Abstract
Generally, predictive biomarker tests are clinically validated on histological formalin-fixed, paraffin-embedded (FFPE) samples. In addition to FFPE samples, cytological samples have also emerged as a useful approach to detect predictive biomarkers. However, as of today, despite the promising results reported in the recent literature, their full implementation in routine clinical practice is still lagging owing to a lack of standardized preparatory protocols, challenging assessments of cyto-histological correlation, and variable inter-observer agreement. The aim of this report was to explore the possibility of implementing a large-scale validation of predictive biomarker testing on cytological material. To this aim, we evaluated the technical feasibility of PD-L1 assessment on a cell block (CB)-derived tissue microarray (cbTMA). Consecutive and unselected CBs prepared from metastatic lymph node fine-needle cytology (FNC) samples were retrospectively collected and used for TMA construction. PD-L1 immunohistochemistry (IHC) was carried out on cbTMA sections with the companion diagnostic kit SP263 assay. TMA contained 33 CB-derived cores. A total of 20 sections were hematoxylin and eosin (H&E) stained. Overall, 29 (88%) samples were visible at least in one H&E-stained slide. Four cases out of five sections stained with the SP263 assay (4/29, 13.8%) showed PD-L1 positivity in neoplastic and/or immune cells; remarkably, no unspecific background was observed. Although our study was based on a limited and non-selected series, our findings do provide proof of concept for the use of cbTMA in predictive biomarker testing on cytological material in large-scale post-clinical trial validation studies, multicenter studies, and quality control programs.
1. Introduction
In the last decade, molecular cytopathology has emerged as a very rapidly evolving field of predictive pathology, with an increasing number of molecular tests performed on a wide range of cytological preparations [1,2]. Indeed, cytology is a first-line diagnostic procedure in many neoplastic settings and, in advanced-stage cancer patients, cytological samples may be the only material available for both diagnosis and molecular biomarker testing for targeted therapies.
Methodologically, the high quality of nucleic acids extracted from cytological specimens and the steady upgrading of multiplexed, highly sensitive molecular assays with minimal nucleic acid input have all contributed to the ongoing development of molecular cytopathology [3,4,5,6]. Despite such great advances in the field and the many advantages of cytological material, especially in hard-to-reach tumors, histological formalin-fixed and paraffin-embedded (FFPE) samples continue to prevail over cytological samples for the selection of targeted treatments. As of today, the persistent paucity of cytological testing in routine pathology practice is largely due to a few unmet analytical challenges affecting the whole gamut of the analytical phases. Among these are a lack of homogeneous tissue handling and processing protocols, discrepant cyto-histological correlation, and interpretation of results. Indeed, unless these challenges are met through the careful validation of predictive biomarker testing on cytological material, this excellent resource will most likely continue to remain underused.
As immunotherapy has emerged as one of the most promising cancer treatments, the process of technical and clinical validation of predictive biomarkers for immune-checkpoint inhibitors on cytological specimens is plainly foreseeable [7,8,9,10,11,12]. Not surprisingly, the last couple of years have witnessed a plethora of studies evaluating the feasibility of assessing PD-L1 expression on cytological samples, particularly in terms of adequacy rate, level of PD-L1 expression, and clinical outcomes [8,9,13,14]. Building on such compelling research and going beyond PD-L1, our research team and others have also very recently demonstrated the technical feasibility of assessing tumor mutational burden (TMB) and mismatch repair deficiency (dMMR) by immunohistochemistry (IHC) testing [11,12,13,14] on cytology material processed as cell blocks (CBs), as evidenced by a high concordance rate between cytological and surgical specimens [8,9].
However, despite being globally massive, these data were mostly generated from limited series analysis. Thus, to fulfill the unmet need for a large-scale validation of predictive biomarker testing on cytological material and, consequently, to overcome the underutilization of cytological samples for targeted treatments, we evaluated the technical feasibility of using CB-derived tissue microarrays (cbTMAs) to evaluate PD-L1 expression in various types of tumor-derived cytological samples.
2. Materials and Methods
2.1. Samples
We retrospectively reviewed consecutive and unselected CBs prepared from fine-needle cytology (FNC) samples of metastatic lymph nodes; FNAs were performed at the Cytopathology Division of the University of Naples “Federico II”. All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Declaration of Helsinki.
