Geospatial Data and Google Street View Images for Monitoring Kudzu Vines in Small and Dispersed Areas
Abstract
1. Introduction
1.1. CNNs Applied to Identification of Invasive Species
1.2. Object Detection Using YOLO (You Only Look Once) and YOLOv8
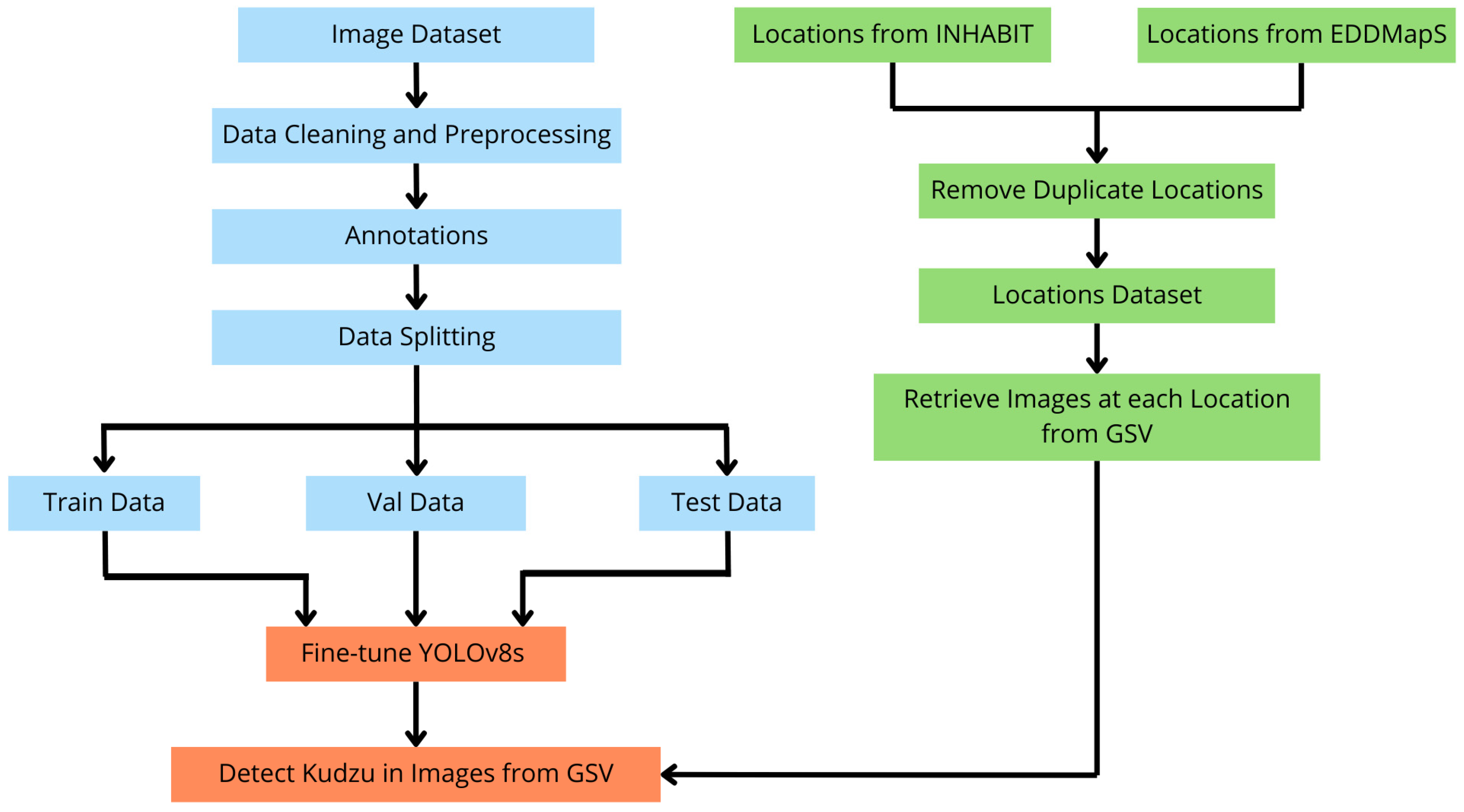
2. Materials and Methods
2.1. Definitions for Training, Validation, and Testing Kudzu Images
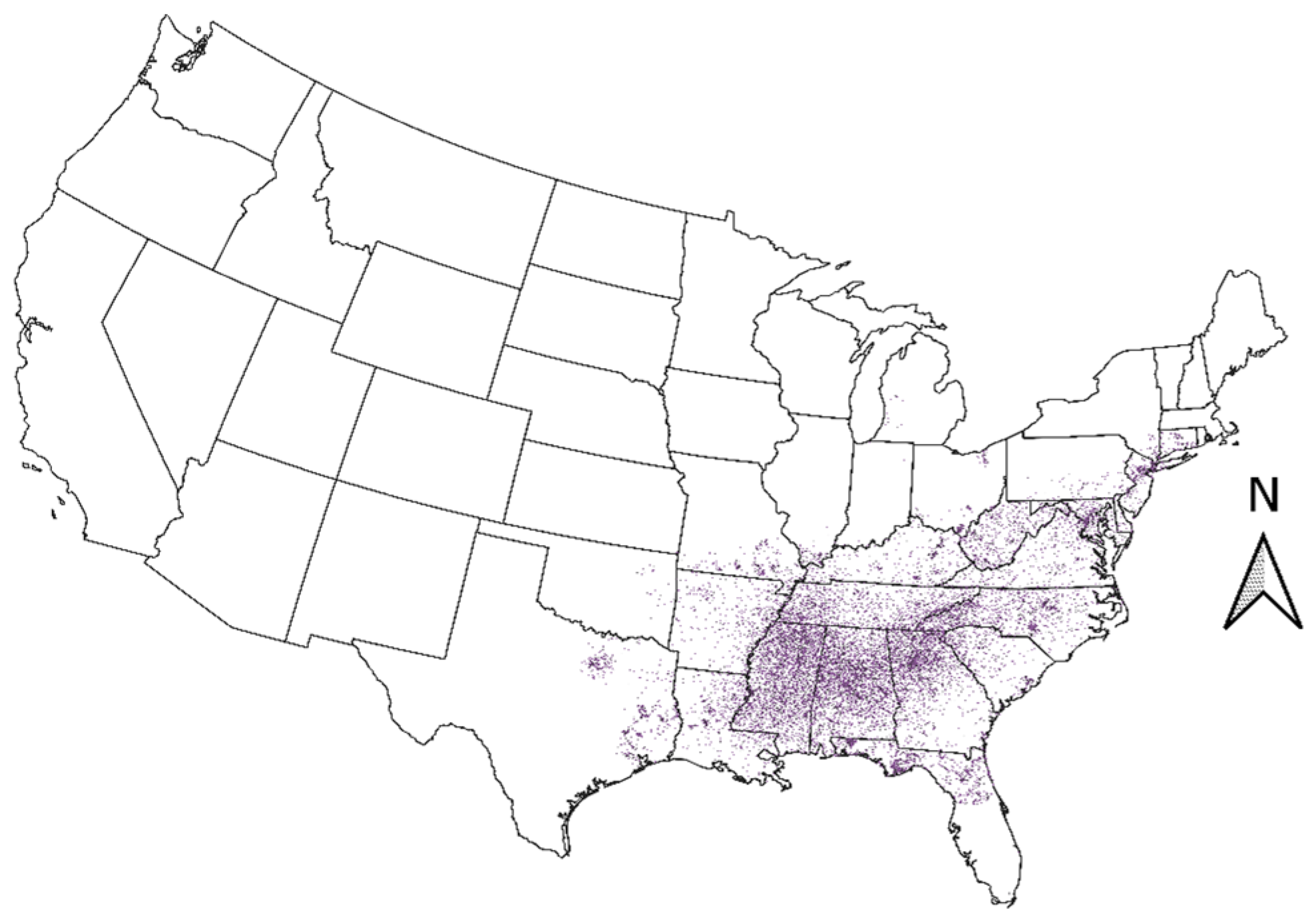
2.2. Geospatial Data
Google Street View (GSV) Images
2.3. YOLOv8 for Kudzu Detection
2.4. Metrics of Evaluation of YOLOv8s Algorithm
2.5. Code Implementation
3. Results
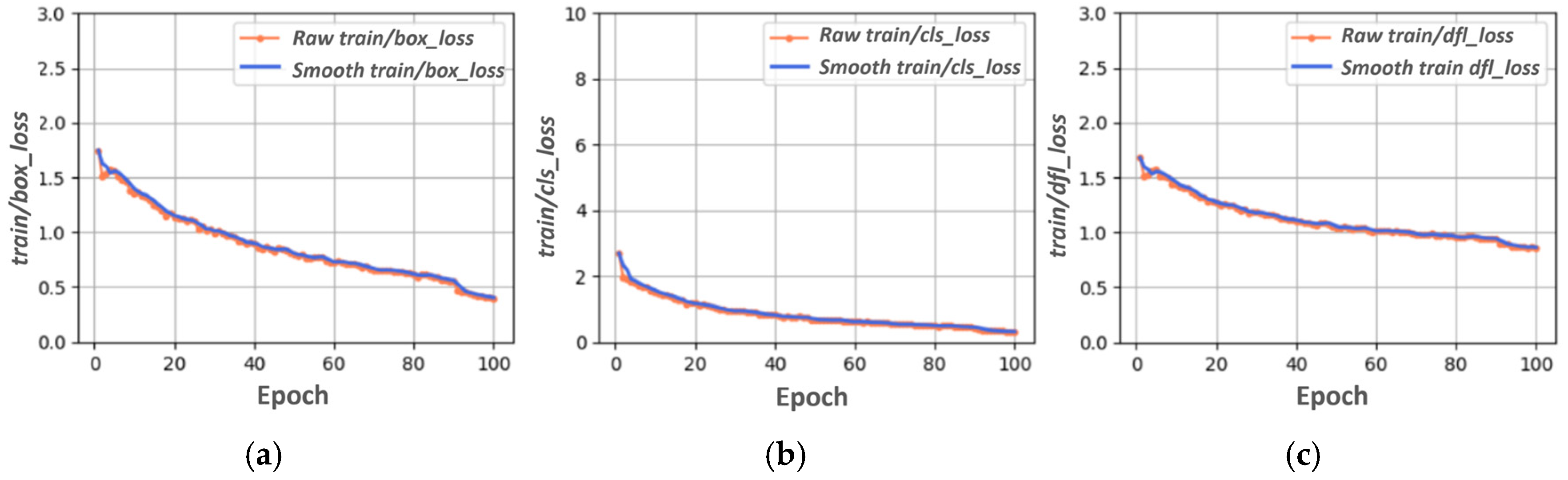
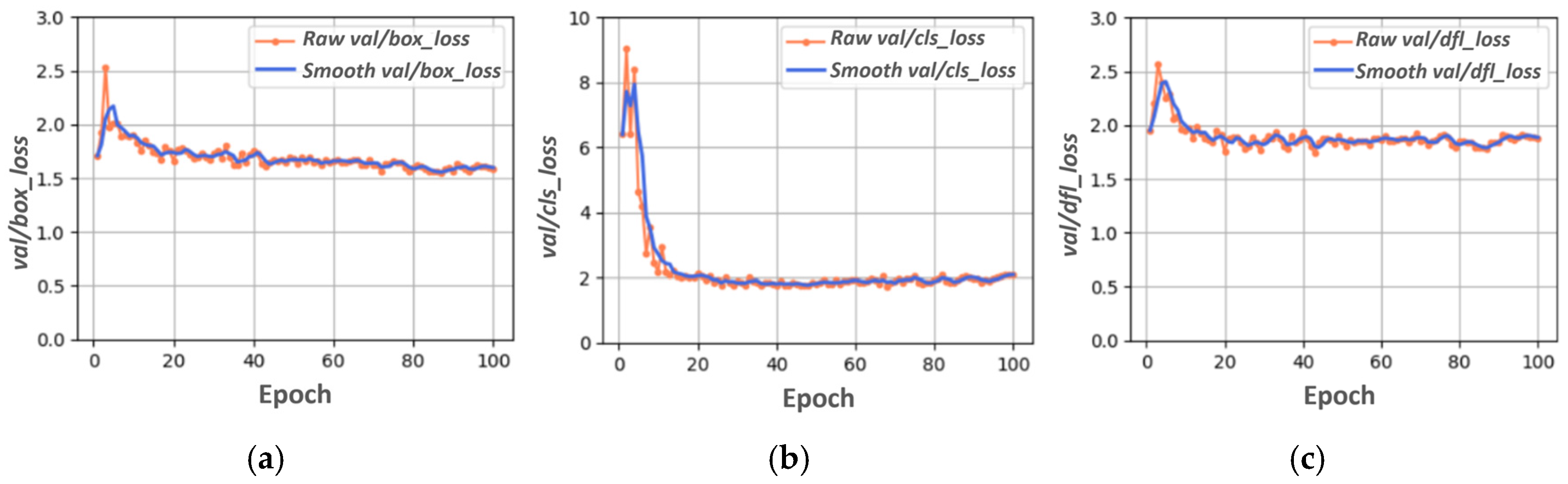
3.1. YOLOV8s Training, Validation, and Testing
3.2. Geospatial Data for Kudzu in the Conterminous United States
Kudzu Image Dataset Retrieval
3.3. YOLOv8s Applied to GSV Images
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| UAV | Unmanned Aerial Vehicle |
| SDM | Species distribution modeling |
| ENM | Ecological niche model |
| AVIRIS | Airborne Visible/Infrared Imaging Spectrometer |
| AOI | Area Of interest |
| USGS | United States Geological Survey |
| GSV | Google Street View |
| API | Application programming interface |
| CNN | Convolutional neural network |
| YOLO | You Only Look Once |
| ReLU | Rectified Linear Unit |
| LiDAR | Light Detection and Ranging |
| SSD | Single Shot MultiBox Detector |
| HOG | Histogram of oriented gradients |
| SIFT | Scale-invariant feature transform |
| CSP | Cross Stage Partial |
| GPS | Global Positioning System |
| AI | Artificial Intelligence |
| DL | Deep learning |
| DFL | Distribution focal loss |
| IoU | Intersection over Union |
| BCE | Binary Cross-Entropy |
| NMS | Non-maximum suppression |
| mAP | Mean average precision |
| AP | Average precision |
Appendix A
