NTRK Gene Expression in Non-Small-Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
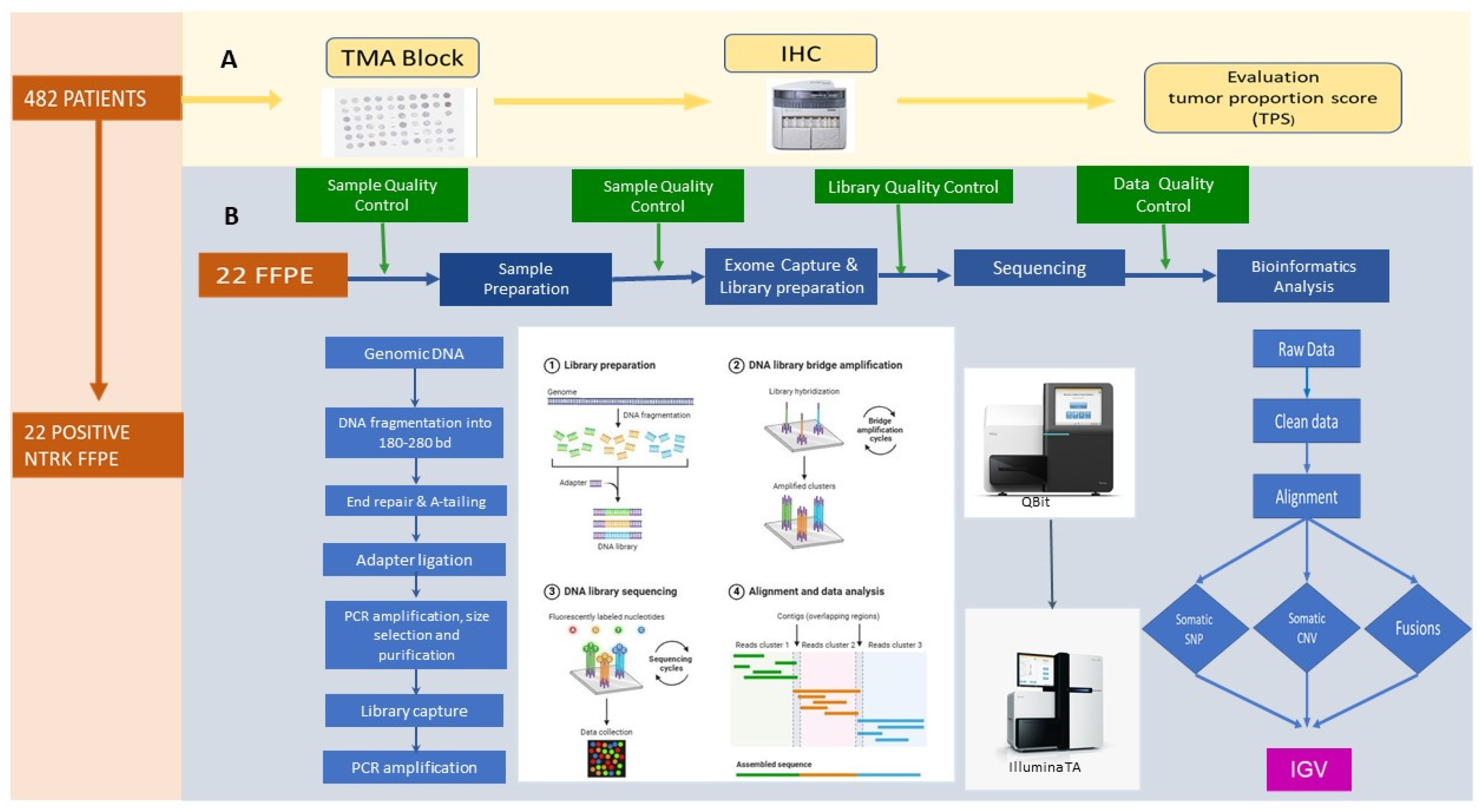
2.1. Construction and Immunohistochemistry of TMA
2.2. Evaluation of TRK Expression by IHC
2.3. Sample Preparation for Whole-Exome Sequencing and Bioinformatics Analysis
2.4. Statistical Methods
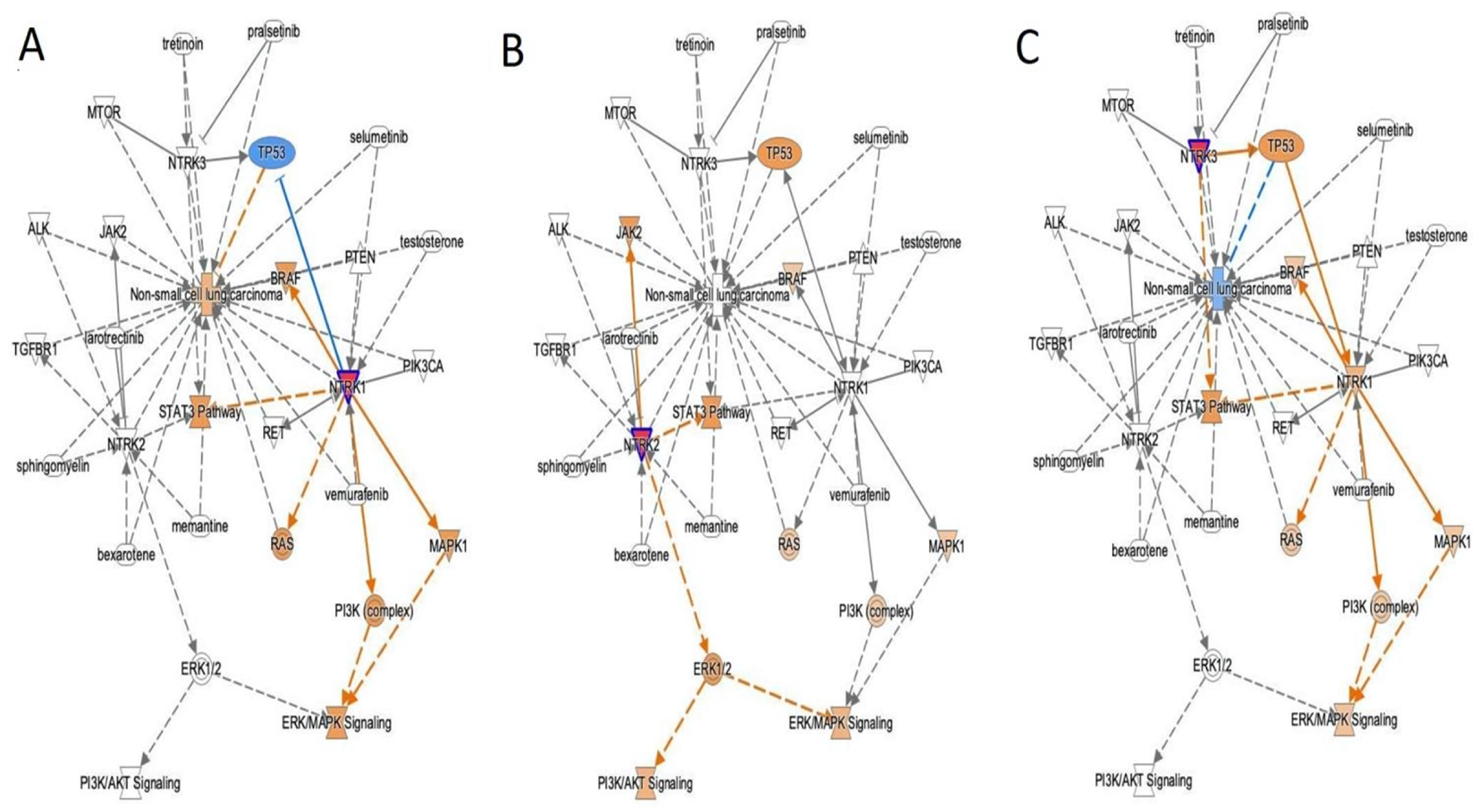
3. Results
3.1. Patient and Tumor Characteristics
3.2. TRK Immunohistochemical Staining
3.3. Associations Between TRK Immunohistochemical Expression in Tumor Cells and Clinicopathological Findings and Patient Prognosis
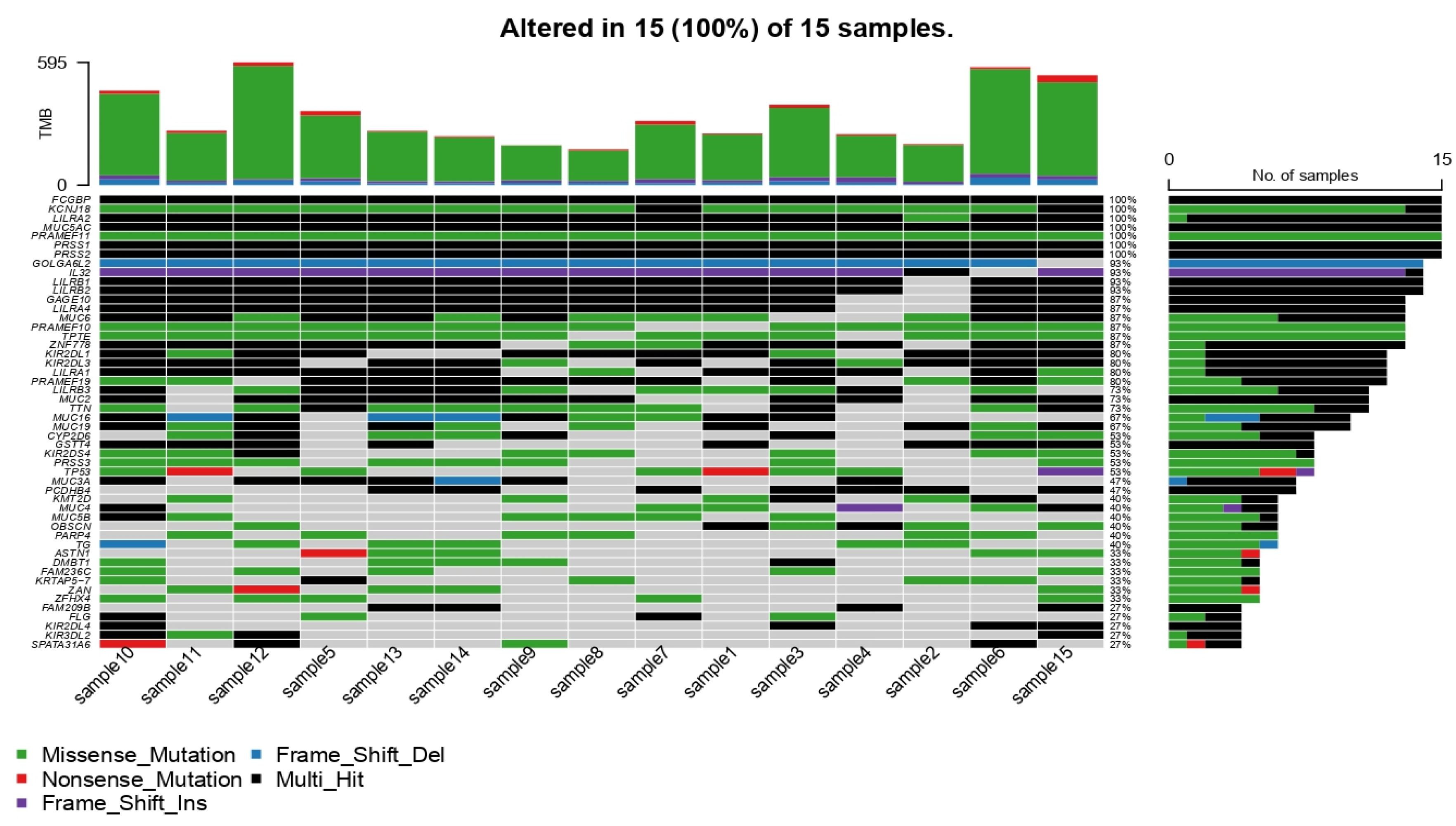
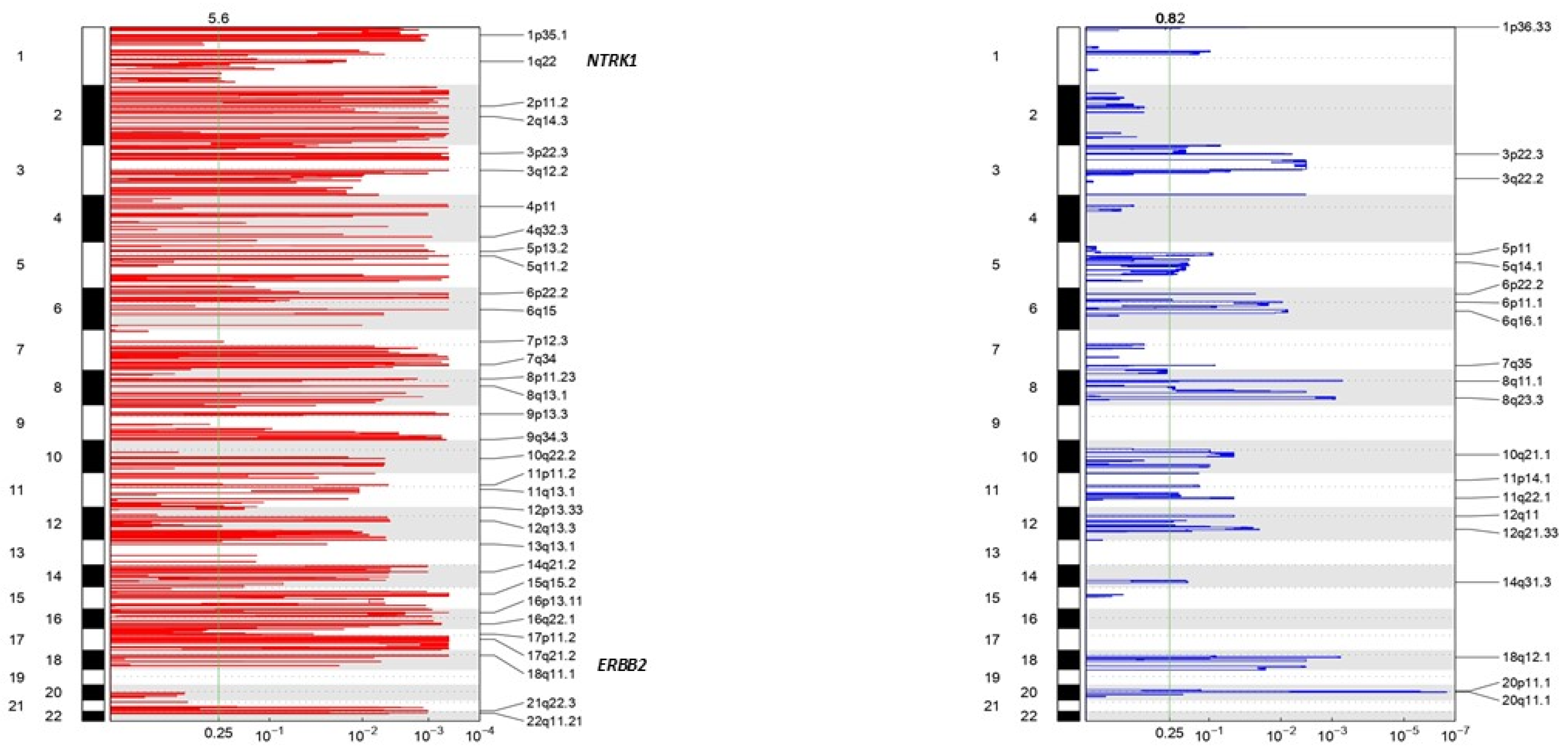
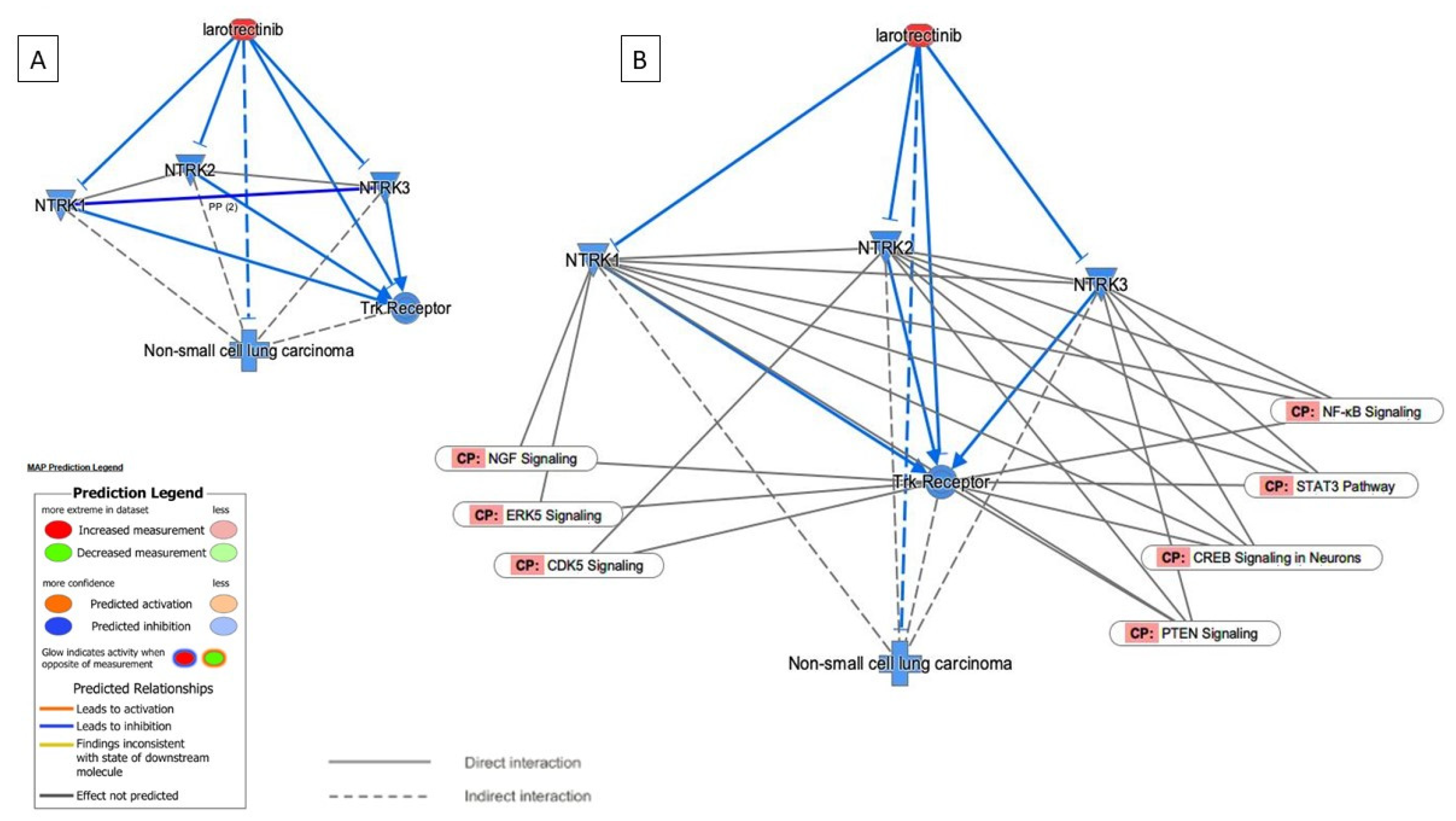
3.4. NTRK Somatic Mutations, CNV, and Fusions Detected by NGS Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amatu, A.; Sartore-Bianchi, A.; Bencardino, K.; Pizzutilo, E.G.; Tosi, F.; Siena, S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 2019, 30, viii5–viii15. [Google Scholar] [CrossRef] [PubMed]
- Ekman, S. How selecting best therapy for metastatic NTRK fusion-positive non-small cell lung cancer? Transl. Lung Cancer Res. 2020, 9, 2535–2544. [Google Scholar] [CrossRef] [PubMed]
- Solomon, J.P.; Benayed, R.; Hechtman, J.F.; Ladanyi, M. Identifying patients with NTRK fusion cancer. Ann. Oncol. 2019, 30, viii16–viii22. [Google Scholar] [CrossRef] [PubMed]
- Okamura, R.; Boichard, A.; Kato, S.; Sicklick, J.K.; Bazhenova, L.; Kurzrock, R. Analysis of NTRK Alterations in Pan-Cancer Adult and Pediatric Malignancies: Implications for NTRK-Targeted Therapeutics. JCO Precis. Oncol. 2018, 2018, PO.18.00183. [Google Scholar] [CrossRef]
- Weiss, L.M.; Funari, V.A. NTRK fusions and Trk proteins: What are they and how to test for them. Hum. Pathol. 2021, 112, 59–69. [Google Scholar] [CrossRef]
- Chou, A.; Fraser, T.; Ahadi, M.; Fuchs, T.; Sioson, L.; Clarkson, A.; Sheen, A.; Singh, N.; Corless, C.L.; Gill, A.J. NTRK gene rearrangements are highly enriched in MLH1/PMS2 deficient, BRAF wild-type colorectal carcinomas-a study of 4569 cases. Mod. Pathol. 2020, 33, 924–932. [Google Scholar] [CrossRef] [PubMed]
