Abstract
Peptidases generate bioactive peptides that can regulate cell signaling and mediate intercellular communication. While the processing of peptide precursors is initiated intracellularly, some modifications by peptidases may be conducted extracellularly. Thimet oligopeptidase (TOP) is a peptidase that processes neuroendocrine peptides with roles in mood, metabolism, and immune responses, among other functions. TOP also hydrolyzes angiotensin I to angiotensin 1–7, which may be involved in the pathophysiology of COVID-19 infection. Although TOP is primarily cytosolic, it can also be associated with the cell plasma membrane or secreted to the extracellular space. Recent work indicates that membrane-associated TOP can be released with extracellular vesicles (EVs) to the extracellular space. Here we briefly summarize the enzyme’s classical function in extracellular processing of neuroendocrine peptides, as well as its more recently understood role in intracellular processing of various peptides that impact human diseases. Finally, we discuss new findings of EV-associated TOP in the extracellular space.
1. Introduction: Thimet Oligopeptidase—Classical Function, Regulation, and Structure
Thimet oligopeptidase (TOP), also called endopeptidase 24.15 (E.C. 3.4.24.15), is a zinc metalloendopeptidase of molecular weight 77 kD [1,2]. TOP is found in humans and other animals, and the enzyme hydrolyzes a number of important neuroendocrine and cardiovascular peptides that help regulate physiological and pathological conditions [2,3,4]. In 1980, Horsthemke and Bauer first characterized TOP as a neutral endopeptidase which degrades gonadotropin-releasing hormone (GnRH, also called LHRH) [5]. This enzyme was primarily detected from soluble extracts of bovine anterior pituitary tissue and was shown to be effective at cleaving GnRH [5]. Later, Orlowski et al. purified the enzyme from rat brain [6] and rat testes [7] and investigated the determination of specificity with synthetic and natural peptides of this soluble metalloendopeptidase [6]. Given the enzyme activity of TOP and its ability to degrade several bioactive peptides, Orlowski et al. proposed a potential function of the metalloendopeptidase in neuropeptide metabolism [6]. The study later found that TOP can cleave and process not only GnRH, but also bradykinin and neurotensin [7].
TOP is regulated at the level of transcription by both positively and negatively acting elements in a cell-specific manner [8,9]. For example, we have shown that expression of TOP is downregulated by estradiol in steroid-sensitive regions of the brain [10,11]. However, there are a number of other potential mechanisms for regulation of TOP levels, activity, and distribution. TOP is dependent on redox state, with activity greatly reduced by dimerization or higher-order oligomerization via intermolecular disulfide linkage of surface cysteine residues [12,13,14]. Although activation of the enzyme by thiol reagents was originally thought to occur only in vitro, more recent studies suggest that S-glutathiolation occurs in vivo [15,16], and that modulation of TOP activity may occur via a redox cascade [17]. Phosphorylation is another post-translational modification that affects TOP activity and possibly function. When TOP is phosphorylated in vitro, its affinity and turnover of GnRH is altered [18], and autophosphorylation following exposure to ATP also affects TOP activity [19]. In addition, compartmentalization and secretion of TOP can be modulated by a variety of factors, including calcium [20] and 14-3-3 epsilon [21]. Other non-conventional modulators of TOP activity have been reported, such as sleep deprivation [22] and mechanical shear [23,24], possibly mediated by G proteins.
The catalytic unit of TOP contains a HEXXH zinc-binding motif in which the two histidine residues serve as ligands to a catalytic Zn2+ ion (Figure 1). Site-directed mutagenesis of the rat enzyme established that His473, Glu474, and His477 make up the HEXXH motif, with Glu502 serving as the third zinc ligand [25]. Based on this active site architecture, TOP is categorized as a gluzincin, in which class thermolysin is the prototypical member [26,27]. Extensive mechanistic studies on thermolysin and related enzymes have shown that glutamate within the HEXXH motif serves as a general acid-base catalyst [26,28,29], and this is also the case for TOP [25]. Enzymes within the group are known to exhibit a bell-shaped pH profile for activity with an optimal pH around 7 [30,31]. A similar bell-shaped profile for the activity of TOP provided additional mechanistic support for its placement within this active site architecture [32].
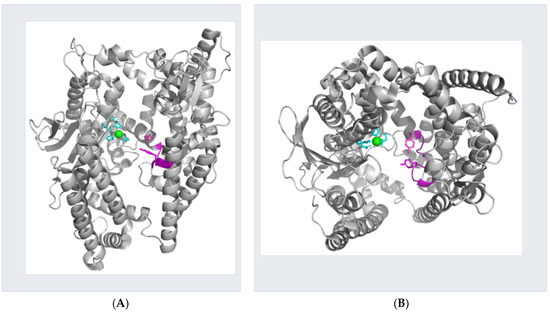
Figure 1.
Crystal structure of thimet oligopeptidase. Ribbon representation of open structure of human thimet oligopeptidase (PDB 1s4b). Side chains of the residues forming the catalytic unit are highlighted in cyan with the metal-ion depicted as a green sphere. The flexible loop, including the side chains of Tyr-605 and Tyr-612, are shown in magenta. (A) View perpendicular to the peptide-binding crevice. (B) View parallel to the peptide-binding crevice. Images were created using PyMOL software (Version 2.5.0).
TOP belongs to the M3 family of peptidases, which includes homologs distributed across organisms from bacteria and fungi to mammals [1]. X-ray structures for 5 members of this family, including TOP, demonstrate that all share a bi-lobal, primarily alpha-helical structure with a 40 Å deep crevice separating the lobes, at the bottom of which resides the active site zinc [3,33,34,35,36]. Depending on the conditions of crystallization, the enzymes appear to have an open [3,33,36] or closed conformation [34,35,37]. While TOP has yet to be crystallized in a closed conformation, several lines of evidence, including those from our laboratory, point to the role of an induced fit as a means for substrate selectivity controlled by repositioning of conserved, catalytic Tyr residues: Tyr residues 605 and 612 in domain II and opposite the enzyme active site have been shown to be critical for efficient catalysis [32,38]. In the open TOP structure these residues are too distant from the active site to interact with a bound substrate. Further, a flexible loop (residues 599–611) is proposed to play a role in proper positioning of the Tyr residues when TOP binds its diverse array of peptide substrates [39]. In addition, substrate specificity is altered by partial denaturation of the enzyme [40]. Recently, the structure of the related enzyme human neurolysin (NLN) was solved in a closed conformation [37]. Comparison to the open form of rat NLN revealed that the secondary structure and fold of the domains were unchanged with structural changes confined to short loops connecting the domains at the bottom of the catalytic cleft. The largest structural change occurring in the loop 600–612 contained conserved Tyr residues.
2. TOP Plays Dual Roles as an Intracellular and Extracellular Peptidase
TOP is expressed in a wide range of mammalian tissues and cell types, with the highest levels of TOP found in the brain, pituitary gland and testis, in keeping with its neuroendocrine function, while lower levels of TOP can be detected in liver, kidney, lung and spleen [2,4,41,42,43,44,45,46,47,48]. The enzyme is primarily cytoplasmic, suggesting its participation in general intracellular degradative processes [2], while there is also significant presence in the nucleus [49,50]. Altogether, intracellular TOP accounts for 75% of the total enzyme [2]. Given its prevalence in the nervous system and its ability to cleave bioactive peptides in vitro, TOP has been hypothesized to play an important role in the metabolism of neuropeptides in the extracellular space [2], i.e., for processing GnRH, bradykinin, and neurotensin in blood (Figure 2). However, it is not clear how TOP reaches the extracellular space. As we discuss in a later section, emerging evidence suggests TOP may be secreted to the extracellular space as a soluble protein or released from the cell on extracellular vesicles.
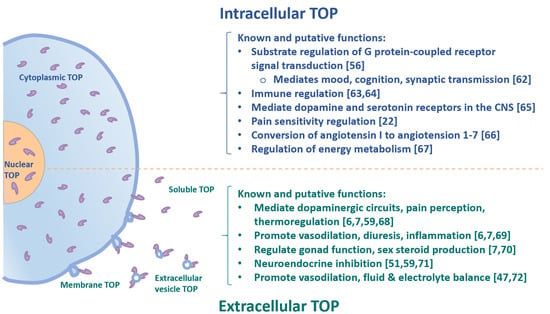
Figure 2.
Schematic overview of intracellular and extracellular TOP and its biological functions.