import math import random from copy import copy import numpy as np import torch.nn as nn from ultralytics.data import build_dataloader, build_yolo_dataset from ultralytics.engine.trainer import BaseTrainer from ultralytics.models import yolo from ultralytics.nn.tasks import DetectionModel from ultralytics.utils import LOGGER, RANK from ultralytics.utils.plotting import plot_images, plot_labels, plot_results from ultralytics.utils.torch_utils import de_parallel, torch_distributed_zero_first
class DetectionTrainer(BaseTrainer):
"""
A class extending the BaseTrainer class for training based on a detection model.
This trainer specializes in object detection tasks, handling the specific requirements for training YOLO models
for object detection.
Attributes:
model (DetectionModel): The YOLO detection model being trained.
data (dict): Dictionary containing dataset information including class names and number of classes.
loss_names (Tuple[str]): Names of the loss components used in training (box_loss, cls_loss, dfl_loss).
Methods:
build_dataset: Build YOLO dataset for training or validation.
get_dataloader: Construct and return dataloader for the specified mode.
preprocess_batch: Preprocess a batch of images by scaling and converting to float.
set_model_attributes: Set model attributes based on dataset information.
get_model: Return a YOLO detection model.
get_validator: Return a validator for model evaluation.
label_loss_items: Return a loss dictionary with labeled training loss items.
progress_string: Return a formatted string of training progress.
plot_training_samples: Plot training samples with their annotations.
plot_metrics: Plot metrics from a CSV file.
plot_training_labels: Create a labeled training plot of the YOLO model.
auto_batch: Calculate optimal batch size based on model memory requirements.
Examples:
>>> from ultralytics.models.yolo.detect import DetectionTrainer
>>> args = dict(model="yolo11n.pt", data="coco8.yaml", epochs=3)