- Lassen, U. How I treat NTRK gene fusion-positive cancers. ESMO Open 2019, 4, e000612. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Patel, M.R. The Challenge and Opportunity of NTRK Inhibitors in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 2916. [Google Scholar] [CrossRef]
- Cocco, E.; Scaltriti, M.; Drilon, A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat. Rev. Clin. Oncol. 2018, 15, 731–747. [Google Scholar] [CrossRef]
- Gatalica, Z.; Xiu, J.; Swensen, J.; Vranic, S. Molecular characterization of cancers with NTRK gene fusions. Mod. Pathol. 2019, 32, 147–153. [Google Scholar] [CrossRef]
- Solomon, J.P.; Linkov, I.; Rosado, A.; Mullaney, K.; Rosen, E.Y.; Frosina, D.; Jungbluth, A.A.; Zehir, A.; Benayed, R.; Drilon, A.; et al. NTRK fusion detection across multiple assays and 33,997 cases: Diagnostic implications and pitfalls. Mod. Pathol. 2020, 33, 38–46. [Google Scholar] [CrossRef]
- Gao, F.; Griffin, N.; Faulkner, S.; Rowe, C.W.; Williams, L.; Roselli, S.; Thorne, R.F.; Ferdoushi, A.; Jobling, P.; Walker, M.M.; et al. The neurotrophic tyrosine kinase receptor TrkA and its ligand NGF are increased in squamous cell carcinomas of the lung. Sci. Rep. 2018, 8, 8135. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, L.F. Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1545–1564. [Google Scholar] [CrossRef] [PubMed]
- Elfving, H.; Brostrom, E.; Moens, L.N.J.; Almlof, J.; Cerjan, D.; Lauter, G.; Nord, H.; Mattsson, J.S.M.; Ullenhag, G.J.; Strell, C.; et al. Evaluation of NTRK immunohistochemistry as a screening method for NTRK gene fusion detection in non-small cell lung cancer. Lung Cancer 2021, 151, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Penault-Llorca, F.; Rudzinski, E.R.; Sepulveda, A.R. Testing algorithm for identification of patients with TRK fusion cancer. J. Clin. Pathol. 2019, 72, 460–467. [Google Scholar] [CrossRef] [PubMed]
- De Winne, K.; Sorber, L.; Lambin, S.; Siozopoulou, V.; Beniuga, G.; Dedeurwaerdere, F.; D’Haene, N.; Habran, L.; Libbrecht, L.; Van Huysse, J.; et al. Immunohistochemistry as a screening tool for NTRK gene fusions: Results of a first Belgian ring trial. Virchows Arch. 2021, 478, 283–291. [Google Scholar] [CrossRef]
- Strohmeier, S.; Brcic, I.; Popper, H.; Liegl-Atzwanger, B.; Lindenmann, J.; Brcic, L. Applicability of pan-TRK immunohistochemistry for identification of NTRK fusions in lung carcinoma. Sci. Rep. 2021, 11, 9785. [Google Scholar] [CrossRef]