CBs were prepared using the Shandon Cytoblock Cell Block Preparation System (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions, as previously described [15]. The original hematoxylin and eosin (H&E)-stained slides were examined by two different pathologists to outline the area of interest with respect to representative cellularity. Generally, a sample is considered eligible for PD-L1 evaluation if it contains a minimum of 100 neoplastic cells. Thus, a low-power microscopic field (10×, LPF) with at least 25–50 cells was deemed satisfactory. After screening of the whole section at 10× for at least 10 LPF, the donor CB cellularity was evaluated as follows: highly cellular (>50% of satisfactory fields); moderately cellular (25–50% of satisfactory fields); poorly cellular (5–25% of satisfactory fields); acellular.
2.2. TMA Construction
The construction of TMA was performed with the fully automated Epredia TMA Grand Master (3DHistech). The TMA control software (TMA Grand Master package) was used to overlap an annotated digital slide with the donor block. According to the digital annotations, one core 2 mm in diameter was automatically punched out from a donor block by the TMA machine and then relocated to a recipient block in a precise alignment. TMA data were automatically archived and stored in a specific file. Subsequently, 4 μm sections were cut from the constructed TMA blocks, H&E-stained, and viewed for quality and cellularity control purposes.
2.3. PD-L1 Immunohistochemistry
PD-L1 IHC was carried out on cbTMA sections with the companion diagnostic kit SP263 assay (Ventana, Tucson, Arizona) on the Ventana’s BenchMark XT platform, following the manufacturer’s instructions. To evaluate non-adjacent cells, non-consecutive sections were immunostained. PD-L1 staining of variable intensity, perceptible at a maximum of 20× magnification, was evaluated by two experienced pathologists in both neoplastic and immune cells. As in routine practice, any partial or complete linear membrane staining that was perceived as distinct from cytoplasmic staining was assessed in neoplastic cells, whereas membrane and/or cytoplasmic staining were assessed in immune cells [16].
3. Results
3.1. Samples
Overall, 33 CBs were collected. Lymph node specimens were derived from cervical (n = 13), axillary (n = 6), mandibular (n = 1), supraclavicular (n = 4), abdominal (n = 4), pectoral (n = 4), and inguinal (n = 1) areas. Cytological diagnoses included metastasis from squamous cell carcinoma (n = 8), breast carcinoma (n = 7), colon carcinoma (n = 4), carcinoma not otherwise specified, NOS (n = 4), carcinoma of gastrointestinal origin (n = 2), pancreatic carcinoma (n = 2), papillary thyroid carcinoma (n = 2), ovarian serous carcinoma (n = 2), medullary thyroid carcinoma (n = 1), and urothelial carcinoma (n = 1). As for the cellularity of CBs, 8 (24.24%) cases were highly cellular, 11 (33.33%) moderately cellular, and 14 (42.42%) were poorly cellular; no acellular CBs were observed.
3.2. TMA
TMA contained 33 CB-derived cores; donor core height ranged from 4.792 to 6.11 mm. A total of 20 sections were H&E-stained. Overall, 29 (88%) samples were visible at least in one H&E-stained slide. Core cellularity content was highly cellular in 4 out of 29 cases (13.8%), moderately cellular in 9 cases (31%), poorly cellular in 11 cases (37.9%), and acellular in 5 (17.3%). Four cases (12%) totally failed to adhere during slide preparation (Figure 1).
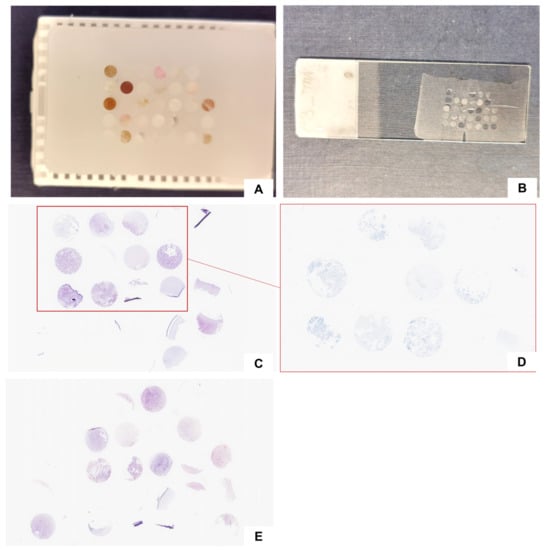
Figure 1.