>>> trainer = DetectionTrainer(overrides=args)
>>> trainer.train()
"""
def build_dataset(self, img_path, mode="train", batch=None):
"""
Build YOLO Dataset for training or validation.
Args:
img_path (str): Path to the folder containing images.
mode (str): `train` mode or `val` mode, users are able to customize different augmentations for each mode.
batch (int, optional): Size of batches, this is for `rect`.
Returns:
(Dataset): YOLO dataset object configured for the specified mode.
"""
gs = max(int(de_parallel(self.model).stride.max() if self.model else 0), 32)
return build_yolo_dataset(self.args, img_path, batch, self.data, mode=mode, rect=mode == "val", stride=gs)
def get_dataloader(self, dataset_path, batch_size=16, rank=0, mode="train"):
"""
Construct and return dataloader for the specified mode.
Args:
dataset_path (str): Path to the dataset.
batch_size (int): Number of images per batch.
rank (int): Process rank for distributed training.
mode (str): 'train' for training dataloader, 'val' for validation dataloader.
Returns:
(DataLoader): PyTorch dataloader object.
"""
assert mode in {"train", "val"}, f"Mode must be 'train' or 'val', not {mode}."
with torch_distributed_zero_first(rank): # init dataset *.cache only once if DDP
dataset = self.build_dataset(dataset_path, mode, batch_size)
shuffle = mode == "train"
if getattr(dataset, "rect", False) and shuffle:
LOGGER.warning("WARNING ⚠ 'rect=True' is incompatible with DataLoader shuffle, setting shuffle=False")
shuffle = False
workers = self.args.workers if mode == "train" else self.args.workers * 2
return build_dataloader(dataset, batch_size, workers, shuffle, rank) # return dataloader
def preprocess_batch(self, batch):
"""
Preprocess a batch of images by scaling and converting to float.
Args:
batch (dict): Dictionary containing batch data with 'img' tensor.
Returns:
(dict): Preprocessed batch with normalized images.