- Haratake, N.; Seto, T. NTRK Fusion-positive Non-small-cell Lung Cancer: The Diagnosis and Targeted Therapy. Clin. Lung Cancer 2021, 22, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Garrido, P.; Hladun, R.; de Alava, E.; Alvarez, R.; Bautista, F.; Lopez-Rios, F.; Colomer, R.; Rojo, F. Multidisciplinary consensus on optimising the detection of NTRK gene alterations in tumours. Clin. Transl. Oncol. 2021, 23, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Farago, A.F.; Taylor, M.S.; Doebele, R.C.; Zhu, V.W.; Kummar, S.; Spira, A.I.; Boyle, T.A.; Haura, E.B.; Arcila, M.E.; Benayed, R.; et al. Clinicopathologic Features of Non-Small-Cell Lung Cancer Harboring an NTRK Gene Fusion. JCO Precis. Oncol. 2018, 2018, PO.18.00037. [Google Scholar] [CrossRef]
- Park, S.J.; More, S.; Murtuza, A.; Woodward, B.D.; Husain, H. New Targets in Non-Small Cell Lung Cancer. Hematol. Oncol. Clin. N. Am. 2017, 31, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Remon, J.; Pignataro, D.; Novello, S.; Passiglia, F. Current treatment and future challenges in ROS1- and ALK-rearranged advanced non-small cell lung cancer. Cancer Treat. Rev. 2021, 95, 102178. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, E.; Feld, E.; Horn, L. Driven by Mutations: The Predictive Value of Mutation Subtype in EGFR-Mutated Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-González, J.P.; Chaves, J.J.; Parra-Medina, R. Multiple mutations in the EGFR gene in lung cancer: A systematic review. Transl. Lung Cancer Res. 2022, 11, 2148–2163. [Google Scholar] [CrossRef]
- Chuang, J.C.; Stehr, H.; Liang, Y.; Das, M.; Huang, J.; Diehn, M.; Wakelee, H.A.; Neal, J.W. ERBB2-Mutated Metastatic Non-Small Cell Lung Cancer: Response and Resistance to Targeted Therapies. J. Thorac. Oncol. 2017, 12, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Sentana-Lledo, D.; Academia, E.; Viray, H.; Rangachari, D.; Kobayashi, S.S.; VanderLaan, P.A.; Costa, D.B. EGFR exon 20 insertion mutations and ERBB2 mutations in lung cancer: A narrative review on approved targeted therapies from oral kinase inhibitors to antibody-drug conjugates. Transl. Lung Cancer Res. 2023, 12, 1590–1610. [Google Scholar] [CrossRef]
