Abstract
Pathogens can be detected electrochemically by measuring guanine oxidation signals generated from RNA or DNA hybridized to a biosensor working electrode. However, the associated limit of detection (LOD) is not sufficiently low for widespread clinical use. Working electrodes employing nanomaterials such as carbon nanotubes successfully reduce the LOD, but nanosensors experience high variability, poor fabrication yield, and high production cost. Our work presented here demonstrates a novel approach for electrochemically detecting low-concentration pathogens and antimicrobial resistance genes that transfers the guanine oxidation source from naturally occurring RNA to synthetic oligonucleotides. In our assay, signal amplification is accomplished by binding RNA from lysed microbial cells to microparticles conjugated with millions of guanine-rich oligonucleotide tags. We employed a sandwich hybridization assay to bind RNA between a screen-printed carbon working electrode conjugated with recognition probes, and a microparticle conjugated with electrochemical oligonucleotide tags. These tags contained a polyguanine detection sequence and an RNA capture sequence on the same oligonucleotide. Single-stranded polyguanine was prefabricated into a quadruplex to enable 8-oxoguanine signals at 0.47 V. This eliminated nonspecific guanine oxidation signals from the RNA, while further reducing the LOD over guanine oxidation. A 70-mer capture sequence was found to be more selective and hybridized faster at room temperature than conventional 20-mer capture sequences. Particle sizes were evaluated from 100 nm to 1.5 µm in diameter, and the larger diameter particles produced greater detection signals. A better performance was obtained by employing magnetic microparticles and magnetically separating magnetic microparticle–RNA complexes from nonspecific materials, such as lysed cell constituents and cell debris, that can interfere with sandwich formation and detection. The high-density magnetic microparticles rested on the electrode surface, causing a portion of the oligonucleotides to adsorb to the working electrode surface.
1. Introduction
Microbial pathogens can be directly measured with an electrochemical biosensor using RNA as a detection target. 16S rRNA is highly stable and can selectively hybridize to a working electrode with a suitable recognition probe [1,2,3]. Guanine molecules on the RNA are typically used as electroactive species due to their relatively low redox potential [4]. RNA with 800 bases would have approximately 200 guanine molecules, assuming 25% of the nucleotides are guanine. However, 106 to 1010 guanine molecules would be required for attaining the necessary limit of detection (LOD) in clinical applications, which limits the use of RNA as a label-free detection target.
2. Technologies
2.1. Electrochemically Detectable Oligonucleotide Tags
A simple method to bring low concentration pathogens and antimicrobial resistance genes to detectable levels is to bind RNA targets with microparticles conjugated with millions of guanine-rich oligonucleotides. This provides orders of magnitude more guanine molecules for detection than direct detection of guanine on RNA. Figure 1 illustrates an oligonucleotide that has a 20-mer polyguanine detection sequence and a 70-mer capture sequence [5]. By eliminating the need for PCR or isothermal amplification, longer capture sequences can be used that are more selective and hybridize faster at room temperature than conventional 20-mer sequences.

Figure 1.
An oligonucleotide with a 20-mer electrochemical polyguanine sequence and 70-mer capture sequence for KPC 16S rRNA.
Figure 2a illustrates how electrochemical oligonucleotide tags are used in a sandwich hybridization assay. The top layer is a magnetic microparticle conjugated with electrochemically detectable oligonucleotide tags with polyguanine and capture sequences. The middle layer is an RNA target, and the bottom layer is a screen-printed carbon electrode conjugated with recognition probes [5]. Each tag contains 20 guanine molecules for detection. A 1.5 µm-diameter microparticle can bind ~106 biotinylated electrochemically detectable oligonucleotide tags to deliver ~20 million guanine molecules for each RNA sandwich. As noted in Table 1, approximately 5%, or ~1,000,000 guanine molecules are sufficiently close to the electrode surface, where guanine can contribute to the oxidation peak either directly or with an electron transport mediator. Additionally, ~1%, or 200,000 guanine molecules are directly below the high-density microparticles, causing the underlying guanine molecules to adsorb to the electrode surface, as illustrated in Figure 2b. Due to their proximity to the electrode, the same concentration of adsorbed guanine molecules should have a stronger signal than guanine molecules bound near the electrode surface.
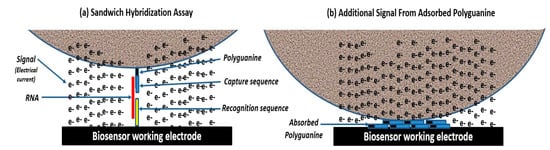
Figure 2.
(a) Sandwich hybridization assay with magnetic microparticles conjugated with millions of electrochemically detectable oligonucleotide tags. (b) The sandwich hybridization assay gains an additional electrochemical signal from oligonucleotide tags that adsorb to the working electrode when high-density magnetic microparticles drop onto the electrode surface.

