R Analysis for Optimizing Enzymatic Saccharification of Watermelon (Citrullus lanatus) Rind †
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Preparation of Watermelon Rinds (Substrate)
2.2.2. Determination of the Most Feasible Enzymes for Enzymatic Hydrolysis of Watermelon Rinds
2.2.3. Optimization of Hydrolysis Condition of Watermelon Rinds by Using RSM
2.2.4. Determination of Total Reduced Sugar
3. Results and Discussion
3.1. Effect of Different Types of Enzyme Treatment
3.2. Screening of Parameters Affecting Saccharification Yield for Enzymatic Hydrolysis of Watermelon Rinds
3.3. Optimization of Immobilization Parameter
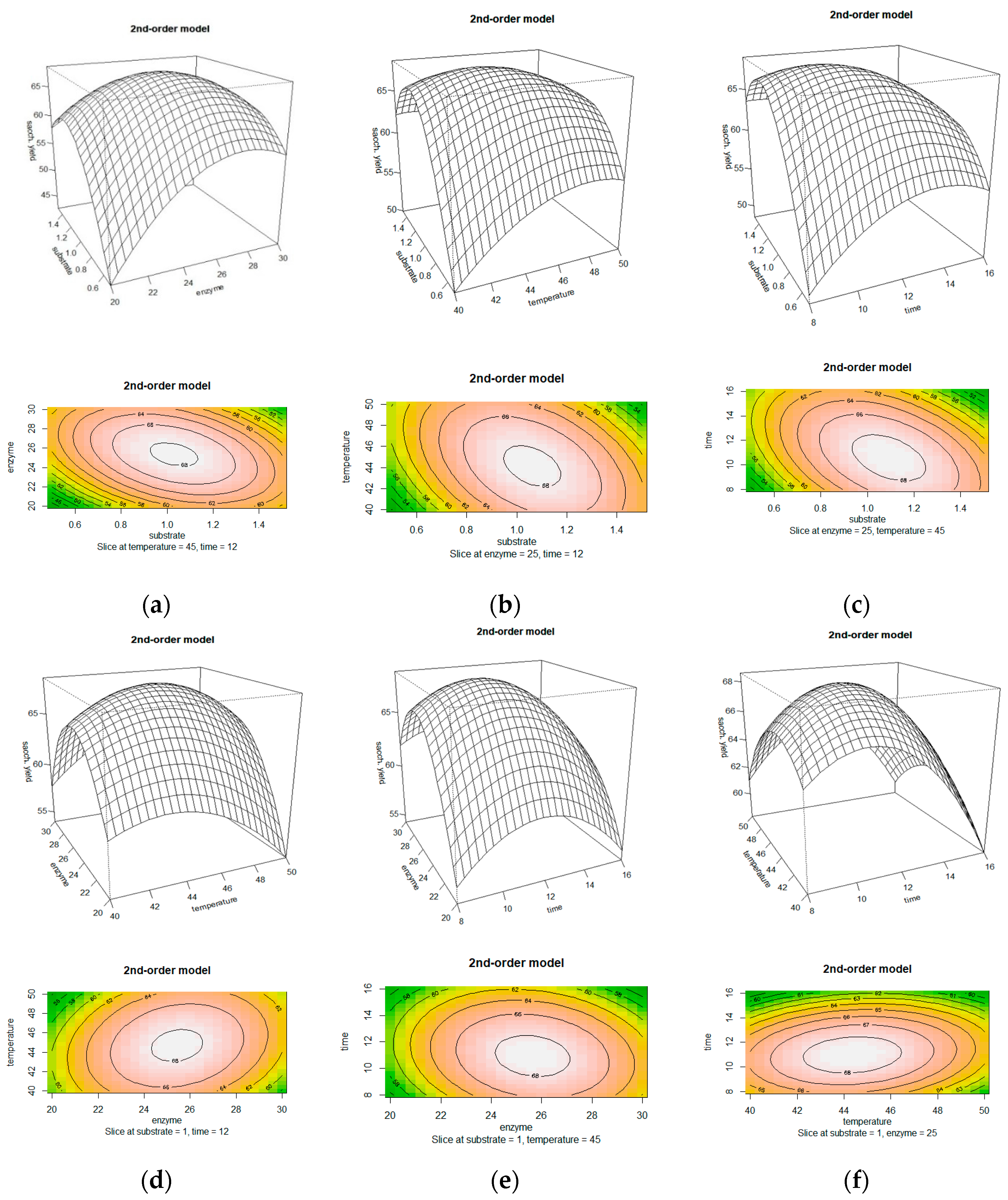
3.4. The Effect of Substrate and Enzyme Loading on Saccharification Yield
3.5. The Effect of Substrate Loading and Incubation Temperature on Saccharification Yield
3.6. The Effect of Substrate Loading and Hydrolysis Time on Saccharification Yield
3.7. The Effect of Enzyme Loading and Incubation Temperature on Saccharification Yield
3.8. The Effect of Enzyme Loading and Hydrolysis Time on Saccharification Yield
3.9. The Effect of Incubation Temperature and Hydrolysis Time on Saccharification Yield
3.10. Attaining Optimum Conditions and Model Validation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amaeze, N.; Okoliegbe, I.; Francis, M. Cellulase production by Aspergillus niger and Saccharomyces cerevisiae using fruit wastes as substrates. Int. J. Appl. Microbiol. Biotechnol. 2015, 3, 36–44. [Google Scholar]
- Kamusoko, R.; Jingura, R.M.; Parawira, W.; Chikwambi, Z. Strategies for valorization of crop residues into biofuels and other value-added products. Biofuels Bioprod. Biorefining 2021, 15, 1950–1964. [Google Scholar] [CrossRef]
- Periyasamy, S.; Isabel, J.B.; Kavitha, S.; Karthik, V.; Mohamed, B.A.; Gizaw, D.G.; Aminabhavi, T.M. Recent advances in consolidated bioprocessing for conversion of lignocellulosic biomass into bioethanol—A review. Chem. Eng. J. 2023, 453, 139783. [Google Scholar] [CrossRef]
- Sampaio, P.S.; Fernandes, P. Machine Learning: A Suitable Method for Biocatalysis. Catalysts 2023, 13, 961. [Google Scholar] [CrossRef]
- Kim, H. Propensity score analysis in non-randomized experimental designs: An overview and a tutorial using R software. New Dir. Child Adolesc. Dev. 2019, 2019, 65–89. [Google Scholar] [CrossRef] [PubMed]