Traditionally, TOP has been classified as a metabolizing enzyme which terminates neuropeptide function through hydrolysis, such that these substrates are unable to bind to their corresponding receptors [51,52]. However, other studies also demonstrate that TOP plays several alternative roles in the physiological metabolism of various neuropeptides [53]. First, it has been reported that TOP can transform inert precursor neuropeptides into their physiologically bioactive forms [41]. Second, the enzyme can convert one peptide into another bioactive peptide, which can either bind to a new receptor or initiate various downstream signaling pathways upon binding to the initial receptor [54]. Additionally, studies have shown that TOP can work as a bio-modulatory enzyme, whereby a peptide product will act in opposition to its original parent peptide [55]. Given its many roles in the metabolism of neuropeptides, TOP may also play an important role in cell regulation. In fact, studies with rat brain tissue have shown that TOP and its relevant peptide substrates are regulators of the G protein-coupled receptor (GPCR)-mediated signal transduction pathways (Figure 2) [56].
As noted, bioactive peptide signals serve as important chemical messengers for the mediation of intercellular communication in multicellular organisms by binding to cell surface receptors of target cells. Since most peptides are initially synthesized as inactive precursors that require further processing, hydrolysis by peptidase enzymes is required to gain full functionality [2,4]. However, peptide processing is not uniquely intracellular, and may also continue in the extracellular milieu. Thus, extracellular peptidases such as TOP play a crucial role in the physiological generation and regulation of bioactive peptide signals. In the extracellular space, TOP may either convert an inert precursor to an active moiety, or proteolyze peptides to generate products unable to activate receptors. TOP is known to hydrolyze only short peptides of fewer than 20 residues in length, and to recognize greatly variable cleavage sequences depending on the specific substrates [51,57,58,59].
Established physiological substrates of TOP have been shown to include a number of important bioactive peptides of the nervous system, reproductive axis, pain nociception, as well as the cardiovascular and renal systems, such as GnRH, neurotensin, somatostatin, opioids, bradykinin, and angiotensin [6,7,42,47,51,55,59,60,61]. TOP can inactivate vasodilatory peptide bradykinin [2], and thus may be involved in cardiovascular homeostasis (Figure 2).
3. Cytosolic TOP Can Process Intracellular Peptides and Serve as a Natural Regulator of Intracellular Signaling
Although the majority of published works have focused on TOP function in the cleavage of a number of bioactive peptides in the extracellular space [6,7,42,47,51,55,59,60,61], its role in processing intracellular bioactive peptides has increasingly been investigated over the past decade [56]. Since TOP is primarily a cytoplasmic enzyme, it is logical that it would act as a natural regulator of cell signaling through processing of intracellular peptides (Figure 2) [56]. Berti et al. analyzed the intracellular peptides in human embryonic kidney 293 cells with a quantitative peptidomimetic approach and confirmed that TOP can cleave intracellular substrates that may be used for antigen presentation [62]. Cunha et al. found that TOP is physiologically relevant in the cleavage of intracellular peptides that help to control signal transduction of G protein-coupled receptor (GPCR) [56]. Significant changes in the intracellular peptide composition may disturb cell homeostasis in certain pathological conditions, as GPCRs constitute many cell surface receptors that mediate certain key processes, including mood, cognition, pain, appetite, and synaptic transmission [63]. In order to study the effects of TOP in vivo, Dos Santos et al. generated TOP knockout mice (KO) [64]. The TOP KO mice showed different intracellular peptide ratios, with altered mRNA expression of serotonin 5HT2a receptor, proteasome beta5, and dopamine D2 receptor, as compared to WT mice. In addition, TOP KO mice exhibit depressive-like behavior, as well as attention and memory retention deficits. This suggests the role of TOP on intracellular peptide metabolism and its consequent effects on receptors of serotonin and dopamine, which are known mediators of neuronal communication in many psychiatric disorders [64]. In addition, Visniauskas et al. reported that sleep deprivation decreases TOP activity in the striatum and hypothalamus, which can potentially alter the processing of neuropeptides, and that processing of opioid peptides by TOP may be related to the increase in pain sensitivity in rats [22].
TOP has also been shown to have a protective role in Alzheimer’s disease (AD) [65,66,67]. Amyloid-β (Aβ) peptide deposition in the brain plays a major role in AD pathology, and intracellular Aβ accumulation is an early-stage event in the disease; it may be a source for extracellular deposits during disease progression [68]. TOP is involved in the processing of amyloid precursor protein (APP) at a beta-secretase cleavage site and in the degradation of Aβ peptide, a major player in AD pathology [69,70]. Of note, the findings of Koike et al. were in contrast to earlier studies, which had excluded TOP as a candidate beta-secretase [71]. Furthermore, TOP has been found in neurofibrillary tangles and senile plaques in AD autoptic tissue [72]. Pollio et al. demonstrated that overexpression of TOP was neuroprotective against Aβ-mediated toxicity in primary cortical neurons [67]. Conversely, knockdown of TOP by RNA interference (RNAi) made neurons more vulnerable to Aβ toxicity [67]. Notably, the authors found an age-dependent increase in TOP expression in the brain tissue of the TgCRND8 transgenic mouse model of amyloid plaque deposition, where TOP co-localized with Aβ plaque [67]. TOP expression was significantly increased in AD brain tissue compared to non-AD controls in humans [67]. Taken together, these findings suggest that increased expression of neuronal TOP may play a protective and compensatory defense role against increased Aβ load seen in early stages of AD pathology [67].
Berti et al. have identified the presence of specific intracellular peptides, which cannot be degraded by TOP [62] in the adipose tissue of Wistar rats [73]. They found that these intracellular peptides may be important in insulin-stimulated glucose uptake in 3T3-L1 adipocytes [73], suggesting the physiological function of intracellular peptides and their potential in metabolic disorders, i.e., obesity and insulin resistance. Very recently, the same group further investigated the involvement of TOP in energy metabolism using TOP KO mice [74]. These animals gained 75% less body weight than controls and did not show non-alcoholic fatty liver steatosis or insulin resistance when compared to their wildtype control mice, after 24 weeks of high fat diet. This work has identified the involvement of TOP in energy metabolism regulation, and TOP-regulated intracellular peptides may be responsible for the regulation of energy metabolism and obesity by an unknown mechanism of action [74].
In antigen-presenting cells, cytosolic peptides/proteins destined for degradation are processed by the proteasome. These peptides will then be transported to the cell surface for antigen presentation. Cytotoxic T lymphocytes (CTLs) recognize peptides that are presented by HLA class I molecules on the cell surface. Kessler et al. reported that cytoplasmic TOP can complement proteasome activity to cleave CLE epitopes [75], resulting in HLA class I binding peptide fragments not compatible for antigenic presentation. Thus, intracellular TOP can affect antigen processing and presentation by its endoproteolytic activity. The relatively new function of intracellular TOP in processing cytosolic peptides provides a new appreciation of this classical enzyme for its role in intracellular peptide regulation of mood, cognition, pain, and psychiatric and neurodegenerative diseases, metabolic diseases, and immunological regulatory functions.
4. Effects of TOP in Intracellular Processing of Angiotensin 1–7 and Its Potential Involvement in COVID-19 Pathogenesis
TOP has long been known to convert angiotensin I in vitro to the biologically active peptide angiotensin 1–7 [76]. More recently, studies have found that TOP is involved in the intracellular conversion of angiotensin I to angiotensin 1–7, especially within the mitochondria and the nucleus [77], therefore contributing to the elevated levels of angiotensin 1–7 in tissues and in the circulation. In addition to TOP, other endopeptidases are also involved in the production of angiotensin 1–7 from angiotensin I [78]. Angiotensin-converting enzyme (ACE)/angiotensin II is a classical signaling pathway in which ACE/ACE2 are carboxypeptidases that can correspondingly conduct catalytic processing of angiotensin I/II to produce angiotensin 1–7 [78]. In addition, other carboxypeptidases that convert angiotensin II to angiotensin 1–7 are carboxypeptidase A and prolylcarboxypeptidase [79,80,81]. All the above endopeptidases and carboxypeptidases may contribute to the processing of angiotensin II to angiotensin 1–7.
Recent studies have found a potential link between angiotensin 1–7 and COVID-19 pathophysiology [82,83]. Patients with severe COVID-19 illness had significantly increased circulating levels of angiotensin 1–7 and decreased angiotensin II [82,83]. The angiotensin 1–7/angiotensin II ratio was 3-fold higher in COVID-19 patients as compared to that in healthy controls, thus suggesting an increased activity of ACE2 or other angiotensin 1–7 forming activity after COVID-19 infection [82]. Although TOP is an important angiotensin 1–7 forming factor, expression or activity of TOP in COVID-19 patients have not been analyzed. Additionally, TOP has been shown to process angiotensin I to form angiotensin 1–7 in the hippocampus of the brain [84,85], the kidneys [86,87,88,89], and the cardiovascular system [90,91,92]. These organs have all been shown to be vulnerable to direct infection by SARS-CoV-2 virus, further implicating the potential role of TOP in COVID-19 pathophysiology [93,94,95,96,97,98,99].
