR Receptor in NK Regulation during Viral Infections. Immuno 2021, 1, 305-331. https://doi.org/10.3390/immuno1030021
Rizzo S, Schiuma G, Beltrami S, Gentili V, Rizzo R, Bortolotti D. Role of KIR Receptor in NK Regulation during Viral Infections. Immuno. 2021; 1(3):305-331. https://doi.org/10.3390/immuno1030021
Chicago/Turabian StyleRizzo, Sabrina, Giovanna Schiuma, Silvia Beltrami, Valentina Gentili, Roberta Rizzo, and Daria Bortolotti. 2021. "Role of KIR Receptor in NK Regulation during Viral Infections" Immuno 1, no. 3: 305-331. https://doi.org/10.3390/immuno1030021
APA StyleRizzo, S., Schiuma, G., Beltrami, S., Gentili, V., Rizzo, R., & Bortolotti, D. (2021). Role of KIR Receptor in NK Regulation during Viral Infections. Immuno, 1(3), 305-331. https://doi.org/10.3390/immuno1030021