- Li, H.; Yan, S.; Liu, Y.; Ma, L.; Liu, X.; Liu, Y.; Cheng, Y. Analysis of NTRK mutation and clinicopathologic factors in lung cancer patients in northeast China. Int. J. Biol. Markers 2020, 35, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Drilon, A.; Laetsch, T.W.; Kummar, S.; DuBois, S.G.; Lassen, U.N.; Demetri, G.D.; Nathenson, M.; Doebele, R.C.; Farago, A.F.; Pappo, A.S.; et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N. Engl. J. Med. 2018, 378, 731–739. [Google Scholar] [CrossRef]
- Ardini, E.; Menichincheri, M.; Banfi, P.; Bosotti, R.; De Ponti, C.; Pulci, R.; Ballinari, D.; Ciomei, M.; Texido, G.; Degrassi, A.; et al. Entrectinib, a Pan-TRK, ROS1, and ALK Inhibitor with Activity in Multiple Molecularly Defined Cancer Indications. Mol. Cancer Ther. 2016, 15, 628–639. [Google Scholar] [CrossRef]
- Hechtman, J.F.; Benayed, R.; Hyman, D.M.; Drilon, A.; Zehir, A.; Frosina, D.; Arcila, M.E.; Dogan, S.; Klimstra, D.S.; Ladanyi, M.; et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am. J. Surg. Pathol. 2017, 41, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Yip, S.; Sorensen, P.H. Methods for Identifying Patients with Tropomyosin Receptor Kinase (TRK) Fusion Cancer. Pathol. Oncol. Res. 2020, 26, 1385–1399. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.P.; Taylor, S.E.; O’Leary, J.J.; Finn, S.P. Molecular testing in oncology: Problems, pitfalls and progress. Lung Cancer 2014, 83, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Zito Marino, F.; Pagliuca, F.; Ronchi, A.; Cozzolino, I.; Montella, M.; Berretta, M.; Errico, M.E.; Donofrio, V.; Bianco, R.; Franco, R. NTRK Fusions, from the Diagnostic Algorithm to Innovative Treatment in the Era of Precision Medicine. Int. J. Mol. Sci. 2020, 21, 3718. [Google Scholar] [CrossRef]
- Solomon, J.P.; Hechtman, J.F. Detection of NTRK Fusions: Merits and Limitations of Current Diagnostic Platforms. Cancer Res. 2019, 79, 3163–3168. [Google Scholar] [CrossRef]
- Marchio, C.; Scaltriti, M.; Ladanyi, M.; Iafrate, A.J.; Bibeau, F.; Dietel, M.; Hechtman, J.F.; Troiani, T.; Lopez-Rios, F.; Douillard, J.Y.; et al. ESMO recommendations on the standard methods to detect NTRK fusions in daily practice and clinical research. Ann. Oncol. 2019, 30, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Durzynska, M.; Michalek, I.M. Pan-TRK immunohistochemistry as a tool in the screening for NTRK gene fusions in cancer patients. Oncol. Clin. Pract. 2024, 20, 15–21. [Google Scholar] [CrossRef]