Prevailing evidence highlights the role of ACE2 as the receptor for both SARS-CoV-1 and SARS-CoV-2 (virus of COVID-19 infection) binding and entry into host cells [100,101]. ACE2 is especially abundant in alveolar epithelial cells and vascular endothelial cells of the lung [78]. In fact, the lethality of infection has been associated with the loss of key regulatory factors in the lung related to viral binding suppression of ACE2 [102]. Tan et al. reported regulatory effects of angiotensin 1–7 in ACE2 expression and found organ-specific down regulation of local ACE2 expression by continuous infusion of angiotensin 1–7 in vivo in rats [103]. However, elevated ACE2 expression has been observed in patients with COVID-19 infection [83,104]. One may, thus, expect that TOP may possibly affect ACE2 expression through processing intracellular angiotensin 1–7, therefore affecting vulnerability of the cells of given tissue/organs to SARS-CoV-2 infection [82,83]. Interestingly, Soria-Castro et al. [105] recently reported that COVID-19 patients showed increased carboxypeptidase A3 (CPA3), which is also known for its function in converting angiotensin II to angiotensin 1–7 [81]. Another enzyme prolylcarboxypeptidase that can catalytically convert angiotensin II to angiotensin 1–7 may also be involved in COVID-19 pathophysiology [106]. Taken together, these findings suggest potential involvement of TOP or other endopeptidases and carboxypeptidases in the pathophysiology of COVID-19 infection is an area worthy of further investigation.
5. Membrane-Associated TOP, and the Novel Membrane Vesicle-Associated TOP in the Extracellular Space
As noted, although a major portion of peptide processing is initiated in the intracellular space, many peptides are further modified and activated in the extracellular milieu by enzymes like TOP [2,4]. Thus, TOP must be localized extracellularly in addition to its intracellular form, either released to the extracellular space as a soluble protein to the extracellular space or associated with the cell plasma membrane on the membrane surface. TOP has also been found in various subcellular locations depending on cell type, including nuclear [49,50], cytosolic, secreted forms [41,42,44,49,50,65,107], and membrane-associated forms [41,42,108], depending on cell types. Acker et al. found that about 20–25% of the total TOP enzyme activity is associated with membrane fractions [41]. Interestingly, TOP has been visualized on the membrane surface of the cell plasma membrane by confocal microscopy [108]. In addition, TOP is secreted to the extracellular milieu [2,4,65,107], and TOP activity has been detected in the media of DHT-treated LNCaP androgen responsive cells [46].
Extracellular vesicles (EVs) are small subcellular membrane blebs which are released from almost all cell types upon cell activation or during various types of programmed cell death, such as apoptosis, pyroptosis, necroptosis, and NETosis [109,110,111,112,113,114,115,116,117]. EVs can be generally classified into three main subgroups based on their size and subcellular origin: exosomes (<100 nm), microvesicles (MVs) (<1 μm), and apoptotic bodies (1–5 μm) [112,116,118,119]. Exosomes are the smallest EVs by size (30–100 nm in diameter) and are secreted from the intracellular space [116]. Membrane microvesicles (MVs) range from 100 to 1000 nm in diameter and bud directly from the cell plasma membrane surface [112,118,120]. Importantly, EVs carry a vast array of intracellular or membrane-associated molecules, such as a variety of biologically active proteins, membrane receptors, lipids, and nuclear DNA or small RNAs from the nucleus, cytoplasm, and cell membranes [109,110,111,112,116,121]. EVs are released by almost all eukaryotic cells [112,118] as well as bacteria [122]. They can serve as shuttles which transfer cargos from parental cells to recipient cells that are either nearby or far away [112,116,118,121,123]. In this way, EVs may modulate the function of target cells in response to physiological or pathological stimuli. In contrast to classical chemical signaling pathways, EVs provide alternative modes of paracrine and endocrine communication via direct cell-to-cell contact or receptor–ligand mediated signaling [116]. EVs have been associated with various disease processes. In fact, EV levels have been shown to be elevated in metabolic diseases, infections, and malignancy [116,124,125,126,127].
Our previous work showed that matrix metalloproteinase superfamily 14 (MMP14), a transmembrane protease, can be released with EVs when macrophages are stimulated by tobacco smoke extract, and the EV-associated MMP14 is biologically active for collagen cleavage [109]. The works from our and other groups have shown that TOP could be visualized on the cell plasma membrane surface by confocal microscopy [108,113]. Interestingly, we also found that membrane-associated TOP can also be released with EVs (Figure 2 and Figure 3) [113], in addition to its soluble form [46], to the extracellular space. We showed that TOP-positive EVs are generated from cultured DU145 prostate cancer cells that were treated without or with dihydrotestosterone, and that TOP-positive EVs are also generated by PC3 prostate cancer cells when stimulated by calcium ionophore A23187 (Figure 3) [113]. Interestingly, EV-associated TOP exhibits considerable enzymatic activity as measured via quenched fluorescence assay [113]. These results suggest that EV-associated TOP might be a previously unrecognized, novel form of extracellular TOP [113]. It is unknown whether the release of EV-associated TOP from the plasma membrane is a regulated process or a random consequence of cell activation or cell death, making this an area worthy of further investigation.
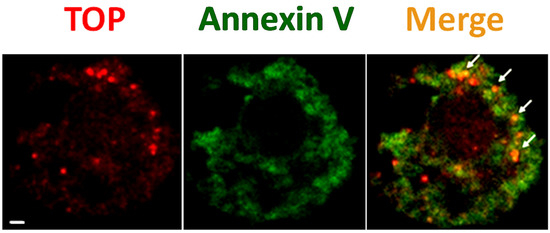
Figure 3.
TOP-positive EVs bud from the plasma membrane surface of A23187-activated cells. PC3 prostate cancer cells were treated with 10 μM calcium ionophore A23187 for 24 h. Cells were then chemically fixed and detected with anti-TOP antibody (red, labeled as “TOP”) and FITC-labeled “Annexin V” (green, which preferentially binds externalized phosphatidylserine). The yellow color in the merged images (“Merge”) demonstrates co-localization of the two labels, which represents TOP-positive EVs. Scale bar 1 μm.
In considering the above findings, one may ask why TOP is found on EVs and what potential advantages this association with EVs may confer to the enzyme. Many physiological substrates of TOP, such as bradykinin, GnRH, and neurotensin, are found either in the extracellular milieu or in the circulation. In order to regulate its physiological substrates, TOP must be accessible to the extracellular space. Previous studies have shown that TOP can be secreted to the extracellular surroundings as a soluble free protein [2,65,107]. Studies of cytokines, which are small proteins that are secreted by immune cells, have short half-lives in the circulation (typically <1 h) [128,129]. Notably, cytokines on the surface of cancer cell derived EVs remain functional and unchanged at 24 h following incubation in plasma at 37 °C, demonstrating that association of cytokines with the EV surface profoundly increases their stability [130]. Icimoto et al. reported that free protein of recombinant TOP is not catalytically stable, while efficient immobilization of TOP in gold nanoparticles can significantly increase stability of the enzyme [131]. Therefore, one may expect that EV-associated extracellular TOP may be more stable in its catalytic activity [113], thus making it a more potent mediator of substrate function [132]. Thus, EV-associated TOP might be more physiologically meaningful than soluble TOP in the pathophysiology of human diseases.
Extracellular vesicles were shown to be important in cancer metastasis [133]. Therefore, EV-associated TOP may be particularly important in cancers of tissues such as that of the prostate. In fact, a number of TOP-sensitive substrates in the proliferation of prostate cancer cells represent an interesting area of study in the field of cancer research. As early-stage prostate cancer proliferation is dependent on androgens, early-stage prostate cancer can thus be effectively treated with androgen-ablation therapy. Beginning in the early 1980s, analogs of TOP substrate GnRH-I had already been used in therapeutic approaches against prostate cancer [134,135] by suppressing the hypothalamic–pituitary–gonadal axis during the early androgen dependent phase. Recent studies have shown that GnRH in fact decreases prostate cancer cell growth [134]. As a crucial modulator of the activities of GnRH and its other physiological substrates, extracellular TOP stands as an important subject of study in further understanding the pathophysiological significance of these hormones in the role of prostate cancer disease progression. Our findings demonstrate a new manifestation of extracellular TOP as being carried by EVs when released from prostate cancer cells [113]. In contrast to the soluble form of TOP, which easily diffuses into the circulation and can be diluted by the large volumes of blood in the circulation, EV-associated TOP may be retained in the tumor microenvironment in relatively high concentration [113,116]. Therefore, the EV-associated format of TOP may enable the enzyme to work more potently on its substrates within a microenvironment, thus contributing to the progression of pathological conditions.