"""
batch["img"] = batch["img"].to(self.device, non_blocking=True).float() / 255
if self.args.multi_scale:
imgs = batch["img"]
sz = (
random.randrange(int(self.args.imgsz * 0.5), int(self.args.imgsz * 1.5 + self.stride))
// self.stride
* self.stride
) # size
sf = sz / max(imgs.shape[2:]) # scale factor
if sf != 1:
ns = [
math.ceil(x * sf / self.stride) * self.stride for x in imgs.shape[2:]
] # new shape (stretched to gs-multiple)
imgs = nn.functional.interpolate(imgs, size=ns, mode="bilinear", align_corners=False)
batch["img"] = imgs
return batch
def set_model_attributes(self):
"""Set model attributes based on dataset information."""
# Nl = de_parallel(self.model).model[-1].nl # number of detection layers (to scale hyps)
# self.args.box *= 3 / nl # scale to layers
# self.args.cls *= self.data["nc"] / 80 * 3 / nl # scale to classes and layers
# self.args.cls *= (self.args.imgsz / 640) ** 2 * 3 / nl # scale to image size and layers
self.model.nc = self.data["nc"] # attach number of classes to model
self.model.names = self.data["names"] # attach class names to model
self.model.args = self.args # attach hyperparameters to model
# TODO: self.model.class_weights = labels_to_class_weights(dataset.labels, nc).to(device) * nc
def get_model(self, cfg=None, weights=None, verbose=True):
"""
Return a YOLO detection model.
Args:
cfg (str, optional): Path to model configuration file.
weights (str, optional): Path to model weights.
verbose (bool): Whether to display model information.
Returns:
(DetectionModel): YOLO detection model.
"""
model = DetectionModel(cfg, nc=self.data["nc"], verbose=verbose and RANK == -1)
if weights:
model.load(weights)
return model
def get_validator(self):
"""Return a DetectionValidator for YOLO model validation."""
self.loss_names = "box_loss", "cls_loss", "dfl_loss"
return yolo.detect.DetectionValidator(
self.test_loader, save_dir=self.save_dir, args=copy(self.args), _callbacks=self.callbacks
)
def label_loss_items(self, loss_items=None, prefix="train"):
"""
Return a loss dict with labeled training loss items tensor.
Args:
loss_items (List[float], optional): List of loss values.
prefix (str): Prefix for keys in the returned dictionary.
Returns:
(Dict | List): Dictionary of labeled loss items if loss_items is provided, otherwise list of keys.
"""
keys = [f"{prefix}/{x}" for x in self.loss_names]
if loss_items is not None:
loss_items = [round(float(x), 5) for x in loss_items] # convert tensors to 5 decimal place floats
return dict(zip(keys, loss_items))
else:
return keys
def progress_string(self):
"""Return a formatted string of training progress with epoch, GPU memory, loss, instances and size."""
return ("\n" + "%11s" * (4 + len(self.loss_names))) % (
"Epoch",
"GPU_mem",
*self.loss_names,
"Instances",
"Size",
)
def plot_training_samples(self, batch, ni):
"""
Plot training samples with their annotations.
Args:
batch (dict): Dictionary containing batch data.
ni (int): Number of iterations.
"""
plot_images(
images=batch["img"],
batch_idx=batch["batch_idx"],
cls=batch["cls"].squeeze(-1),
bboxes=batch["bboxes"],
paths=batch["im_file"],
fname=self.save_dir / f"train_batch{ni}.jpg",
on_plot=self.on_plot,
)
def plot_metrics(self):
"""Plot metrics from a CSV file."""
plot_results(file=self.csv, on_plot=self.on_plot) # save results.png
def plot_training_labels(self):
"""Create a labeled training plot of the YOLO model."""
boxes = np.concatenate([lb["bboxes"] for lb in self.train_loader.dataset.labels], 0)
cls = np.concatenate([lb["cls"] for lb in self.train_loader.dataset.labels], 0)
plot_labels(boxes, cls.squeeze(), names=self.data["names"], save_dir=self.save_dir, on_plot=self.on_plot)
def auto_batch(self):
"""
Get optimal batch size by calculating memory occupation of model.
Returns:
(int): Optimal batch size.
"""
train_dataset = self.build_dataset(self.trainset, mode="train", batch=16)
max_num_obj = max(len(label["cls"]) for label in train_dataset.labels) * 4 # 4 for mosaic augmentation
return super().auto_batch(max_num_obj)
Appendix B

References
- Kohli, R.K. Alien—Plant Invasion in the Anthropocene How Does an Alien Plant Become. Anthr. Sci. 2024, 2, 177–179. [Google Scholar] [CrossRef]
- Tonini, F.; Shoemaker, D.; Petrasova, A.; Harmon, B.; Petras, V.; Cobb, R.C.; Mitasova, H.; Meentemeyer, R.K. Tangible geospatial modeling for collaborative solutions to invasive species management. Environ. Model. Softw. 2017, 92, 176–188. [Google Scholar] [CrossRef]
- Lee, J.G.; Kang, M. Geospatial Big Data: Challenges and Opportunities. Big Data Res. 2015, 2, 74–81. [Google Scholar] [CrossRef]
- Young, N.E.; Jarnevich, C.S.; Sofaer, H.R.; Pearse, I.; Sullivan, J.; Engelstad, P.; Stohlgren, T.J. A modeling workflow that balances automation and human intervention to inform invasive plant management decisions at multiple spatial scales. PLoS ONE 2020, 15, e0229253. [Google Scholar] [CrossRef]
- Valavi, R.; Guillera-Arroita, G.; Lahoz-Monfort, J.J.; Elith, J. Predictive performance of presence-only species distribution models: A benchmark study with reproducible code. Ecol. Monogr. 2022, 92, e01486. [Google Scholar] [CrossRef]