Cell-block-derived TMA construction: TMA paraffin block (A) and an unstained section (B); two representative hematoxylin and eosin-stained sections (section numbers 5 (C) and 20 (E)) and the corresponding PD-L1-stained section (D).
4. PD-L1 Immunohistochemistry
Overall, five sections were stained with the companion diagnostic kit SP263 assay. Four cases (4/29, 13.8%) showed PD-L1 positivity; in particular, case ID 5 showed positivity in neoplastic component, cases ID 8 and ID 31 in immune cells, and case ID 25 in both neoplastic and immune cells (Figure 2). The positive signal was clean, and no unspecific background was observed. The characteristics of each sample are summarized in Table 1.
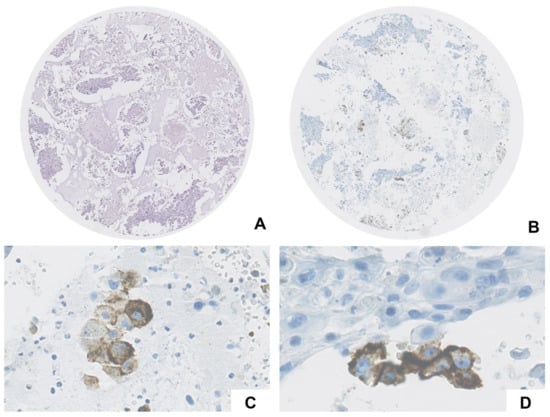
Figure 2.
Case ID 25: metastasis from squamous cell carcinoma (A,B) showed positivity in both immune (cytoplasmic granular staining in macrophages, (C)) and neoplastic cells (complete linear membrane staining, (D)).

Table 1.
Sample characteristics.
5. Discussion
In clinical settings in which FFPE samples are available, TMAs are widely used for the multiplex analysis and validation of tumor markers on histological material. However, this approach is hardly ever applied to cytological specimens [17,18,19]. Early experiences carried out exclusively on fluid cytology demonstrated that TMAs are also efficiently applicable to CB specimens. Indeed, these early studies demonstrated that TMA is highly feasible, especially in samples with higher cellularity [16], and that cbTMAs can accurately display the same cellularity, composition of cells, staining patterns, and intensity of the donor CB [18]. In line with this early research, a recent study which compared the efficiency of conventional smears with cell-block preparations demonstrated that IHC, in combinations with cytology microarrays (CMAs) prepared from CBs, was able to confirm and categorize malignant cells in body fluids as accurately as conventional smears [19].
To the best of our knowledge, no prior experiences have explored the potential use of cbTMA in the validation of predictive biomarkers on cytological specimens. From a technical point of view, our data confirm the suitability of using CB material for TMA construction. As previously reported, to exploit the full potential of this cost- and time-effective technique, cytopathologists should make sure to prevent the loss of material during H&E staining and core fall-off during IHC heating procedures [17]. Although preliminary, our study did show some very promising results. For instance, 88% of samples were visible in at least one H&E- and PD-L1-stained slide. Notably, the TMA, prepared from consecutive and unselected CBs, showed a high degree of cellular variability, reflecting a core height ranging from 4.792 to 6.11 mm. On the other hand, cbTMA displayed a higher percentage of poorly cellular and acellular cores compared to the donor CB cellularity. Moreover, core alignment was difficult to obtain. Indeed, we hypothesize that selecting a homogeneous series of highly cellular CBs could yield better outcomes.
We also evaluated the technical feasibility of PD-L1 assessment on cell block (CB)-derived tissue microarray. In this regard, we observed positive neoplastic and/or immune cells in four cases (13.8%), including three metastases from squamous cell carcinomas and one metastasis from breast carcinoma. We speculate that the low percentage of PD-L1-positive cases was likely due to the use of unselected tumor types. Given the small number of analyzed samples, we were not able to evaluate the concordance rate between PD-L1 expression in TMA cores with that in paired donor CBs, thereby warranting further investigation. However, we were able to observe a clean positive signal showing no non-specific background staining.
In addition to being useful for IHC predictive biomarker testing, we speculate that cbTMA may also be suitable for molecular testing. Indeed, research has shown that punches taken from donor blocks can be re-punched directly into tubes and used for molecular analysis using PCR-based approaches. By doing so, cytopathologists could have the means not only to compare IHC with molecular data in large-scale analyses, but also to investigate rare subpopulations identified during preliminary cbTMA analysis [20,21].