In addition to TOP, there are other cell membrane peptidases that are involved in the inactivation or activation of extracellular hormones, cytokines, regulatory peptides, paracrine peptides, and neuropeptides [136]. A notable peptidase is dipeptidyl peptidase IV (DPPIV), which is also a cell surface peptidase [136]. Kandzija et al. recently reported that placental EVs can carry bioactive DPPIV and these DPPIV-positive EVs are elevated in patients with gestational diabetes mellitus [137]. The EV-associated bioactive DPPIV can cleave glucagon-like peptide-1, which is known to regulate glucose-dependent insulin secretion and, therefore, contributing to diabetes [137]. Furthermore, ACE2 is a carboxypeptidase which has also been localized to the cell membrane surface. Several recent studies [89,138,139,140] have reported that ACE2 can be released with EVs (EVs and exosomes), and that these ACE2-associated EVs can competitively bind SARS-CoV-2 virus spike protein. In turn, EV-carried ACE2 may serve as a novel therapeutic strategy for COVID-19 prevention and/or treatment [139]. All in all, there is now direct evidence of at least three peptidases (TOP, DPPIV, and ACE2) which can be associated with EVs. Therefore, EV-harboring peptidases might be a common natural phenomenon. Whether other peptidases are associated with EVs would be a worthwhile area for further investigation [137].
6. Conclusions
Thimet oligopeptidase (TOP) has classically been considered a cytoplasmic enzyme whose substrates are mainly extracellular peptides. Later studies revealed an intracellular role for TOP. Our results indicate that TOP is released on extracellular vesicles from prostate cancer cells, consistent with findings concerning peptidases on EVs from other tissues.
Taken together, the wide range of locales and substrates for TOP suggests potential for the enzyme’s role in regulation and treatment of a variety of natural and pathological states.
7. Summary
Since TOP was first characterized over 40 years ago, its roles in processing peptides in the extracellular and intracellular space have been elucidated with biochemical, cellular, and molecular approaches, for its potential involvement in neuroendocrinology, behavior, metabolic and cardiovascular diseases, cancer, and infectious diseases. Novel findings about membrane-associated TOP may provide insights for potential diagnosis and therapeutic strategies in a wide variety of diseases.
Author Contributions
Conceptualization, Y.L. and A.J.W.; methodology, Y.L. and L.A.B.; software, J.A.S.; validation, Y.L. and A.J.W.; formal analysis, Y.L.; investigation, Y.L., L.A.B. and J.A.S.; resources, A.J.W.; data curation, A.J.W.; writing—original draft preparation, Y.L.; writing—review and editing, L.A.B., J.A.S. and A.J.W.; visualization, Y.L., L.A.B. and J.A.S.; supervision, A.J.W.; project administration, A.J.W.; funding acquisition, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
Research supported by funding from Wellesley College.
Acknowledgments
We thank all colleagues and collaborators for their support of this work. We also thank the following individuals for their generous donation of prostate cancer cells and enzyme when we conducted TOP-related works: Angelo Poletti (University of Milano), Marc Glucksman (Rosalind Franklin University) and Marc J. Tetel (Wellesley College).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rawlings, N.D.; Barrett, A.J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995, 248, 183–228. [Google Scholar] [CrossRef] [PubMed]
- Shrimpton, C.N.; Smith, A.I.; Lew, R.A. Soluble metalloendopeptidases and neuroendocrine signaling. Endocr. Rev. 2002, 23, 647–664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ray, K.; Hines, C.S.; Coll-Rodriguez, J.; Rodgers, D.W. Crystal structure of human thimet oligopeptidase provides insight into substrate recognition, regulation, and localization. J. Biol. Chem. 2004, 279, 20480–20489. [Google Scholar] [CrossRef]
- Ferro, E.S.; Gewehr, M.C.F.; Navon, A. Thimet Oligopeptidase Biochemical and Biological Significances: Past, Present, and Future Directions. Biomolecules 2020, 10, 1229. [Google Scholar] [CrossRef]
- Horsthemke, B.; Bauer, K. Characterization of a nonchymotrypsin-like endopeptidase from anterior pituitary that hydrolyzes luteining hormone-releasing hormone at the tyrosyl-glycine and histidyl-tryptophan bonds. Biochemistry 1980, 19, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, M.; Michaud, C.; Chu, T.G. A soluble metalloendopeptidase from rat brain. Purification of the enzyme and determination of specificity with synthetic and natural peptides. Eur. J. Biochem. 1983, 135, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, M.; Reznik, S.; Ayala, J.; Pierotti, A.R. Endopeptidase 24.15 from rat testes. Isolation of the enzyme and its specificity toward synthetic and natural peptides, including enkephalin-containing peptides. Biochem. J. 1989, 261, 951–958. [Google Scholar] [CrossRef] [PubMed]
- McCool, S.; Pierotti, A.R. Expression of the thimet oligopeptidase gene is regulated by positively and negatively acting elements. DNA Cell Biol. 2000, 19, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Morrison, L.S.; Pierotti, A.R. Thimet oligopeptidase expression is differentially regulated in neuroendocrine and spermatid cell lines by transcription factor binding to SRY (sex-determining region Y), CAAT and CREB (cAMP-response-element-binding protein) promoter consensus sequences. Biochem. J. 2003, 376, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Cyr, N.E.; Kua, L.H.; Bruce, L.A.; Chadwick, J.G.; Tetel, M.J.; Wolfson, A.J. Nuclear Thimet oligopeptidase is coexpressed with oestrogen receptor alpha in hypothalamic cells and regulated by oestradiol in female mice. J. Neuroendocrinol. 2010, 22, 936–943. [Google Scholar] [CrossRef]
- Bruce, L.A.; Cyr, N.E.; Qiao, J.W.; Defries, C.C.; Tetel, M.J.; Wolfson, A.J. Neuropeptidase activity is down-regulated by estradiol in steroid-sensitive regions of the hypothalamus in female mice. Neuropeptides 2012, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Tisljar, U.; Barrett, A.J. Thiol-dependent metallo-endopeptidase characteristics of Pz-peptidase in rat and rabbit. Biochem. J. 1990, 267, 531–533. [Google Scholar] [CrossRef]
- Shrimpton, C.N.; Glucksman, M.J.; Lew, R.A.; Tullai, J.W.; Margulies, E.H.; Roberts, J.L.; Smith, A.I. Thiol activation of endopeptidase EC 3.4.24.15. A novel mechanism for the regulation of catalytic activity. J. Biol. Chem. 1997, 272, 17395–17399. [Google Scholar] [CrossRef] [PubMed]
- Sigman, J.A.; Sharky, M.L.; Walsh, S.T.; Pabon, A.; Glucksman, M.J.; Wolfson, A.J. Involvement of surface cysteines in activity and multimer formation of thimet oligopeptidase. Protein Eng. 2003, 16, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Demasi, M.; Piassa Filho, G.M.; Castro, L.M.; Ferreira, J.C.; Rioli, V.; Ferro, E.S. Oligomerization of the cysteinyl-rich oligopeptidase EP24.15 is triggered by S-glutathionylation. Free Radic. Biol. Med. 2008, 44, 1180–1190. [Google Scholar] [CrossRef]