- Callen, S.T.; Miller, A.J. Signatures of niche conservatism and niche shift in the North American kudzu (Pueraria montana) invasion. Divers. Distrib. 2015, 21, 853–863. [Google Scholar] [CrossRef]
- Bradley, B.A.; Wilcove, D.S.; Oppenheimer, M. Climate change increases risk of plant invasion in the Eastern United States. Biol. Invasions 2010, 12, 1855–1872. [Google Scholar] [CrossRef]
- Evans, A.E.; Jarnevich, C.S.; Beaury, E.M.; Engelstad, P.S.; Teich, N.B.; LaRoe, J.M.; Bradley, B.A. Shifting hotspots: Climate change projected to drive contractions and expansions of invasive plant abundance habitats. Divers. Distrib. 2024, 30, 41–54. [Google Scholar] [CrossRef]
- Hoffberg, S.L.; Mauricio, R. The persistence of invasive populations of kudzu near the northern periphery of its range in New York City determined from historical data. J. Torrey Bot. Soc. 2016, 143, 437–442. [Google Scholar] [CrossRef]
- Jensen, T.; Hass, F.S.; Akbar, M.S.; Petersen, P.H.; Arsanjani, J.J. Employing machine learning for detection of invasive species using sentinel-2 and aviris data: The case of Kudzu in the United States. Sustainability 2020, 12, 3544. [Google Scholar] [CrossRef]
- Liang, W.; Abidi, M.; Carrasco, L.; McNelis, J.; Tran, L.; Li, Y.; Grant, J. Mapping vegetation at species level with high-resolution multispectral and lidar data over a large spatial area: A case study with Kudzu. Remote Sens. 2020, 12, 609. [Google Scholar] [CrossRef]
- Meyer, M.d.F.; Gonçalves, J.A.; Cunha, J.F.R.; Ramos, S.C.d.C.e.S.; Bio, A.M.F. Application of a Multispectral UAS to Assess the Cover and Biomass of the Invasive Dune Species Carpobrotus edulis. Remote Sens. 2023, 15, 2411. [Google Scholar] [CrossRef]
- Xu, Z.; Hu, H.; Wang, T.; Zhao, Y.; Zhou, C.; Xu, H.; Mao, X. Identification of growth years of Kudzu root by hyperspectral imaging combined with spectral–spatial feature tokenization transformer. Comput. Electron. Agric. 2023, 214, 108332. [Google Scholar] [CrossRef]
- Kotowska, D.; Pärt, T.; Żmihorski, M. Evaluating Google Street View for tracking invasive alien plants along roads. Ecol. Indic. 2021, 121, 107020. [Google Scholar] [CrossRef]
- Nesse, K.; Airt, L. How Useful Is GSV As an Environmental Observation Tool? An Analysis of the Evidence So Far. SSRN Electron. J. 2017. [Google Scholar] [CrossRef]
- Rousselet, J.; Imbert, C.-E.; Dekri, A.; Garcia, J.; Goussard, F.; Vincent, B.; Denux, O.; Robinet, C.; Dorkeld, F.; Roques, A.; et al. Assessing Species Distribution Using Google Street View: A Pilot Study with the Pine Processionary Moth. PLoS ONE 2013, 8, e74918. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Primoy, D.; Fagúndez, J. Assessment of the distribution and recent spread of the invasive grass Cortaderia selloana in Industrial Sites in Galicia, NW Spain. Flora 2019, 259, 151465. [Google Scholar] [CrossRef]
- Perrett, A.; Pollard, H.; Barnes, C.; Schofield, M.; Qie, L.; Bosilj, P.; Brown, J.M. DeepVerge: Classification of roadside verge biodiversity and conservation potential. Comput. Environ. Urban Syst. 2023, 102, 101968. [Google Scholar] [CrossRef]
- Shen, M.; Tang, M.; Jiao, W.; Li, Y. Kudzu invasion and its influential factors in the southeastern United States. Int. J. Appl. Earth Obs. Geoinf. 2024, 130, 103872. [Google Scholar] [CrossRef]
- Jarnevich, C.S.; LaRoe, J.; Engelstad, P.; Hays, B.; Henderson, G.; Williams, D.; Shadwell, K.; Pearse, I.S.; Prevey, J.S.; Sofaer, H.R. INHABIT Species Potential Distribution Across the Contiguous United States; Version 3.0; U.S. Geological Survey Data Release; U.S. Geological Survey: Reston, VA, USA, 2023. [CrossRef]
- Zaka, M.M.; Samat, A. Advances in Remote Sensing and Machine Learning Methods for Invasive Plants Study: A Comprehensive Review. Remote Sens. 2024, 16, 3781. [Google Scholar] [CrossRef]
- Kaya, A.; Seydi, A.; Catal, C.; Yalin, H.; Temucin, H. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Czerminski, R.; Jamieson, A.C. Neural Networks and Deep Learning. In The Machine Age of Customer Insight; Emerald Publishing Limited: Leeds, UK, 2021; pp. 91–101. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Cohen, J.G.; Lewis, M.J. Development of an Automated Monitoring Platform for Invasive Plants in a Rare Great Lakes Ecosystem Using Uncrewed Aerial Systems and Convolutional Neural Networks. In Proceedings of the 2020 International Conference on Unmanned Aircraft Systems (ICUAS), Athens, Greece, 1–4 September 2020; pp. 1553–1564. [Google Scholar] [CrossRef]
- Kattenborn, T.; Eichel, J.; Schmidtlein, S.; Wiser, S.; Burrows, L.; Fassnacht, F.E. Convolutional Neural Networks accurately predict cover fractions of plant species and communities in Unmanned Aerial Vehicle imagery. Remote Sens. Ecol. Conserv. 2020, 6, 472–486. [Google Scholar] [CrossRef]
- Kun, X.; Wei, W.; Sun, Y.; Wang, Y.; Xin, Q. Mapping fine-spatial-resolution vegetation spring phenology from individual Landsat images using a convolutional neural network. Int. J. Remote Sens. 2023, 44, 3059–3081. [Google Scholar] [CrossRef]