- Poshina, D.N.; Raik, S.V.; Poshin, A.N.; Skorik, Y.A. Accessibility of chitin and chitosan in enzymatic hydrolysis: A review. Polym. Degrad. Stab. 2018, 156, 269–278. [Google Scholar] [CrossRef]
- Kassim, M.A.; Hussin, A.H.; Meng, T.K.; Kamaludin, R.; Zaki MS, I.M.; Zakaria WZ, E.W. Valorisation of watermelon (Citrullus lanatus) rind waste into bioethanol: An optimization and kinetic studies. Int. J. Environ. Sci. Technol. 2022, 14, 8671–8680. [Google Scholar] [CrossRef]
- Hasem, N.H.; Mohamad Fuzi SF, Z.; Kormin, F.; Abu Bakar, M.F.; Sabran, S.F. Extraction and partial characterization of durian rind pectin. IOP Conf. Ser. Earth Environ. Sci. 2019, 269, 012019. [Google Scholar] [CrossRef]
- Suhag, M.; Kumar, A.; Singh, J. Saccharification and fermentation of pretreated banana leaf waste for ethanol production. SN Appl. Sci. 2020, 2, 1448. [Google Scholar] [CrossRef]
- Sahariah, P.; Másson, M. Antimicrobial chitosan and chitosan derivatives: A review of the structure–activity relationship. Biomacromolecules 2017, 18, 3846–3868. [Google Scholar] [CrossRef]
- Gama, R. A Lignocellulolytic Enzyme System for Fruit Waste Degradation: Commercial Enzyme Mixture Synergy and Bioreactor Design. Ph.D. Dissertation, Rhodes University, Grahamstown, South Africa, 2013. [Google Scholar]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, A. Effect of particle size on the kinetics of enzymatic hydrolysis of microcrystalline cotton cellulose: A modeling and simulation study. Appl. Biochem. Biotechnol. 2019, 187, 800–816. [Google Scholar] [CrossRef] [PubMed]
- Payne, C.M.; Knott, B.C.; Mayes, H.B.; Hansson, H.; Himmel, M.E.; Sandgren, M.; Beckham, G.T. Fungal cellulases. Chem. Rev. 2015, 115, 1308–1448. [Google Scholar] [CrossRef]
- Alrumman, S.A. Enzymatic saccharification and fermentation of cellulosic date palm wastes to glucose and lactic acid. Braz. J. Microbiol. 2016, 47, 110–119. [Google Scholar] [CrossRef]
- Gao, Z.; Alshehri, K.; Li, Y.; Qian, H.; Sapsford, D.; Cleall, P.; Harbottle, M. Advances in biological techniques for sustainable lignocellulosic waste utilization in biogas production. Renew. Sustain. Energy Rev. 2022, 170, 112995. [Google Scholar] [CrossRef]
- Copley, S.D. Shining a light on enzyme promiscuity. Curr. Opin. Struct. Biol. 2017, 47, 167–175. [Google Scholar] [CrossRef]