- Malvezzi, A.; Higa, P.M.; Amaral, A.T.D.; Silva, G.M.; Gozzo, F.C.; Ferro, E.S.; Castro, L.M.; de Rezende, L.; Monteiro, G.; Demasi, M. The cysteine-rich protein thimet oligopeptidase as a model of the structural requirements for S-glutathiolation and oxidative oligomerization. PLoS ONE 2012, 7, e39408. [Google Scholar] [CrossRef]
- Icimoto, M.Y.; Ferreira, J.C.; Yokomizo, C.H.; Bim, L.V.; Marem, A.; Gilio, J.M.; Oliveira, V.; Nantes, I.L. Redox modulation of thimet oligopeptidase activity by hydrogen peroxide. FEBS Open Bio 2017, 7, 1037–1050. [Google Scholar] [CrossRef]
- Tullai, J.W.; Cummins, P.M.; Pabon, A.; Roberts, J.L.; Lopingco, M.C.; Shrimpton, C.N.; Smith, A.I.; Martignetti, J.A.; Ferro, E.S.; Glucksman, M.J. The neuropeptide processing enzyme EC 3.4.24.15 is modulated by protein kinase A phosphorylation. J. Biol. Chem. 2000, 275, 36514–36522. [Google Scholar] [CrossRef]
- Portaro, F.C.; Hayashi, M.A.; Silva, C.L.; de Camargo, A.C. Free ATP inhibits thimet oligopeptidase (EC 3.4.24.15) activity, induces autophosphorylation in vitro, and controls oligopeptide degradation in macrophage. Eur. J. Biochem. 2001, 268, 887–894. [Google Scholar] [CrossRef]
- Oliveira, V.; Garrido, P.A.; Rodrigues, C.C.; Colquhoun, A.; Castro, L.M.; Almeida, P.C.; Shida, C.S.; Juliano, M.A.; Juliano, L.; Camargo, A.C.; et al. Calcium modulates endopeptidase 24.15 (EC 3.4.24.15) membrane association, secondary structure and substrate specificity. FEBS J. 2005, 272, 2978–2992. [Google Scholar] [CrossRef]
- Carreño, F.R.; Goñi, C.N.; Castro, L.M.; Ferro, E.S. 14-3-3 epsilon modulates the stimulated secretion of endopeptidase 24.15. J. Neurochem. 2005, 93, 10–25. [Google Scholar] [CrossRef]
- Visniauskas, B.; Simoes, P.S.R.; Dalio, F.M.; Naffah-Mazzacoratti, M.D.G.; Oliveira, V.; Tufik, S.; Chagas, J.R. Sleep deprivation changes thimet oligopeptidase (THOP1) expression and activity in rat brain. Heliyon 2019, 5, e02896. [Google Scholar] [CrossRef] [PubMed]
- Cotter, E.J.; von Offenberg Sweeney, N.; Coen, P.M.; Birney, Y.A.; Glucksman, M.J.; Cahill, P.A.; Cummins, P.M. Regulation of endopeptidases EC3.4.24.15 and EC3.4.24.16 in vascular endothelial cells by cyclic strain: Role of Gi protein signaling. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Guinan, A.F.; Rochfort, K.D.; Fitzpatrick, P.A.; Walsh, T.G.; Pierotti, A.R.; Phelan, S.; Murphy, R.P.; Cummins, P.M. Shear stress is a positive regulator of thimet oligopeptidase (EC3.4.24.15) in vascular endothelial cells: Consequences for MHC1 levels. Cardiovasc. Res. 2013, 99, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Cummins, P.M.; Pabon, A.; Margulies, E.H.; Glucksman, M.J. Zinc coordination and substrate catalysis within the neuropeptide processing enzyme endopeptidase EC 3.4.24.15. Identification of active site histidine and glutamate residues. J. Biol. Chem. 1999, 274, 16003–16009. [Google Scholar] [CrossRef]
- Cerda-Costa, N.; Gomis-Ruth, F.X. Architecture and function of metallopeptidase catalytic domains. Protein Sci. Publ. Protein Soc. 2014, 23, 123–144. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Thomas, P.D.; Huang, X.; Bateman, A.; Finn, R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018, 46, D624–D632. [Google Scholar] [CrossRef]
- Auld, D.S. Catalytic Mechanisms for Metallopeptidases; Argonne National Lab.: Argonne, IL, USA, 2007. [Google Scholar]
- Matthews, B.W. Structural basis of the action of thermolysin and related zinc peptidases. Acc. Chem. Res. 1988, 21, 333–340. [Google Scholar] [CrossRef]
- Auld, D.S.; Vallee, B.L. Kinetics of carboxypeptidase A. The pH dependence of tripeptide hydrolysis catalyzed by zinc, cobalt, and manganese enzymes. Biochemistry 1970, 9, 4352–4359. [Google Scholar] [CrossRef]
- Izquierdo-Martin, M.; Stein, R.L. Mechanistic studies on the inhibition of thermolysin by a peptide hydroxamic acid. J. Am. Chem. Soc 1992, 114, 325–331. [Google Scholar] [CrossRef]
- Sigman, J.A.; Edwards, S.R.; Pabon, A.; Glucksman, M.J.; Wolfson, A.J. pH dependence studies provide insight into the structure and mechanism of thimet oligopeptidase (EC 3.4.24.15). FEBS Lett. 2003, 545, 224–228. [Google Scholar] [CrossRef]
- Brown, C.K.; Madauss, K.; Lian, W.; Beck, M.R.; Tolbert, W.D.; Rodgers, D.W. Structure of neurolysin reveals a deep channel that limits substrate access. Proc. Natl. Acad. Sci. USA 2001, 98, 3127–3132. [Google Scholar] [CrossRef] [PubMed]
- Comellas-Bigler, M.; Lang, R.; Bode, W.; Maskos, K. Crystal structure of the E. coli dipeptidyl carboxypeptidase Dcp: Further indication of a ligand-dependent hinge movement mechanism. J. Mol. Biol. 2005, 349, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, B.; Teixeira, P.F.; Berntsson, R.P.; Murcha, M.W.; Branca, R.M.; Radomiljac, J.D.; Regberg, J.; Svensson, L.M.; Bakali, A.; Langel, U.; et al. Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, E3761–E3769. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Rajagopalan, K.; Sadre-Bazzaz, K.; Moreau, M.; Klessig, D.F.; Tong, L. Structure of the Arabidopsis thaliana TOP2 oligopeptidase. Acta Crystallographica. Sect. F Struct. Biol. Commun. 2014, 70, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.F.; Masuyer, G.; Pinho, C.M.; Branca, R.M.M.; Kmiec, B.; Wallin, C.; Wärmländer, S.; Berntsson, R.P.; Ankarcrona, M.; Gräslund, A.; et al. Mechanism of Peptide Binding and Cleavage by the Human Mitochondrial Peptidase Neurolysin. J. Mol. Biol. 2018, 430, 348–362. [Google Scholar] [CrossRef]
- Machado, M.F.; Rioli, V.; Dalio, F.M.; Castro, L.M.; Juliano, M.A.; Tersariol, I.L.; Ferro, E.S.; Juliano, L.; Oliveira, V. The role of Tyr605 and Ala607 of thimet oligopeptidase and Tyr606 and Gly608 of neurolysin in substrate hydrolysis and inhibitor binding. Biochem. J. 2007, 404, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Bruce, L.A.; Sigman, J.A.; Randall, D.; Rodriguez, S.; Song, M.M.; Dai, Y.; Elmore, D.E.; Pabon, A.; Glucksman, M.J.; Wolfson, A.J. Hydrogen bond residue positioning in the 599–611 loop of thimet oligopeptidase is required for substrate selection. FEBS J. 2008, 275, 5607–5617. [Google Scholar] [CrossRef] [PubMed]
- Sigman, J.A.; Patwa, T.H.; Tablante, A.V.; Joseph, C.D.; Glucksman, M.J.; Wolfson, A.J. Flexibility in substrate recognition by thimet oligopeptidase as revealed by denaturation studies. Biochem. J. 2005, 388, 255–261. [Google Scholar] [CrossRef][Green Version]
- Acker, G.R.; Molineaux, C.; Orlowski, M. Synaptosomal membrane-bound form of endopeptidase-24.15 generates Leu-enkephalin from dynorphin1-8, alpha- and beta-neoendorphin, and Met-enkephalin from Met-enkephalin-Arg6-Gly7-Leu8. J. Neurochem. 1987, 48, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.G.; Orlowski, M. Soluble metalloendopeptidase from rat brain: Action on enkephalin-containing peptides and other bioactive peptides. Endocrinology 1985, 116, 1418–1425. [Google Scholar] [CrossRef]
- Moody, T.W.; Mayr, C.A.; Gillespie, T.J.; Davis, T.P. Neurotensin is metabolized by endogenous proteases in prostate cancer cell lines. Peptides 1998, 19, 253–258. [Google Scholar] [CrossRef]