- Shen, M.; Tang, M.; Li, Y. Phenology and spectral unmixing-based invasive kudzu mapping: A case study in knox county, tennessee. Remote Sens. 2021, 13, 4551. [Google Scholar] [CrossRef]
- Zhou, X.; Xin, Q.; Dai, Y.; Li, W. A deep-learning-based experiment for benchmarking the performance of global terrestrial vegetation phenology models. Glonal Ecol. Biogeogr. 2021, 30, 2178–2199. [Google Scholar] [CrossRef]
- Lake, T.A.; Runquist, R.D.B.; Moeller, D.A. Deep learning detects invasive plant species across complex landscapes using Worldview-2 and Planetscope satellite imagery. Remote Sens. Ecol. Conserv. 2022, 8, 875–889. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Tang, F.; Zhang, D.; Zhao, X. Efficiently deep learning for monitoring Ipomoea cairica (L.) sweets in the wild. Math. Biosci. Eng. 2021, 18, 1121–1135. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Y.-Z.; Su, G.-Y.; Tian, H.-K.; Zhang, S.; Sun, Z.-Y.; Yang, L.; Wan, F.-H.; Qian, W.-Q. MmNet: Identifying Mikania micrantha Kunth in the wild via a deep Convolutional Neural Network. J. Integr. Agric. 2020, 19, 1292–1300. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, Z.; Liu, H.; Wu, J.; Lin, W. Individual Tree Species Identification for Complex Coniferous and Broad-Leaved Mixed Forests Based on Deep Learning Combined with UAV LiDAR Data and RGB Images. Forests 2024, 15, 293. [Google Scholar] [CrossRef]
- Deneu, B.; Servajean, M.; Bonnet, P.; Botella, C.; Munoz, F.; Joly, A. Convolutional neural networks improve species distribution modelling by capturing the spatial structure of the environment. PLoS Comput. Biol. 2021, 17, e1008856. [Google Scholar] [CrossRef] [PubMed]
- Botella, C.; Joly, A.; Bonnet, P. Species distribution modeling based on the automated identification of citizen observations. Appl. Plant Sci. 2018, 6, e1029. [Google Scholar] [CrossRef]
- Sermanet, P.; Eigen, D.; Zhang, X.; Mathieu, M.; Fergus, R.; LeCun, Y. OverFeat: Integrated Recognition, Localization and Detection using Convolutional Networks. In Proceedings of the 2nd International Conference on Learning Representations, ICLR 2014—Conference Track Proceedings, Banff, AB, Canada, 14–16 April 2013. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 1137–1149. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You Only Look Once: Unified, Real-Time Object Detection. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–26 June 2005; pp. 886–893. [Google Scholar] [CrossRef]
- Lowe, D.G. Object recognition from local scale-invariant features. In Proceedings of the IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999; Volume 2, pp. 1150–1157. [Google Scholar] [CrossRef]
- Zhen, T. Integrating Deep Learning and Google Street View for Novel Weed Mapping. Master’s Thesis, UC Davis, Davis, CA, USA, 2022. Available online: https://escholarship.org/uc/item/8pd05123 (accessed on 20 January 2025).
- Salahin, S.M.T.U.; Mehnaz, F.; Zaman, A.; Barua, K.; Mahbub, D.M.S. Detecting and Mapping of Roadside Trees from Google Street View. SSRN Electron. J. 2024. [Google Scholar] [CrossRef]
- Velasquez-Camacho, L.; Etxegarai, M.; de-Miguel, S. Implementing Deep Learning algorithms for urban tree detection and geolocation with high-resolution aerial, satellite, and ground-level images. Comput. Environ. Urban Syst. 2023, 105, 102025. [Google Scholar] [CrossRef]
- Gautam, D.; Mawardi, Z.; Elliott, L.; Loewensteiner, D.; Whiteside, T.; Brooks, S. Detection of Invasive Species (Siam Weed) Using Drone-Based Imaging and YOLO Deep Learning Model. Remote Sens. 2025, 17, 120. [Google Scholar] [CrossRef]
- Terven, J.; Córdova-Esparza, D.M.; Romero-González, J.A. A Comprehensive Review of YOLO Architectures in Computer Vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Yaseen, M. What Is YOLOv8: An In-Depth Exploration of the Internal Features of the Next-Generation Object Detector. arXiv 2024, arXiv:2408.15857. Available online: http://arxiv.org/abs/2408.15857 (accessed on 25 February 2025).
- Haritha, I.V.S.L.; Harshini, M.; Patil, S.; Philip, J. Real Time Object Detection using YOLO Algorithm. In Proceedings of the 6th International Conference on Electronics, Communication and Aerospace Technology, ICECA 2022—Proceedings, Coimbatore, India, 1–3 December 2022; pp. 1465–1468. [Google Scholar] [CrossRef]
- Aboyomi, D.D.; Daniel, C. A Comparative Analysis of Modern Object Detection Algorithms: YOLO vs. SSD vs. Faster R-CNN. ITEJ Inf. Technol. Eng. J. 2023, 8, 96–106. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Z.; Lei, L.; Wang, X.; Guo, X. Agricultural Greenhouses Detection in High-Resolution Satellite Images Based on Convolutional Neural Networks: Comparison of Faster R-CNN, YOLO v3 and SSD. Sensors 2020, 20, 4938. [Google Scholar] [CrossRef]
- Tan, L.; Huangfu, T.; Wu, L.; Chen, W. Comparison of YOLO v3, Faster R-CNN, and SSD for Real-Time Pill Identification. Preprint 2021. Available online: https://www.researchsquare.com/article/rs-668895/v1 (accessed on 22 April 2025).