- Pramanik, S.K.; Mahmud, S.; Paul, G.K.; Jabin, T.; Naher, K.; Uddin, M.S.; Saleh, M.A. Fermentation optimization of cellulase production from sugarcane bagasse by Bacillus pseudomycoides and molecular modeling study of cellulase. Curr. Res. Microb. Sci. 2021, 2, 100013. [Google Scholar] [CrossRef]
- Zou, X.; Jiang, X.; Wen, Y.; Wu, S.; Nadege, K.; Ninette, I.; Wang, X. Enzymatic synthesis of structured lipids enriched with conjugated linoleic acid and butyric acid: Strategy consideration and parameter optimization. Bioprocess Biosyst. Eng. 2020, 43, 273–282. [Google Scholar] [CrossRef]
- Breen, E.J.; Tan, W.; Khan, A. The statistical value of raw fluorescence signal in Luminex xMAP based multiplex immunoassays. Sci. Rep. 2016, 6, 26996. [Google Scholar] [CrossRef]
- Behera, S.K.; Meena, H.; Chakraborty, S.; Meikap, B.C. Application of response surface methodology (RSM) for optimization of leaching parameters for ash reduction from low-grade coal. Int. J. Min. Sci. Technol. 2018, 28, 621–629. [Google Scholar] [CrossRef]
- Sariman, S.N.S.A.; Zakaria, W.Z.E.W.; Serri, N.A. The optimization of fructose oleate ester in packed bed reactor (PBR) using R analysis. AIP Conf. Proc. 2023, 2785, 040007. [Google Scholar] [CrossRef]
- Rivera, E.C.; Rabelo, S.C.; dos Reis Garcia, D.; da Costa, A.C. Enzymatic hydrolysis of sugarcane bagasse for bioethanol production: Determining optimal enzyme loading using neural networks. J. Chem. Technol. Biotechnol. 2010, 85, 983–992. [Google Scholar] [CrossRef]
- Nadar, S.S.; Rao, P.; Rathod, V.K. Enzyme assisted extraction of biomolecules as an approach to novel extraction technology: A review. Food Res. Int. 2018, 108, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Kennes, D.; Abubackar, H.N.; Diaz, M.; Veiga, M.C.; Kennes, C. Bioethanol production from biomass: Carbohydrate vs. syngas fermentation. J. Chem. Technol. Biotechnol. 2016, 91, 304–317. [Google Scholar] [CrossRef]
- Pandiyan, K.; Tiwari, R.; Singh, S.; Nain, P.K.; Rana, S.; Arora, A.; Nain, L. Optimization of enzymatic saccharification of alkali pretreated Parthenium sp. using response surface methodology. Enzym. Res. 2014, 2014, 764898. [Google Scholar] [CrossRef][Green Version]
- Zhu, J.Y.; Pan, X. Efficient sugar production from plant biomass: Current status, challenges, and future directions. Renew. Sustain. Energy Rev. 2022, 164, 112583. [Google Scholar] [CrossRef]