- Patel, P.G.; Selvarajah, S.; Guérard, K.P.; Bartlett, J.M.S.; Lapointe, J.; Berman, D.M.; Okello, J.B.A.; Park, P.C. Reliability and performance of commercial RNA and DNA extraction kits for FFPE tissue cores. PLoS ONE 2017, 12, e0179732. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Overfield, C.J.; Edgar, M.A.; Wessell, D.E.; Wilke, B.K.; Garner, H.W. NTRK-rearranged spindle cell neoplasm of the lower extremity: Radiologic-pathologic correlation. Skelet. Radiol. 2022, 51, 1707–1713. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, M.; Huang, T.; Liao, W.; Xu, M.; Gu, J. GeneFuse: Detection and visualization of target gene fusions from DNA sequencing data. Int. J. Biol. Sci. 2018, 14, 843–848. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Dębski, K.J.; Dabrowski, M.; Czarnecka, A.M.; Szczylik, C. Gene set enrichment analysis and ingenuity pathway analysis of metastatic clear cell renal cell carcinoma cell line. Am. J. Physiol. Ren. Physiol. 2016, 311, F424–F436. [Google Scholar] [CrossRef]
- Giannos, P.; Kechagias, K.S.; Bowden, S.; Tabassum, N.; Paraskevaidi, M.; Kyrgiou, M. PCNA in Cervical Intraepithelial Neoplasia and Cervical Cancer: An Interaction Network Analysis of Differentially Expressed Genes. Front. Oncol. 2021, 11, 779042. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Yao, F.; Xiang, C.; Zhao, J.; Shang, Z.; Guo, L.; Ding, W.; Ma, S.; Yu, A.; Shao, J.; et al. Identification of NTRK gene fusions in lung adenocarcinomas in the Chinese population. J. Pathol. Clin. Res. 2021, 7, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Heydt, C.; Wölwer, C.B.; Velazquez Camacho, O.; Wagener-Ryczek, S.; Pappesch, R.; Siemanowski, J.; Rehker, J.; Haller, F.; Agaimy, A.; Worm, K.; et al. Detection of gene fusions using targeted next-generation sequencing: A comparative evaluation. BMC Med. Genom. 2021, 14, 62. [Google Scholar] [CrossRef]