- Pierotti, A.; Dong, K.W.; Glucksman, M.J.; Orlowski, M.; Roberts, J.L. Molecular cloning and primary structure of rat testes metalloendopeptidase EC 3.4.24.15. Biochemistry 1990, 29, 10323–10329. [Google Scholar] [CrossRef] [PubMed]
- Pineau, C.; McCool, S.; Glucksman, M.J.; Jegou, B.; Pierotti, A.R. Distribution of thimet oligopeptidase (E.C. 3.4.24.15) in human and rat testes. J. Cell Sci. 1999, 112 Pt 20, 3455–3462. [Google Scholar] [CrossRef] [PubMed]
- Swanson, T.A.; Kim, S.I.; Myers, M.; Pabon, A.; Philibert, K.D.; Wang, M.; Glucksman, M.J. The role of neuropeptide processing enzymes in endocrine (prostate) cancer: EC 3.4.24.15 (EP24.15). Protein Pept. Lett. 2004, 11, 471–478. [Google Scholar] [CrossRef] [PubMed]
- McKie, N.; Dando, P.M.; Rawlings, N.D.; Barrett, A.J. Thimet oligopeptidase: Similarity to ‘soluble angiotensin II-binding protein’ and some corrections to the published amino acid sequence of the rat testis enzyme. Biochem. J. 1993, 295 Pt 1, 57–60. [Google Scholar] [CrossRef]
- Molina, H.M.; Carmona, A.K.; Kouyoumdjian, M.; Borges, D.R. Thimet oligopeptidase EC 3.4.24.15 is a major liver kininase. Life Sci. 2000, 67, 509–520. [Google Scholar] [CrossRef]
- Fontenele-Neto, J.D.; Massarelli, E.E.; Gurgel Garrido, P.A.; Beaudet, A.; Ferro, E.S. Comparative fine structural distribution of endopeptidase 24.15 (EC3.4.24.15) and 24.16 (EC3.4.24.16) in rat brain. J. Comp. Neurol. 2001, 438, 399–410. [Google Scholar] [CrossRef]
- Massarelli, E.E.; Casatti, C.A.; Kato, A.; Camargo, A.C.; Bauer, J.A.; Glucksman, M.J.; Roberts, J.L.; Hirose, S.; Ferro, E.S. Differential subcellular distribution of neurolysin (EC 3.4.24.16) and thimet oligopeptidase (EC 3.4.24.15) in the rat brain. Brain Res. 1999, 851, 261–265. [Google Scholar] [CrossRef]
- Dahms, P.; Mentlein, R. Purification of the main somatostatin-degrading proteases from rat and pig brains, their action on other neuropeptides, and their identification as endopeptidases 24.15 and 24.16. Eur. J. Biochem. 1992, 208, 145–154. [Google Scholar] [CrossRef]
- Montiel, J.L.; Cornille, F.; Roques, B.P.; Noble, F. Nociceptin/orphanin FQ metabolism: Role of aminopeptidase and endopeptidase 24.15. J. Neurochem. 1997, 68, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Grum-Tokars, V.; Swanson, T.A.; Cotter, E.J.; Cahill, P.A.; Roberts, J.L.; Cummins, P.M.; Glucksman, M.J. Novel roles of neuropeptide processing enzymes: EC3.4.24.15 in the neurome. J. Neurosci. Res. 2003, 74, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, J.P.; Alvarez Gonzalez, M.L.; Gerard, A.; Franchimont, P. Gonadotropin releasing hormone inhibitory autofeedback by subproducts antagonist at N-methyl-D-aspartate receptors: A model of autocrine regulation of peptide secretion. Endocrinology 1994, 134, 1589–1592. [Google Scholar] [CrossRef] [PubMed]
- Chappell, M.C.; Modrall, J.G.; Diz, D.I.; Ferrario, C.M. Novel aspects of the renal renin-angiotensin system: Angiotensin-(1-7), ACE2 and blood pressure regulation. Contrib. Nephrol. 2004, 143, 77–89. [Google Scholar] [CrossRef]
- Cunha, F.M.; Berti, D.A.; Ferreira, Z.S.; Klitzke, C.F.; Markus, R.P.; Ferro, E.S. Intracellular peptides as natural regulators of cell signaling. J. Biol. Chem. 2008, 283, 24448–24459. [Google Scholar] [CrossRef]
- Checler, F.; Barelli, H.; Dauch, P.; Dive, V.; Vincent, B.; Vincent, J.P. Neurolysin: Purification and assays. Methods Enzymol. 1995, 248, 593–614. [Google Scholar] [CrossRef]
- Checler, F.; Emson, P.C.; Vincent, J.P.; Kitabgi, P. Inactivation of neurotensin by rat brain synaptic membranes. Cleavage at the Pro10-Tyr11 bond by endopeptidase 24.11 (enkephalinase) and a peptidase different from proline-endopeptidase. J. Neurochem. 1984, 43, 1295–1301. [Google Scholar] [CrossRef]
- Mentlein, R.; Dahms, P. Endopeptidases 24.16 and 24.15 are responsible for the degradation of somatostatin, neurotensin, and other neuropeptides by cultivated rat cortical astrocytes. J. Neurochem. 1994, 62, 27–36. [Google Scholar] [CrossRef]
- Barrett, A.J.; Brown, M.A. Chicken liver Pz-peptidase, a thiol-dependent metallo-endopeptidase. Biochem. J. 1990, 271, 701–706. [Google Scholar] [CrossRef]
- Dando, P.M.; Brown, M.A.; Barrett, A.J. Human thimet oligopeptidase. Biochem. J. 1993, 294 Pt 2, 451–457. [Google Scholar] [CrossRef]
- Berti, D.A.; Morano, C.; Russo, L.C.; Castro, L.M.; Cunha, F.M.; Zhang, X.; Sironi, J.; Klitzke, C.F.; Ferro, E.S.; Fricker, L.D. Analysis of intracellular substrates and products of thimet oligopeptidase in human embryonic kidney 293 cells. J. Biol. Chem. 2009, 284, 14105–14116. [Google Scholar] [CrossRef] [PubMed]
- Kobilka, B.K.; Deupi, X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 2007, 28, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.B.D.; Franco, R.D.; Camarini, R.; Munhoz, C.D.; Eichler, R.A.S.; Gewehr, M.C.F.; Reckziegel, P.; Llanos, R.P.; Dale, C.S.; Silva, V.; et al. Thimet Oligopeptidase (EC 3.4.24.15) Key Functions Suggested by Knockout Mice Phenotype Characterization. Biomolecules 2019, 9, 382. [Google Scholar] [CrossRef] [PubMed]
- Ferro, E.S.; Tullai, J.W.; Glucksman, M.J.; Roberts, J.L. Secretion of metalloendopeptidase 24.15 (EC 3.4.24.15). DNA Cell Biol. 1999, 18, 781–789. [Google Scholar] [CrossRef]
- Carrarini, C.; Russo, M.; Dono, F.; Di Pietro, M.; Rispoli, M.G.; Di Stefano, V.; Ferri, L.; Barbone, F.; Vitale, M.; Thomas, A.; et al. A Stage-Based Approach to Therapy in Parkinson’s Disease. Biomolecules 2019, 9, 388. [Google Scholar] [CrossRef] [PubMed]
- Pollio, G.; Hoozemans, J.J.; Andersen, C.A.; Roncarati, R.; Rosi, M.C.; van Haastert, E.S.; Seredenina, T.; Diamanti, D.; Gotta, S.; Fiorentini, A.; et al. Increased expression of the oligopeptidase THOP1 is a neuroprotective response to Abeta toxicity. Neurobiol. Dis. 2008, 31, 145–158. [Google Scholar] [CrossRef] [PubMed]
- LaFerla, F.M.; Green, K.N.; Oddo, S. Intracellular amyloid-beta in Alzheimer’s disease. Nat. Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef]
- Koike, H.; Seki, H.; Kouchi, Z.; Ito, M.; Kinouchi, T.; Tomioka, S.; Sorimachi, H.; Saido, T.C.; Maruyama, K.; Suzuki, K.; et al. Thimet oligopeptidase cleaves the full-length Alzheimer amyloid precursor protein at a beta-secretase cleavage site in COS cells. J. Biochem. 1999, 126, 235–242. [Google Scholar] [CrossRef]
- Yamin, R.; Malgeri, E.G.; Sloane, J.A.; McGraw, W.T.; Abraham, C.R. Metalloendopeptidase EC 3.4.24.15 is necessary for Alzheimer’s amyloid-beta peptide degradation. J. Biol. Chem. 1999, 274, 18777–18784. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.M.; Tummolo, D.M.; Spruyt, M.A.; Jacobsen, J.S.; Sonnenberg-Reines, J. Evaluation of cathepsins D and G and EC 3.4.24.15 as candidate beta-secretase proteases using peptide and amyloid precursor protein substrates. J. Neurochem. 1996, 66, 2436–2445. [Google Scholar] [CrossRef] [PubMed]
- Conn, K.J.; Pietropaolo, M.; Ju, S.T.; Abraham, C.R. Monoclonal antibodies against the human metalloprotease EC 3.4.24.15 label neurofibrillary tangles in Alzheimer’s disease brain. J. Neurochem. 1996, 66, 2011–2018. [Google Scholar] [CrossRef]