In conclusion, although our study was based on a limited and non-selected series, it does provide a proof of concept that cbTMA can be used for biomarker testing validation on cytological material (Figure 2). Furthermore, the possible application of this novel technique in the framework of large-scale post-clinical trials validation studies, multicenter studies, and quality-control programs clearly suggests the huge and yet unexplored potential of cytological material in the “multiverse” of predictive pathology (Figure 3).
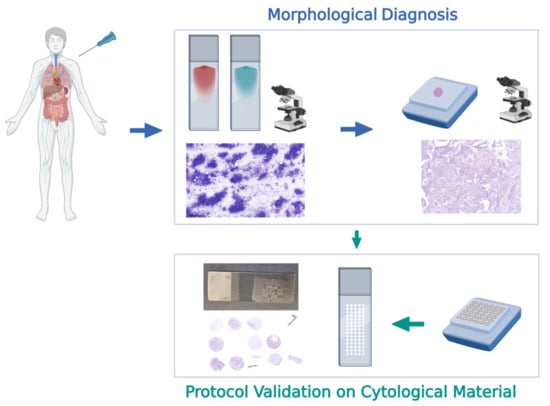
Figure 3.
Flow chart of a novel approach for biomarker testing validation on cytological material, based on the use of cell-block-derived tissue microarray (cbTMA).
Author Contributions
Conceptualization, U.M., G.T. and E.V.; methodology, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; software, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; validation, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; formal Analysis, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; investigation, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; data curation, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; writing—original draft preparation, G.A. and E.V.; writing—review and editing, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; visualization, A.I., G.A., P.P., U.M., C.B., G.T. and E.V.; supervision, U.M., G.T. and E.V.; project administration, U.M., G.T. and E.V.; funding acquisition, G.T. All authors have read and agreed to the published version of the manuscript.
Funding
1. Monitoraggio ambientale, studio ed approfondimento della salute della popolazione residente in aree a rischio—In attuazione della D.G.R. Campania n.180/2019 to G.T. 2. POR Campania FESR 2014–2020 Progetto “Sviluppo di Approcci Terapeutici Innovativi per patologie Neoplastiche resistenti ai trattamenti—SATIN” to G.T.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and due to the retrospective nature does not require approval by the Institutional Review Board (or Ethics Committee).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We thank Paola Merolla for editing the manuscript.
Conflicts of Interest
Umberto Malapelle has received personal fees (as consultant and/or speaker bureau) from Boehringer Ingelheim, Roche, MSD, Amgen, Thermo Fisher Scientific, Eli Lilly, Diaceutics, GSK, Merck, and AstraZeneca, unrelated to the current work. Giancarlo Troncone reports personal fees (as speaker bureau or advisor) from Roche, MSD, Pfizer, Boehringer Ingelheim, Eli Lilly, BMS, GSK, Menarini, AstraZeneca, Amgen, and Bayer, unrelated to the current work. Elena Vigliar has received personal fees (as consultant and/or speaker bureau) from Diaceutics, unrelated to the current work. The other authors have nothing to disclose.