- Mettler, M.S.; Mushrif, S.H.; Paulsen, A.D.; Javadekar, A.D.; Vlachos, D.G.; Dauenhauer, P.J. Revealing pyrolysis chemistry for biofuels production: Conversion of cellulose to furans and small oxygenates. Energy Environ. Sci. 2012, 5, 5414–5424. [Google Scholar] [CrossRef]
| Df | Sum Sq | Mean Sq | F-Value | Prob (>F) | |
|---|---|---|---|---|---|
| Substrate loading | 4 | 1975.6 | 489.4 | 945.7 | 7.63 × 10−13 *** |
| Enzyme loading | 4 | 1764.8 | 441.2 | 516.9 | 1.54 × 10−11 *** |
| Temperature | 4 | 3141.4 | 785.3 | 274.7 | 3.55 × 10−10 *** |
| Hydrolysis time | 4 | 3682 | 920.5 | 331.1 | 1.41 × 10−10 *** |
| Polynomial Coefficient | Std. Error | t-Value | p-Value Prob(>|t|) | |
|---|---|---|---|---|
| (Intercept) | 68.2766 | 0.8047 | 84.8478 | <2.2 × 10−16 *** |
| 1.5425 | 0.5690 | 2.7109 | 0.0161 * | |
| 1.3650 | 0.5690 | 2.3989 | 0.0298 * | |
| −0.5925 | 0.5690 | −1.0413 | 0.3142 | |
| −2.0366 | 0.5690 | −3.5793 | 0.0027 ** | |
| −6.1125 | 0.9855 | −6.2021 | 1.693 × 10−5 *** | |
| −4.5750 | 0.9855 | −4.6421 | 0.0003192 *** | |
| −5.5550 | 0.9855 | −5.6365 | 4.734 × 10−5 *** | |
| 1.8925 | 0.9855 | 1.9203 | 0.0740 | |
| −1.8050 | 0.9855 | −1.8315 | 0.0869 | |
| 1.0550 | 0.9855 | 1.0705 | 0.3013 | |
| −9.5229 | 0.7527 | −12.6513 | 2.092 × 10−9 *** | |
| −7.2341 | 0.7527 | −9.6106 | 8.404 × 10−8 *** | |
| −3.2804 | 0.7527 | −4.3581 | 0.0005622 *** | |
| −4.3141 | 0.7527 | −5.7314 | 3.971 × 10−5 *** | |
| Lack of fit | 1.6594 | 0.3000 | ||
| 0.96 | ||||
| 0.9226 | ||||
| F-statistic | 25.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan Zakaria, W.Z.E.; Yusof, K.; Serri, N.A. R Analysis for Optimizing Enzymatic Saccharification of Watermelon (Citrullus lanatus) Rind. Eng. Proc. 2025, 84, 5. https://doi.org/10.3390/engproc2025084005
Wan Zakaria WZE, Yusof K, Serri NA. R Analysis for Optimizing Enzymatic Saccharification of Watermelon (Citrullus lanatus) Rind. Engineering Proceedings. 2025; 84(1):5. https://doi.org/10.3390/engproc2025084005
Chicago/Turabian StyleWan Zakaria, Wan Zafira Ezza, Khairunisa Yusof, and Noor Aziah Serri. 2025. "R Analysis for Optimizing Enzymatic Saccharification of Watermelon (Citrullus lanatus) Rind" Engineering Proceedings 84, no. 1: 5. https://doi.org/10.3390/engproc2025084005
APA StyleWan Zakaria, W. Z. E., Yusof, K., & Serri, N. A. (2025). R Analysis for Optimizing Enzymatic Saccharification of Watermelon (Citrullus lanatus) Rind. Engineering Proceedings, 84(1), 5. https://doi.org/10.3390/engproc2025084005