Table 1.
Guanine molecules available for detection per RNA target.
2.2. Magnetic Particle Conjugates
Magnetic microparticles performed better than nanoparticles or nonmagnetic microparticles due to their higher surface area for binding oligonucleotides, greater contact on working electrodes for adsorbing guanine molecules from oligonucleotide tags, and better response to magnetic fields for separating conjugates from nonspecific materials. Table 2 indicates the relative signal achieved from 10,000 targets with the protocol described below using different microparticle sizes [6]. Larger microparticles with more tags generated a greater electrical current peak. Polyguanine oligonucleotides that were prefabricated into quadruplexes produced an 8-oxoguanine oxidation peak at 0.47 V relative to a baseline signal that was greater than the guanine oxidation peak at 0.90 V from single-stranded oligonucleotides [7]. This allowed a lower LOD to be achieved, and also eliminated nonspecific guanine oxidation signals from RNA.

Table 2.
Relative 8-oxoguanine signal from 104 targets/mL from different-sized magnetic microparticles.
2.3. Filter Concentration and Magnetic Separation
Filter concentration with lysis and magnetic separation provided 2 logs more targets for detection, as noted in Table 3 [6,8,9]. Concentrating large volumes of 1 mL or greater is important to use when targets in the samples are heterogeneous and could clump to nonspecific materials. Sample volumes, filter pore size, lysis solution, and the magnetic separation protocol were optimized to attain a capture yield of ~90%.

Table 3.
Estimate of the detection targets available from direct detection and filter concentration with magnetic separation.
3. Materials and Methods
We determined the presence of Klebsiella pneumoniae Carbapenemase (KPC)-producing bacteria by detecting KPC 16S rRNA [6]. A total of 28 samples were evaluated, comprising 22 KPC-producing samples with Klebsiella pneumoniae ATCC BAA 1705, Klebsiella pneumoniae ATCC BAA-2814, or E. coli ATCC BAA 2340, four non-KPC-producing samples with Klebsiella pneumoniae ATCC 13883, and two samples with no bacteria. The bacteria were prepared in tryptic soy agar medium (Becton, Dickinson (BD), Franklin Lakes, NJ, USA) and serial diluted in broth medium and commercial urine (Sigma-Aldrich, St. Louis, MO, USA) to 104 cfu/mL. For the experiments, 1 mL samples were concentrated with a 0.45 μm filter (Pall, Port Washington, NY, USA), re-suspended in biology-grade water, and incubated at room temperature for 15 min. The bacteria were then lysed in 200 μL lysis buffer consisting of 2 M guanidinium thiocyanate (GTC), 80 mM β-mercaptoethanol (BME), 25 mM sodium citrate, 20 μg/mL of glycogen (pH 6), and 5 μL dimethyl sulfoxide (1%), then incubated at room temperature (RT) for 5 min. The solution was mixed for 5 min with 7 μL of 1.5 μm streptavidin-coated magnetic microparticles (Bangs Laboratories, Fishers, IN, USA) which were conjugated with the biotinylated electrochemical oligonucleotides illustrated Figure 1 (Integrated DNA Technologies (IDT), San Jose, CA, USA) that were prefabricated into guanine quadruplexes, then incubated at RT for 10 min. The samples were placed in a magnetic separation microtiter (Epigentek, Farmingdale, NY, USA), a magnetic field was applied for 2 min, then the supernatant was discarded. The magnet field was removed, and the magnetic particle complexes were washed with 100 μL of 80 mM sodium acetate (pH 9). The magnetic particle complexes were then re-suspended in sodium acetate and allowed to hybridize for 10 min at RT on a streptavidin-coated carbon working electrode (DropSens, Asturias, Spain) conjugated with capture probes to enable sandwich structures to form. A potentiostat (PalmSens, Houten, The Netherlands) was connected, and a square-wave voltammetry scan produced a peak current at ~0.47 V from 8-oxoguanine oxidation in sodium acetate (pH 9). The net 8-oxoguanine oxidation signal was determined to be the difference between the 8-oxoguanine oxidation signal in sodium acetate and a baseline sodium acetate signal. The study protocol took 45 min to perform.
4. Results and Discussion
The results are summarized in Table 4. All 22 KPC-producing samples generated a positive signal. Of the six non-KPC-producing samples, five reported true negative. One false negative was detected, which was most likely caused by poor sensor calibration.