- Rudzinski, E.R.; Lockwood, C.M.; Stohr, B.A.; Vargas, S.O.; Sheridan, R.; Black, J.O.; Rajaram, V.; Laetsch, T.W.; Davis, J.L. Pan-Trk Immunohistochemistry Identifies NTRK Rearrangements in Pediatric Mesenchymal Tumors. Am. J. Surg. Pathol. 2018, 42, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. Non-Small Cell Lung Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 497–530. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, N.K.D.; Lee, S.-H.; Kim, S.T.; Park, S.H.; Park, J.O.; Park, Y.S.; Lim, H.Y.; Kang, W.K.; Park, W.Y.; et al. NTRK gene amplification in patients with metastatic cancer. Precis. Future Med. 2017, 1, 129–137. [Google Scholar] [CrossRef][Green Version]
- Olmedo, M.E.; Cervera, R.; Cabezon-Gutierrez, L.; Lage, Y.; Corral de la Fuente, E.; Gomez Rueda, A.; Mielgo-Rubio, X.; Trujillo, J.C.; Counago, F. New horizons for uncommon mutations in non-small cell lung cancer: BRAF, KRAS, RET, MET, NTRK, HER2. World J. Clin. Oncol. 2022, 13, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Overbeck, T.R.; Reiffert, A.; Schmitz, K.; Rittmeyer, A.; Körber, W.; Hugo, S.; Schnalke, J.; Lukat, L.; Hugo, T.; Hinterthaner, M.; et al. NTRK Gene Fusions in Non-Small-Cell Lung Cancer: Real-World Screening Data of 1068 Unselected Patients. Cancers 2023, 15, 2966. [Google Scholar] [CrossRef] [PubMed]
- Kheder, E.S.; Hong, D.S. Emerging Targeted Therapy for Tumors with NTRK Fusion Proteins. Clin. Cancer Res. 2018, 24, 5807–5814. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, I.; Rachagani, S.; Hauke, R.; Krishn, S.R.; Paknikar, S.; Seshacharyulu, P.; Karmakar, S.; Nimmakayala, R.K.; Kaushik, G.; Johansson, S.L.; et al. MUC5AC interactions with integrin β4 enhances the migration of lung cancer cells through FAK signaling. Oncogene 2016, 35, 4112–4121. [Google Scholar] [CrossRef] [PubMed]
- Gjerstorff, M.F.; Pøhl, M.; Olsen, K.E.; Ditzel, H.J. Analysis of GAGE, NY-ESO-1 and SP17 cancer/testis antigen expression in early stage non-small cell lung carcinoma. BMC Cancer 2013, 13, 466. [Google Scholar] [CrossRef] [PubMed]
- Oshita, H.; Nishino, R.; Takano, A.; Fujitomo, T.; Aragaki, M.; Kato, T.; Akiyama, H.; Tsuchiya, E.; Kohno, N.; Nakamura, Y.; et al. RASEF is a novel diagnostic biomarker and a therapeutic target for lung cancer. Mol. Cancer Res. 2013, 11, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Maat, W.; Beiboer, S.H.; Jager, M.J.; Luyten, G.P.; Gruis, N.A.; van der Velden, P.A. Epigenetic regulation identifies RASEF as a tumor-suppressor gene in uveal melanoma. Invest. Ophthalmol. Vis. Sci. 2008, 49, 1291–1298. [Google Scholar] [CrossRef]