- Sanchez, S.A.; Romero, H.J.; Morales, A.D. A review: Comparison of performance metrics of pretrained models for object detection using the TensorFlow framework. IOP Conf. Ser. Mater. Sci. Eng. 2020, 844, 012024. [Google Scholar] [CrossRef]
- Jocher, G.; Qiu, J.; Chaurasia, A. Ultralytics YOLO; Version 8.0.0; Ultralytics: Frederick, MD, USA, 2023. [Google Scholar]
- Li, R.; He, Y.; Li, Y.; Qin, W.; Abbas, A.; Ji, R.; Li, S.; Wu, Y.; Sun, X.; Yang, J. Identification of cotton pest and disease based on CFNet-VoV-GCSP-LSKNet-YOLOv8s: A new era of precision agriculture. Front. Plant Sci. 2024, 15, 1348402. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common Objects in Context. In Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014, Proceedings, Part V; Springer: Cham, Switzerland; pp. 740–755. [CrossRef]
- Wang, G.; Chen, Y.; An, P.; Hong, H.; Hu, J.; Huang, T. UAV-YOLOv8: A Small-Object-Detection Model Based on Improved YOLOv8 for UAV Aerial Photography Scenarios. Sensors 2023, 23, 7190. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.; Amran, G.A.; Tahmid, S.M.A.; Alabrah, A.; AL-Bakhrani, A.A.; Ali, A. A Deep-Learning-Based Model for the Detection of Diseased Tomato Leaves. Agronomy 2024, 14, 1593. [Google Scholar] [CrossRef]
- Wang, Y.; Yi, C.; Huang, T.; Liu, J. Research on Intelligent Recognition for Plant Pests and Diseases Based on Improved YOLOv8 Model. Appl. Sci. 2024, 14, 5353. [Google Scholar] [CrossRef]
- Ahmed, S.; Revolinski, S.R.; Maughan, P.W.; Savic, M.; Kalin, J.; Burke, I.C. Deep learning–based detection and quantification of weed seed mixtures. Weed Sci. 2024, 72, 655–663. [Google Scholar] [CrossRef]
- Diao, Z.; Ma, S.; Zhang, D.; Zhang, J.; Guo, P.; He, Z.; Zhao, S.; Zhang, B. Algorithm for Corn Crop Row Recognition during Different Growth Stages Based on ST-YOLOv8s Network. Agronomy 2024, 14, 1466. [Google Scholar] [CrossRef]
- Venverloo, T.; Duarte, F. Towards real-time monitoring of insect species populations. Sci. Rep. 2024, 14, 18727. [Google Scholar] [CrossRef] [PubMed]
- Kathir, M.; Balaji, V.; Ashwini, K. Animal Intrusion Detection Using Yolo V8. In Proceedings of the 2024 10th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 14–15 March 2024; Volume 1, pp. 206–211. [Google Scholar] [CrossRef]
- Dwyer, B.; Nelson, J.; Hansen, T. Roboflow, Version 1.0. 2024. Available online: https://roboflow.com (accessed on 25 February 2025).
- Shahinfar, S.; Meek, P.; Falzon, G. ‘How many images do I need?’ Understanding how sample size per class affects deep learning model performance metrics for balanced designs in autonomous wildlife monitoring. Ecol. Inform. 2020, 57, 101085. [Google Scholar] [CrossRef]
- Bullock, J.; Cuesta-Lazaro, C.; Quera-Bofarull, A. XNet: A convolutional neural network (CNN) implementation for medical X-Ray image segmentation suitable for small datasets. In Proceedings of the SPIE 10953, Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 19–21 February 2019. [Google Scholar] [CrossRef]
- Foroughi, F.; Chen, Z.; Wang, J. A CNN-Based System for Mobile Robot Navigation in Indoor Environments via Visual Localization with a Small Dataset. World Electr. Veh. J. 2021, 12, 134. [Google Scholar] [CrossRef]
- Gholamy, A.; Kreinovich, V.; Kosheleva, O. Why 70/30 or 80/20 Relation Between Training and Testing Sets: A Pedagogical Explanation. Departmental Technical Reports (CS). 2018. Available online: https://scholarworks.utep.edu/cs_techrep/1209 (accessed on 25 February 2025).
- Zhao, Y.; Zou, Y.; Wang, L.; Su, R.; He, Q.; Jiang, K.; Chen, B.; Xing, Y.; Liu, T.; Zhang, H.; et al. Tropical Rainforest Successional Processes can Facilitate Successfully Recovery of Extremely Degraded Tropical Forest Ecosystems Following Intensive Mining Operations. Front. Environ. Sci. 2021, 9, 701210. [Google Scholar] [CrossRef]
- EDDMapS. Early Detection & Distribution Mapping System. The University of Georgia—Center for Invasive Species and Ecosystem Health. Available online: http://www.eddmaps.org/ (accessed on 25 February 2025).
- Ali, A.H.; Abdulazeez, A.M. Transfer Learning in Machine Learning: A Review of Methods and Applications. Indones. J. Comput. Sci. 2024, 13, 4227–4259. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Ren, W.; Zhang, Z.; Jia, Z.; Wang, L.; Tan, T. Focal and Efficient IOU Loss for Accurate Bounding Box Regression. Neurocomputing 2021, 506, 146–157. [Google Scholar] [CrossRef]
- Charoenphakdee, N.; Vongkulbhisal, J.; Chairatanakul, N.; Sugiyama, M. On Focal Loss for Class-Posterior Probability Estimation: A Theoretical Perspective. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 5198–5207. [Google Scholar] [CrossRef]
- Li, X.; Lv, C.; Wang, W.; Li, G.; Yang, L.; Yang, J. Generalized Focal Loss: Towards Efficient Representation Learning for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 3139–3153. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Guillén, L.A. Accuracy Assessment in Convolutional Neural Network-Based Deep Learning Remote Sensing Studies—Part 1: Literature Review. Remote Sens. 2021, 13, 2450. [Google Scholar] [CrossRef]
- Ozenne, B.; Subtil, F.; Maucort-Boulch, D. The precision--recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 2015, 68, 855–859. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From Precision, Recall and F-Measure to ROC, Informedness, Markedness and Correlation. arxiv 2020, arXiv:2010.16061. Available online: https://arxiv.org/abs/2010.16061v1 (accessed on 25 February 2025).
- Olson, D.L.; Delen, D. Advanced data mining techniques. In Advanced Data Mining Techniques; Springer Nature: Dordrecht, The Netherlands, 2008; pp. 1–180. [Google Scholar] [CrossRef]
- Sobti, A.; Arora, C.; Balakrishnan, M. Object Detection in Real-Time Systems: Going beyond Precision. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision, WACV 2018, Lake Tahoe, NV, USA, 12–15 March 2018; pp. 1020–1028. [Google Scholar] [CrossRef]
- Khow, Z.J.; Tan, Y.F.; Karim, H.A.; Rashid, H.A.A. Improved YOLOv8 Model for a Comprehensive Approach to Object Detection and Distance Estimation. IEEE Access 2024, 12, 63754–63767. [Google Scholar] [CrossRef]
- Loewenstein, N.J.; Enloe, S.F.; Everest, J.W.; Miller, J.H.; Ball, D.M.; Patterson, M.G. History and Use of Kudzu in the Southeastern United States—Alabama Cooperative Extension System. System ACE. Available online: https://www.aces.edu/blog/topics/forestry-wildlife/the-history-and-use-of-kudzu-in-the-southeastern-united-states/ (accessed on 15 April 2025).