References
- Troncone, G.; Roy-Chowdhuri, S. Modern cytopathology: An evolving field. Cytopathology 2021, 32, 560–561. [Google Scholar] [CrossRef] [PubMed]
- Angerilli, V.; Galuppini, F.; Pagni, F.; Fusco, N.; Malapelle, U.; Fassan, M. The role of the pathologist in the next-generation era of tumor molecular characterization. Diagnostics 2021, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Pepe, F.; Sgariglia, R.; Nacchio, M.; Russo, G.; Conticelli, F.; Girolami, I.; Eccher, A.; Bellevicine, C.; Vigliar, E.; et al. Next generation sequencing in cytology. Cytopathology 2021, 32, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Wei, S. Overview of molecular testing of cytology specimens. Acta Cytol. 2020, 64, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Spatz, A. Making cytology specimens solid materials for testing predictive marker of immunotherapy in NSCLC. Oncotarget 2018, 9, 35472. [Google Scholar] [CrossRef] [PubMed]
- Bellevicine, C.; Troncone, G. The cytopathologist’s expanding role in the 2018 updated molecular testing guidelines for lung cancer. Cancer Cytopathol. 2018, 126, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Tsao, M.S.; Kerr, K.M.; Kockx, M.; Beasley, M.-B.; Borczuk, A.C.; Botling, J.; Bubendorf, L.; Chirieac, L.; Chen, G.; Chou, T.-Y.; et al. PD-L1 Immunohistochemistry comparability study in real-life clinical samples: Results of blueprint phase 2 project. J. Thorac. Oncol. 2018, 13, 1302–1311. [Google Scholar] [CrossRef] [PubMed]
- Gosney, J.R.; Boothman, A.M.; Ratcliffe, M.; Kerr, K.M. Cytology for PD-L1 testing: A systematic review. Lung Cancer 2020, 141, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Iaccarino, A.; Salatiello, M.; Migliatico, I.; De Luca, C.; Gragnano, G.; Russo, M.; Bellevicine, C.; Malapelle, U.; Troncone, G.; Vigliar, E. PD-L1 and beyond: Immuno-oncology in cytopathology. Cytopathology 2021, 32, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Alborelli, I.; Bratic Hench, I.; Chijioke, O.; Savic Prince, S.; Bubendorf, L.; Leuenberger, L.P.; Tolnay, M.; Leonards, K.; Quagliata, L.; Jermann, P.; et al. Robust assessment of tumor mutational burden in cytological specimens from lung cancer patients. Lung Cancer 2020, 149, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Pepe, F.; Pisapia, P.; Gristina, V.; Rocco, D.; Micheli, M.; Iaccarino, A.; Tufano, R.; Gragnano, G.; Russo, G.; De Luca, C.; et al. Tumor mutational burden cytological samples: A pilot study. Cancer Cytopathol. 2021, 129, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, E.M.; Landon, G.; Broaddus, R.R.; Roy-Chowdhuri, S. Evaluating mismatch repair/microsatellite instability status using cytology effusion specimens to determine eligibility for immunotherapy. Arch Pathol. Lab. Med. 2021, 145, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Roy-Chowdhuri, S. Immunocytochemistry of cytology specimens for predictive biomarkers in lung cancer. Transl. Lung Cancer Res. 2020, 9, 898–905. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Nambirajan, A.; Borczuk, A.; Chen, G.; Minami, Y.; Moreira, A.L.; Motoi, N.; Papotti, M.; Rekhtman, N.; Russell, P.A.; et al. Immunocytochemistry for predictive biomarker testing in lung cancer cytology. Cancer Cytopathol. 2019, 127, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Vigliar, E.; Iaccarino, A.; Campione, S.; Campanino, M.R.; Clery, E.; Pisapia, P.; De Luca, C.; Bellevicine, C.; Malapelle, U.; De Dominicis, G.; et al. PD-L1 expression in cell-blocks of non-small cell lung cancer: The impact of prolonged fixation. Diagn Cytopathol. 2020, 48, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Ventana PD-L1 (SP263) Assay. Available online: https://www.rochebiomarkers.be/content/media/Files/PD-L1_SP263_package_insert.pdf (accessed on 7 January 2022).
- Wen, C.H.; Su, Y.C.; Wang, S.L.; Yang, S.F.; Chai, C.Y. Application of the microarray technique to cell blocks. Acta Cytol. 2007, 51, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Pu, R.T.; Giordano, T.J.; Michael, C.W. Utility of cytology microarray constructed from effusion cell blocks for immunomarker validation. Cancer 2008, 114, 300–306. [Google Scholar] [CrossRef][Green Version]
- Bandyopadhyay, A.; Bhattacharyya, S.; Roy, S.; Majumdar, K.; Bose, K.; Boler, A.K. Cytology microarray on cell block preparation: A novel diagnostic approach in fluid cytology. J. Cytol. 2019, 36, 79–83. [Google Scholar]
- Lacombe, A.; Carafa, V.; Schneider, S.; Sticker-Jantscheff, M.; Tornillo, L.; Eppenberger-Castori, S. Re-punching tissue microarrays is possible: Why can this be useful and how to do it. Microarrays 2015, 4, 245–254. [Google Scholar] [CrossRef]
- Vassella, E.; Galván, J.A.; Zlobec, I. Tissue microarray technology for molecular applications: Investigation of cross-contamination between tissue samples obtained from the same punching device. Microarrays 2015, 4, 188–195. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).