- Berti, D.A.; Russo, L.C.; Castro, L.M.; Cruz, L.; Gozzo, F.C.; Heimann, J.C.; Lima, F.B.; Oliveira, A.C.; Andreotti, S.; Prada, P.O.; et al. Identification of intracellular peptides in rat adipose tissue: Insights into insulin resistance. Proteomics 2012, 12, 2668–2681. [Google Scholar] [CrossRef]
- Gewehr, M.C.F.; Teixeira, A.A.S.; Santos, B.A.C.; Biondo, L.A.; Gozzo, F.C.; Cordibello, A.M.; Eichler, R.A.S.; Reckziegel, P.; Da Silva, R.N.O.; Dos Santos, N.B.; et al. The Relevance of Thimet Oligopeptidase in the Regulation of Energy Metabolism and Diet-Induced Obesity. Biomolecules 2020, 10, 321. [Google Scholar] [CrossRef] [PubMed]
- Kessler, J.H.; Khan, S.; Seifert, U.; Le Gall, S.; Chow, K.M.; Paschen, A.; Bres-Vloemans, S.A.; de Ru, A.; van Montfoort, N.; Franken, K.L.; et al. Antigen processing by nardilysin and thimet oligopeptidase generates cytotoxic T cell epitopes. Nat. Immunol. 2011, 12, 45–53. [Google Scholar] [CrossRef]
- Chappell, M.C.; Allred, A.J.; Ferrario, C.M. Pathways of angiotensin-(1-7) metabolism in the kidney. Nephrol. Dial. Transplant. 2001, 16 (Suppl. 1), 22–26. [Google Scholar] [CrossRef]
- Chappell, M.C. The Angiotensin-(1-7) Axis: Formation and Metabolism Pathways. In Angiotensin-(1-7): A Comprehensive Review; Santos, R.A.S., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–26. [Google Scholar]
- Santos, R.A.S.; Sampaio, W.O.; Alzamora, A.C.; Motta-Santos, D.; Alenina, N.; Bader, M.; Campagnole-Santos, M.J. The ACE2/Angiotensin-(1-7)/MAS Axis of the Renin-Angiotensin System: Focus on Angiotensin-(1-7). Physiol. Rev. 2018, 98, 505–553. [Google Scholar] [CrossRef] [PubMed]
- Carey, R.M. Newly discovered components and actions of the renin-angiotensin system. Hypertension 2013, 62, 818–822. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reudelhuber, T.L. A place in our hearts for the lowly angiotensin 1-7 peptide? Hypertension 2006, 47, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.A. Angiotensin-(1-7). Hypertension 2014, 63, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.L.V.; da Silva, F.A.; Bolais-Ramos, L.; de Oliveira, G.C.; Ribeiro, R.C.; Pereira, D.A.A.; Annoni, F.; Diniz, M.M.L.; Silva, T.G.F.; Zivianni, B.; et al. Increased circulating levels of angiotensin-(1-7) in severely ill COVID-19 patients. medRxiv 2021. [Google Scholar] [CrossRef]
- van Lier, D.; Kox, M.; Santos, K.; van der Hoeven, H.; Pillay, J.; Pickkers, P. Increased blood angiotensin converting enzyme 2 activity in critically ill COVID-19 patients. ERJ Open Res. 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.G.; Souza, L.L.; Becari, C.; Duarte, D.A.; Camacho, F.R.; Oliveira, J.A.; Gomes, M.D.; Oliveira, E.B.; Salgado, M.C.; Garcia-Cairasco, N.; et al. Angiotensin II-independent angiotensin-(1-7) formation in rat hippocampus: Involvement of thimet oligopeptidase. Hypertension 2013, 62, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, K.; Chan, D.; Watermeyer, T. The cognitive consequences of the COVID-19 epidemic: Collateral damage? Brain Commun. 2020, 2, fcaa069. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.A.; Cruz-Diaz, N.; Marshall, A.C.; Pirro, N.T.; Su, Y.; Gwathmey, T.M.; Rose, J.C.; Chappell, M.C. An angiotensin-(1-7) peptidase in the kidney cortex, proximal tubules, and human HK-2 epithelial cells that is distinct from insulin-degrading enzyme. Am. J. Physiol. Renal. Physiol. 2015, 308, F594–F601. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Diaz, N.; Wilson, B.A.; Pirro, N.T.; Brosnihan, K.B.; Marshall, A.C.; Chappell, M.C. Identification of dipeptidyl peptidase 3 as the Angiotensin-(1-7) degrading peptidase in human HK-2 renal epithelial cells. Peptides 2016, 83, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.A.; Nautiyal, M.; Gwathmey, T.M.; Rose, J.C.; Chappell, M.C. Evidence for a mitochondrial angiotensin-(1-7) system in the kidney. Am. J. Physiol. Renal. Physiol. 2016, 310, F637–F645. [Google Scholar] [CrossRef]
- Khan, S.; Chen, L.; Yang, C.R.; Raghuram, V.; Khundmiri, S.J.; Knepper, M.A. Does SARS-CoV-2 Infect the Kidney? J. Am. Soc. Nephrol. 2020, 31, 2746–2748. [Google Scholar] [CrossRef]
- Chappell MC, T.E.; Brosnihan, K.B.; Ferrario, C.M. Conversion of angiotensin I to angiotensin-(1-7) by thimet oligopeptidase (EC 3.4. 24.15) in vascular smooth muscle cells. J. Vasc. Med. Biol. 1994, 5, 129–137. [Google Scholar]
- Suski, M.; Gębska, A.; Olszanecki, R.; Stachowicz, A.; Uracz, D.; Madej, J.; Korbut, R. Influence of atorvastatin on angiotensin I metabolism in resting and TNF-α-activated rat vascular smooth muscle cells. J. Renin-Angiotensin-Aldosterone Syst. 2014, 15, 378–383. [Google Scholar] [CrossRef]
- Chung, M.K.; Zidar, D.A.; Bristow, M.R.; Cameron, S.J.; Chan, T.; Harding, C.V., 3rd; Kwon, D.H.; Singh, T.; Tilton, J.C.; Tsai, E.J.; et al. COVID-19 and Cardiovascular Disease: From Bench to Bedside. Circ. Res. 2021, 128, 1214–1236. [Google Scholar] [CrossRef]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; Weizman, O.E.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J. Exp. Med. 2021, 218. [Google Scholar] [CrossRef]
- Rhea, E.M.; Logsdon, A.F.; Hansen, K.M.; Williams, L.M.; Reed, M.J.; Baumann, K.K.; Holden, S.J.; Raber, J.; Banks, W.A.; Erickson, M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2021, 24, 368–378. [Google Scholar] [CrossRef]
- Ramani, A.; Müller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Müller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 targets neurons of 3D human brain organoids. Embo. J. 2020, 39, e106230. [Google Scholar] [CrossRef]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef]
- Vijayan, A.; Humphreys, B.D. SARS-CoV-2 in the kidney: Bystander or culprit? Nat. Rev. Nephrol. 2020, 16, 703–704. [Google Scholar] [CrossRef]
- Wu, L.; O’Kane, A.M.; Peng, H.; Bi, Y.; Motriuk-Smith, D.; Ren, J. SARS-CoV-2 and cardiovascular complications: From molecular mechanisms to pharmaceutical management. Biochem. Pharmacol. 2020, 178, 114114. [Google Scholar] [CrossRef]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Jha, N.K.; Jeyaraman, M.; Rachamalla, M.; Ojha, S.; Dua, K.; Chellappan, D.K.; Muthu, S.; Sharma, A.; Jha, S.K.; Jain, R.; et al. Current Understanding of Novel Coronavirus: Molecular Pathogenesis, Diagnosis, and Treatment Approaches. Immuno 2021, 1, 4. [Google Scholar] [CrossRef]
- Kuriakose, J.; Montezano, A.C.; Touyz, R.M. ACE2/Ang-(1-7)/Mas1 axis and the vascular system: Vasoprotection to COVID-19-associated vascular disease. Clin. Sci. 2021, 135, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Wu, J.; Ma, H. Regulation of angiotensin-converting enzyme 2 and Mas receptor by Ang-(1-7) in heart and kidney of spontaneously hypertensive rats. J. Renin-Angiotensin-Aldosterone Syst. 2011, 12, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.; Cheng, M.; Lee, T.; McGeer, A.; Sweet, D.; Tran, K.; Lee, T.; Murthy, S.; Boyd, J.; Singer, J.; et al. Sustained Dysregulation of the Plasma Renin-angiotensin System in Acute COVID-19. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Soria-Castro, R.; Meneses-Preza, Y.G.; Rodríguez López, G.M.; Romero-Ramírez, S.; Sosa-Hernandez, V.A.; Cervantes-Díaz, R.; Pérez-Fragoso, A.; Torres-Ruíz, J.J.; Gómez-Martín, D.; Campillo-Navarro, M.; et al. Severe COVID-19 is marked by dysregulated serum levels of carboxypeptidase A3 and serotonin. medRxiv 2021. [Google Scholar] [CrossRef]