- Le, T.; Gerber, D.E. ALK alterations and inhibition in lung cancer. Semin. Cancer Biol. 2017, 42, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Dong, Y.; Tao, H.; Shi, L.; Tong, L.; Tang, J.; Zhang, S.; Liu, Z. ALK-rearranged squamous cell carcinoma of the lung. Thorac. Cancer 2021, 12, 1106–1114. [Google Scholar] [CrossRef]
- Song, Z.; Yu, X.; Zhang, Y. Clinicopathological characteristics and survival of ALK, ROS1 and RET rearrangements in non-adenocarcinoma non-small cell lung cancer patients. Cancer Biol. Ther. 2017, 18, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Murugan, S.; Lindberg, J.; Chellappa, V.; Shen, X.; Pawitan, Y.; Vu, T.N. Fusion Gene Detection Using Whole-Exome Sequencing Data in Cancer Patients. Front. Genet. 2022, 13, 820493. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.A.; Ely, H.A.; Shoemaker, R.; Boomer, A.; Culver, B.P.; Hoskins, I.; Haimes, J.D.; Walters, R.D.; Fernandez, D.; Stahl, J.A.; et al. Detecting Gene Rearrangements in Patient Populations Through a 2-Step Diagnostic Test Comprised of Rapid IHC Enrichment Followed by Sensitive Next-Generation Sequencing. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 513–523. [Google Scholar] [CrossRef] [PubMed]



| Clinical Correlation of NTRK | TRK IHC | |||
|---|---|---|---|---|
| Negative | Positive | p-Value | ||
| Cohort: 482 patients | 460/95.4% | 22/4.6% | ||
| Age | <70 | 258/53.5% | 12/2.4% | 0.887 |
| ≥70 | 202/41.9% | 10/2.1% | ||
| Sex | Male | 251/52% | 12/2.5% | 0.999 |
| Female | 209/43.4% | 10/2.1% | ||
| Histology | AdCa | 289/60% | 3/0.6% | 0.001 |
| SqCC | 143/30% | 16/3.3% | ||
| LCC | 28/5.8% | 3/0.6% | ||
| pT (TNM8) | pT1 | 170/35.3% | 5/1% | 0.175 |
| pT2–T4 | 290/60.2% | 17/3.5% | ||
| Stage (TNM8) | IA | 172/35.7% | 7/1.4% | 0.78 |
| IB | 183/38% | 8/1.7% | ||
| IIA | 46/9.5% | 3/0.6% | ||
| IIB | 51/10.6% | 4/0,8% | ||
| IIIA | 6/1.2% | 0/0% | ||
| IIIB | 0/0% | 0/0% | ||
| IV | 2/0.4% | 0/0% | ||
| Grade | G1 | 92/19.1% | 0/0% | 0.036 |
| G2 | 174/36.1% | 8/1.7% | ||
| G3 | 194/40.2% | 14/2.9% | ||
| Overall survival | Dead (10 years) | 338/70.1% | 16/3.3% | 0.938 |
