Portable Chest X-ray Synthetic Image Generation for the COVID-19 Screening †
Abstract
:1. Introduction
2. Methodology
2.1. Approaches for Data Augmentation
2.2. Approaches for Screening Tasks
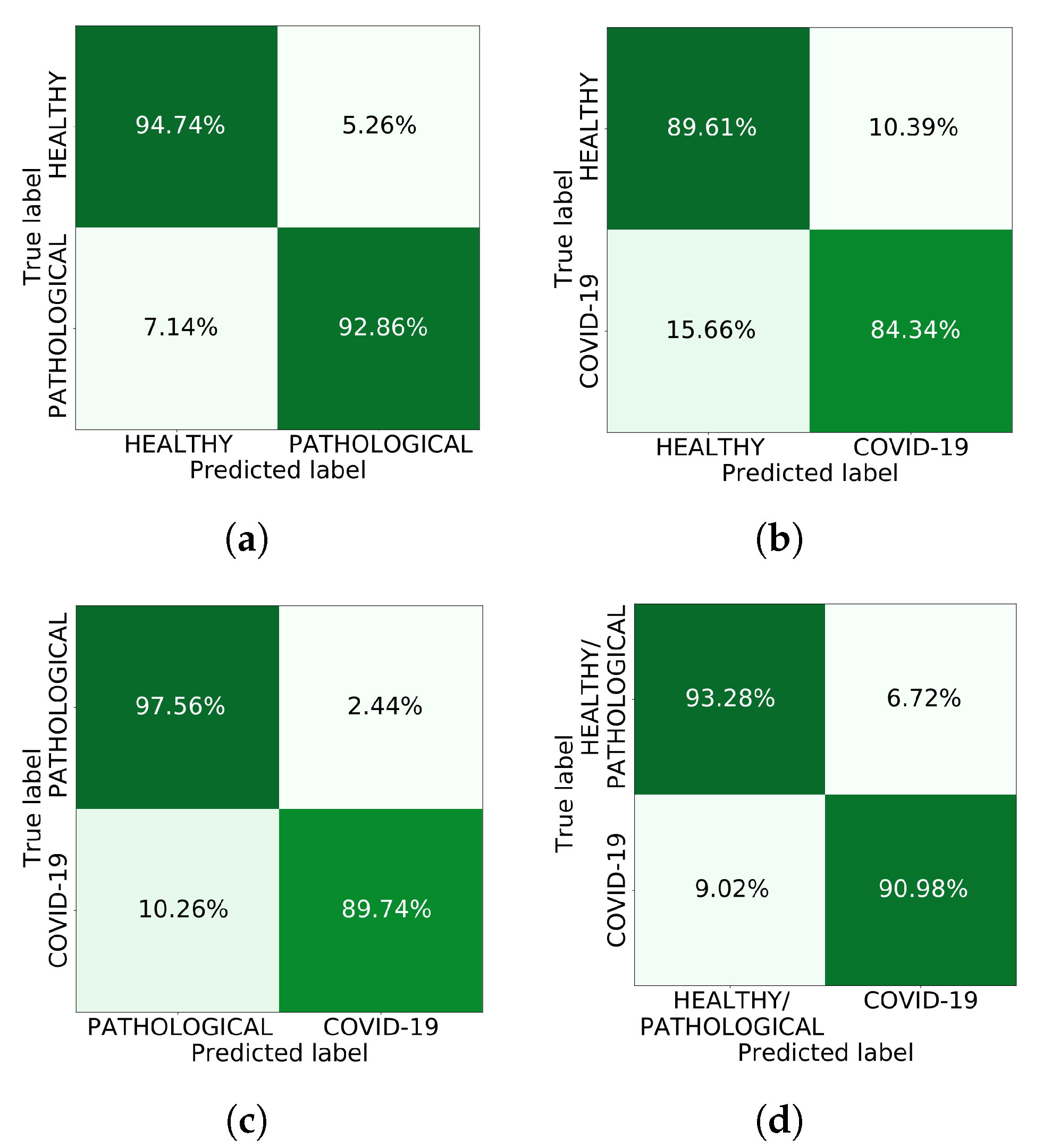
3. Results and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Pollard, C.A.; Morran, M.P.; Nestor-Kalinoski, A.L. The COVID-19 pandemic: A global health crisis. Physiol. Genom. 2020, 52, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Kooraki, S.; Hosseiny, M.; Myers, L.; Gholamrezanezhad, A. Coronavirus (COVID-19) outbreak: What the department of radiology should know. J. Am. Coll. Radiol. 2020, 17, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Vidal, P.L.; de Moura, J.; Novo, J.; Ortega, M. Multi-stage transfer learning for lung segmentation using portable X-ray devices for patients with COVID-19. Expert Syst. Appl. 2021, 173, 114677. [Google Scholar] [CrossRef] [PubMed]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative adversarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- de Moura, J.; Novo, J.; Ortega, M. Fully automatic deep convolutional approaches for the analysis of Covid-19 using chest X-ray images. medRxiv 2020. [Google Scholar] [CrossRef]
- Morís, D.I.; de Moura, J.; Novo, J.; Ortega, M. Cycle Generative Adversarial Network Approaches to Produce Novel Portable Chest X-Rays Images for Covid-19 Diagnosis. In Proceedings of the ICASSP 2021—2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Toronto, ON, Canada, 6–11 June 2021; pp. 1060–1064. [Google Scholar] [CrossRef]
- De Moura, J.; García, L.R.; Vidal, P.F.L.; Cruz, M.; López, L.A.; Lopez, E.C.; Novo, J.; Ortega, M. Deep convolutional approaches for the analysis of covid-19 using chest x-ray images from portable devices. IEEE Access 2020, 8, 195594–195607. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morís, D.I.; de Moura, J.; Novo, J.; Ortega, M. Portable Chest X-ray Synthetic Image Generation for the COVID-19 Screening. Eng. Proc. 2021, 7, 6. https://doi.org/10.3390/engproc2021007006
Morís DI, de Moura J, Novo J, Ortega M. Portable Chest X-ray Synthetic Image Generation for the COVID-19 Screening. Engineering Proceedings. 2021; 7(1):6. https://doi.org/10.3390/engproc2021007006
Chicago/Turabian StyleMorís, Daniel I., Joaquim de Moura, Jorge Novo, and Marcos Ortega. 2021. "Portable Chest X-ray Synthetic Image Generation for the COVID-19 Screening" Engineering Proceedings 7, no. 1: 6. https://doi.org/10.3390/engproc2021007006
APA StyleMorís, D. I., de Moura, J., Novo, J., & Ortega, M. (2021). Portable Chest X-ray Synthetic Image Generation for the COVID-19 Screening. Engineering Proceedings, 7(1), 6. https://doi.org/10.3390/engproc2021007006