- QGIS. 2024. Available online: http://qgis.org (accessed on 22 April 2025).
- Jarnevich, C.S.; Young, N.E.; Thomas, C.C.; Grissom, P.; Backer, D.; Frid, L. Assessing ecological uncertainty and simulation model sensitivity to evaluate an invasive plant species’ potential impacts to the landscape. Sci. Rep. 2020, 10, 19069. [Google Scholar] [CrossRef] [PubMed]
- Streetview Request and Response|Street View Static API|Google for Developers. Available online: https://developers.google.com/maps/documentation/streetview/request-streetview (accessed on 11 March 2025).

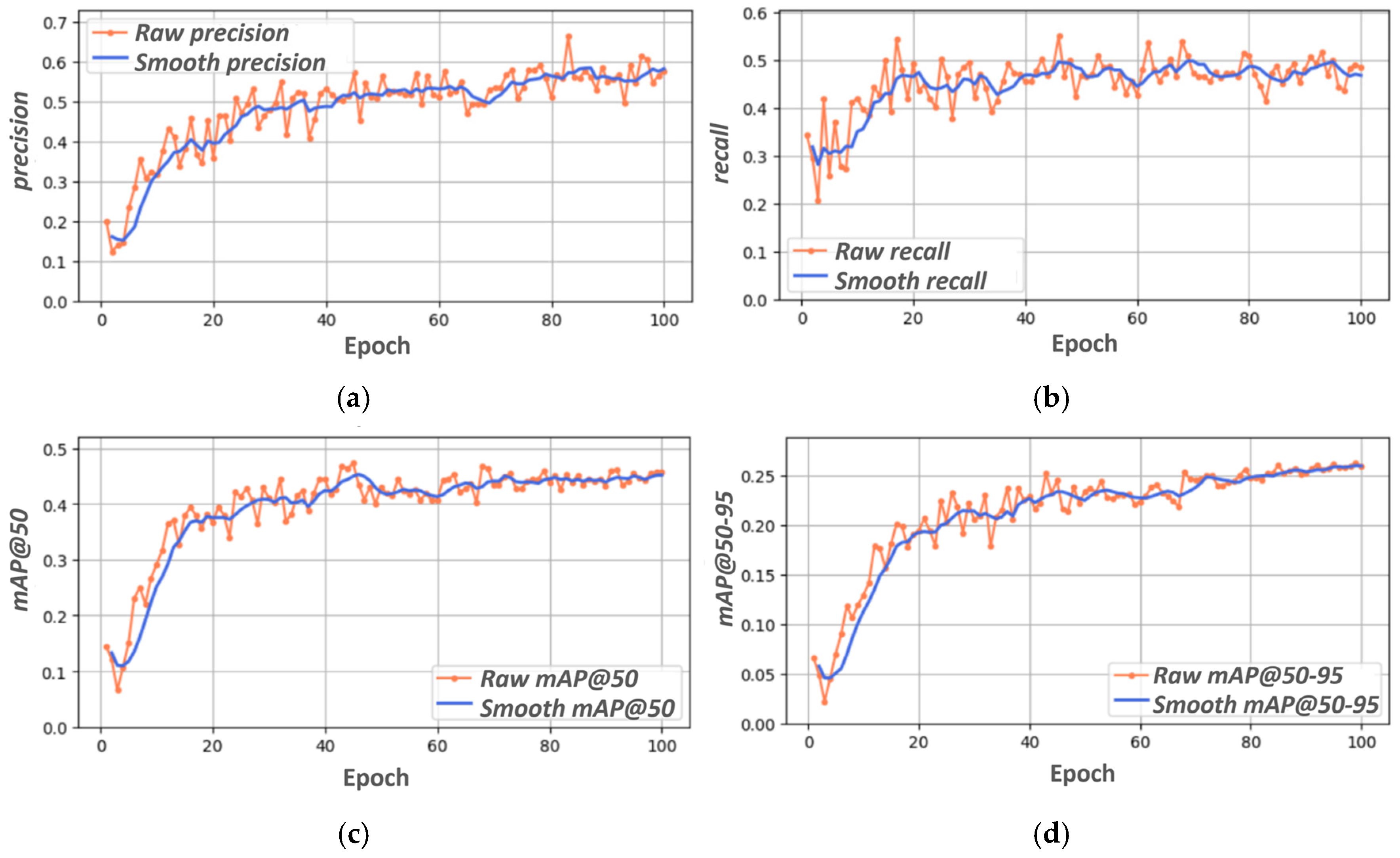


| Metric | Value |
|---|---|
| precision | 0.564 |
| recall | 0.488 |
| mAP@50 | 0.458 |
| mAP@50–95 | 0.263 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Closa-Tarres, A.; Rojano, F.; Strager, M.P. Geospatial Data and Google Street View Images for Monitoring Kudzu Vines in Small and Dispersed Areas. Earth 2025, 6, 40. https://doi.org/10.3390/earth6020040
Closa-Tarres A, Rojano F, Strager MP. Geospatial Data and Google Street View Images for Monitoring Kudzu Vines in Small and Dispersed Areas. Earth. 2025; 6(2):40. https://doi.org/10.3390/earth6020040
Chicago/Turabian StyleClosa-Tarres, Alba, Fernando Rojano, and Michael P. Strager. 2025. "Geospatial Data and Google Street View Images for Monitoring Kudzu Vines in Small and Dispersed Areas" Earth 6, no. 2: 40. https://doi.org/10.3390/earth6020040
APA StyleClosa-Tarres, A., Rojano, F., & Strager, M. P. (2025). Geospatial Data and Google Street View Images for Monitoring Kudzu Vines in Small and Dispersed Areas. Earth, 6(2), 40. https://doi.org/10.3390/earth6020040