| Alive (10 years) | 122/25.3% | 6/1.2% | ||
| Cases | Fusion Partner | Partner 1 | Partner 2 | Histology | NTRK-IHC | Other Somatic Mutations |
|---|---|---|---|---|---|---|
| Sample 1 | AFAP1–NTRK2 | Chr4 | Chr9 | Adca | +1 | TP53 *, PIK3CA |
| Sample 3 | ALK–KCNQ5 | Chr2 | Chr6 | SqCC | - | |
| Sample 4 | ALK–CTLC | Chr2 | Chr17 | Adca | - | |
| Sample 5 | ALK–HIP1 | Chr2 | Chr7 | SqCC | - | PTEN |
| Sample 6 | ALK–HIP1 | Chr2 | Chr7 | SqCC | - | |
| Sample 6 | ALK–KCNQ5 | Chr2 | Chr6 | SqCC | - | |
| Sample 7 | TPM3–NTRK1 | Chr1 | Chr1 | SqCC | +2 | |
| Sample 7 | ALK–VCL | Chr2 | Chr10 | SqCC | - | |
| Sample 8 | ALK–HIP1 | Chr2 | Chr7 | SqCC | - | APC |
| Sample 8 | ROS1–CEP85L | Chr6 | Chr6 | SqCC | - | |
| Sample 9 | PCM1–NRG1 | Chr8 | Chr8 | Adca | - | |
| Sample 10 | RASEF–NTRK2 | Chr9 | Chr9 | SqCC | +3 | |
| Sample 10 | MET–KIF5B | Chr7 | Chr10 | SqCC | - | |
| Sample 12 | RET–SPECC1L | Chr10 | Chr22 | SqCC | - | NTRK1, RET |
| Sample 13 | ROS1–GOPC | Chr6 | Chr6 | LCC | - | BRAF |
| Sample 13 | MET–CAPZA2 | Chr7 | Chr7 | LCC | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutierrez-Herrera, J.; Montero-Fernandez, M.A.; Kokaraki, G.; De Petris, L.; Maia Falcão, R.; Molina-Centelles, M.; Guijarro, R.; Ekman, S.; Ortiz-Villalón, C. NTRK Gene Expression in Non-Small-Cell Lung Cancer. J. Respir. 2025, 5, 2. https://doi.org/10.3390/jor5010002
Gutierrez-Herrera J, Montero-Fernandez MA, Kokaraki G, De Petris L, Maia Falcão R, Molina-Centelles M, Guijarro R, Ekman S, Ortiz-Villalón C. NTRK Gene Expression in Non-Small-Cell Lung Cancer. Journal of Respiration. 2025; 5(1):2. https://doi.org/10.3390/jor5010002
Chicago/Turabian StyleGutierrez-Herrera, Jair, M. Angeles Montero-Fernandez, Georgia Kokaraki, Luigi De Petris, Raul Maia Falcão, Manuel Molina-Centelles, Ricardo Guijarro, Simon Ekman, and Cristian Ortiz-Villalón. 2025. "NTRK Gene Expression in Non-Small-Cell Lung Cancer" Journal of Respiration 5, no. 1: 2. https://doi.org/10.3390/jor5010002
APA StyleGutierrez-Herrera, J., Montero-Fernandez, M. A., Kokaraki, G., De Petris, L., Maia Falcão, R., Molina-Centelles, M., Guijarro, R., Ekman, S., & Ortiz-Villalón, C. (2025). NTRK Gene Expression in Non-Small-Cell Lung Cancer. Journal of Respiration, 5(1), 2. https://doi.org/10.3390/jor5010002