- Silva-Aguiar, R.P.; Peruchetti, D.B.; Rocco, P.R.M.; Schmaier, A.H.; PMR, E.S.; Martins, M.A.; Carvalho, V.F.; Pinheiro, A.A.S.; Caruso-Neves, C. Role of the renin-angiotensin system in the development of severe COVID-19 in hypertensive patients. Am. J. Physiol. Lung Cell Mol. Physiol. 2020, 319, L596–L602. [Google Scholar] [CrossRef] [PubMed]
- Garrido, P.A.; Vandenbulcke, F.; Ramjaun, A.R.; Vincent, B.; Checler, F.; Ferro, E.; Beaudet, A. Confocal microscopy reveals thimet oligopeptidase (EC 3.4.24.15) and neurolysin (EC 3.4.24.16) in the classical secretory pathway. DNA Cell Biol. 1999, 18, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Crack, P.J.; Wu, T.J.; Cummins, P.M.; Ferro, E.S.; Tullai, J.W.; Glucksman, M.J.; Roberts, J.L. The association of metalloendopeptidase EC 3.4.24.15 at the extracellular surface of the AtT-20 cell plasma membrane. Brain Res. 1999, 835, 113–124. [Google Scholar] [CrossRef]
- Li, C.J.; Liu, Y.; Chen, Y.; Yu, D.; Williams, K.J.; Liu, M.L. Novel proteolytic microvesicles released from human macrophages after exposure to tobacco smoke. Am. J. Pathol. 2013, 182, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, Y.; Wei, W.; Liu, M.L. Extracellular vesicles in autoimmune vasculitis—Little dirts light the fire in blood vessels. Autoimmun. Rev. 2019, 18, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bruce, L.; Wolfson, A. Extracellular Thimet Oligopeptidase is Released with Extracellular Vesicles from Human Prostate Cancer Cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Paine, M.S.; Brooks, A.M.; McCubrey, J.A.; Renegar, R.H.; Wang, R.; Terrian, D.M. Senescence-associated exosome release from human prostate cancer cells. Cancer Res 2008, 68, 7864–7871. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Liu, M.L. Extracellular vesicles and lupus nephritis—New insights into pathophysiology and clinical implications. J. Autoimmun. 2020, 115, 102540. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zhao, Y.; Wu, X.; Zhang, N.; Song, H.; Wei, W.; Liu, M.L. Recent advances in Extracellular Vesicles and their involvements in vasculitis. Free Radic. Biol. Med. 2021, 171, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Williams, K.J.; Werth, V.P. Microvesicles in Autoimmune Diseases. Adv. Clin. Chem. 2016, 77, 125–175. [Google Scholar] [CrossRef]
- Latifkar, A.; Hur, Y.H.; Sanchez, J.C.; Cerione, R.A.; Antonyak, M.A. New insights into extracellular vesicle biogenesis and function. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Liu, M.L.; Williams, K.J. Microvesicles: Potential markers and mediators of endothelial dysfunction. Curr. Opin. Endocrinol. Diabetes Obes. 2012, 19, 121–127. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Cai, Q.; He, B.; Weiberg, A.; Buck, A.H.; Jin, H. Small RNAs and extracellular vesicles: New mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog. 2019, 15, e1008090. [Google Scholar] [CrossRef]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Stepanian, A.; Bourguignat, L.; Hennou, S.; Coupaye, M.; Hajage, D.; Salomon, L.; Alessi, M.C.; Msika, S.; de Prost, D. Microparticle increase in severe obesity: Not related to metabolic syndrome and unchanged after massive weight loss. Obesity (Silver Spring) 2013, 21, 2236–2243. [Google Scholar] [CrossRef]
- Eguchi, A.; Lazic, M.; Armando, A.M.; Phillips, S.A.; Katebian, R.; Maraka, S.; Quehenberger, O.; Sears, D.D.; Feldstein, A.E. Circulating adipocyte-derived extracellular vesicles are novel markers of metabolic stress. J. Mol. Med. 2016, 94, 1241–1253. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, G.; Liu, M.L. Microvesicles as Emerging Biomarkers and Therapeutic Targets in Cardiometabolic Diseases. Genom. Proteom. Bioinform. 2018, 16, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Fang, Q.H.; Liu, M.L.; Lin, J.N. Current understanding of the role of Adipose-derived Extracellular Vesicles in Metabolic Homeostasis and Diseases: Communication from the distance between cells/tissues. Theranostics 2020, 10, 7422–7435. [Google Scholar] [CrossRef]
- Zhang, J.M.; An, J. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 2007, 45, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Tarrant, J.M. Blood cytokines as biomarkers of in vivo toxicity in preclinical safety assessment: Considerations for their use. Toxicol. Sci. 2010, 117, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.Y.; Lee, W.; Kenny, H.A.; Dang, L.H.; Ellis, L.M.; Jonasch, E.; Lengyel, E.; Naora, H. Cancer-derived small extracellular vesicles promote angiogenesis by heparin-bound, bevacizumab-insensitive VEGF, independent of vesicle uptake. Commun. Biol. 2019, 2, 386. [Google Scholar] [CrossRef]
- Icimoto, M.Y.; Mendes Brito, A.M.; Ramos, M.P.C.; Oliveira, V.; Nantes-Cardoso, I.L. Increased Stability of Oligopeptidases Immobilized on Gold Nanoparticles. Catalysts 2020, 10, 78. [Google Scholar] [CrossRef]
- Ko, S.Y.; Naora, H. Extracellular Vesicle Membrane-Associated Proteins: Emerging Roles in Tumor Angiogenesis and Anti-Angiogenesis Therapy Resistance. Int. J. Mol. Sci. 2020, 21, 5418. [Google Scholar] [CrossRef]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Labrie, F. GnRH agonists and the rapidly increasing use of combined androgen blockade in prostate cancer. Endocr.-Relat. Cancer 2014, 21, R301–R317. [Google Scholar] [CrossRef] [PubMed]
- Plosker, G.L.; Brogden, R.N. Leuprorelin. A review of its pharmacology and therapeutic use in prostatic cancer, endometriosis and other sex hormone-related disorders. Drugs 1994, 48, 930–967. [Google Scholar] [CrossRef]
- Mentlein, R. Cell-surface peptidases. Int. Rev. Cytol. 2004, 235, 165–213. [Google Scholar] [CrossRef] [PubMed]
- Kandzija, N.; Zhang, W.; Motta-Mejia, C.; Mhlomi, V.; McGowan-Downey, J.; James, T.; Cerdeira, A.S.; Tannetta, D.; Sargent, I.; Redman, C.W.; et al. Placental extracellular vesicles express active dipeptidyl peptidase IV; levels are increased in gestational diabetes mellitus. J. Extracell. Vesicles 2019, 8, 1617000. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour, M.; Rezaie, J.; Nouri, M.; Panahi, Y. The role of extracellular vesicles in COVID-19 virus infection. Infect. Genet. Evol. 2020, 85, 104422. [Google Scholar] [CrossRef]
- Zhang, Q.; Jeppesen, D.K.; Higginbotham, J.N.; Franklin, J.L.; Crowe, J.E., Jr.; Coffey, R.J. Angiotensin-converting Enzyme 2-containing Small Extracellular Vesicles and Exomeres Bind the Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein. Gastroenterology 2021, 160, 958–961.e953. [Google Scholar] [CrossRef]
- Cocozza, F.; Névo, N.; Piovesana, E.; Lahaye, X.; Buchrieser, J.; Schwartz, O.; Manel, N.; Tkach, M.; Théry, C.; Martin-Jaular, L. Extracellular vesicles containing ACE2 efficiently prevent infection by SARS-CoV-2 Spike protein-containing virus. J. Extracell. Vesicles 2020, 10, e12050. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).