Table 4.
Summary of the samples tested for Klebsiella pneumoniae carbapenemase (KPC) RNA.
Figure 3 illustrates 8-oxoguanine oxidation peaks from a positive KPC-producing K. pneumoniae (upper curve) and a negative non-KPC-producing K. pneumoniae (lower curve). A detection threshold of 0.2 μA was determined from the baseline sodium acetate signal variability plus three standard deviations.
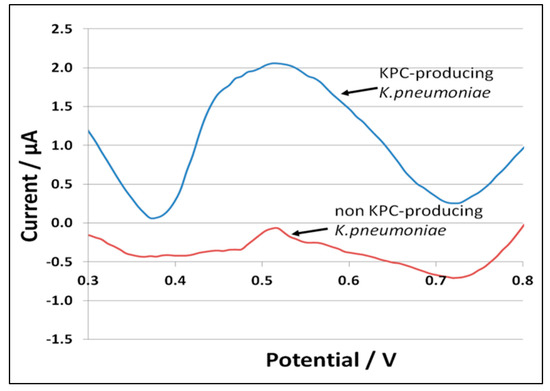
Figure 3.
8-oxoguanine oxidation peaks from positive KPC-producing K. pneumoniae (upper curve) and negative non-KPC-producing K. pneumoniae (lower curve).
This study of ours demonstrated the first use of the electrochemically detectable oligonucleotide tags in a sandwich hybridization assay. Although the study was conducted using urine samples, the protocol can be used for other sample types, such as whole blood, by adjusting the sample preparation steps for lysing, filtration, and magnetic separation.
The concentrations used in the study were 10,000 targets/mL. Positive samples were also attained from 100 target/mL concentrations. The limit of detection will be improved by using sample volumes greater than 1 mL and microparticle diameters larger than 1.5 µm. A modified protocol that directly detected electrochemically detectable oligonucleotide tags eluted from magnetic microparticles was previously used for potable water after filter concentrating 10 L samples [8].
The platform has the added capability of multiplexing a large number of targets. Multiplexing is planned for detecting 32 different targets from a sample. Each individual target will have a microparticle conjugate, and eight sensor working electrodes will be conjugated with a blend of four recognition probes. The working electrodes will be capable of detecting four different targets bound with either polyG, polyA, polyT, or polyC, which form oxidation peaks at different redox potentials.
Electrochemical detection with oligonucleotide tags is ideally suited for the rapid, mobile detection of complex infections that are caused by a large number of different pathogens and can require different treatments based on antimicrobial resistance. For example, sepsis is a life-threatening organ dysfunction caused by a dysregulated host response to infection. There are currently 1,700,000 US sepsis cases/year (30–50 million global cases), and 350,000 US sepsis deaths/year (11 million global cases) [10]. Prior to COVID-19, sepsis accounted for more than 50% of hospital deaths, and mortality increased dramatically with greater disease severity: 10–40% mortality due to sepsis, and 40–80% mortality due to septic shock [11,12,13]. Septic patients also represent a disproportionately high burden of hospital utilization. The average length of stay (LOS) for sepsis patients in US hospitals is approximately 75% greater than for most other conditions, and the mean LOS dramatically increases with sepsis severity, from 4.5 days to 6.5 days for sepsis and 16.5 days for septic shock [14,15,16]. A total of 60% of sepsis patients are re-hospitalized within 12 months, and they exhibit a high incidence of permanent organ damage, cognitive impairment, and physical disability [17]. The annual financial impact of sepsis on the US healthcare system is $24 billion in medical costs, with an additional $1.6 billion in litigation payments from misdiagnosis [18,19]. It has been estimated that if the US achieved earlier sepsis identification and evidenced-based treatment, there would be 92,000 fewer deaths annually, 1.25 million fewer days of hospital stays annually, and reductions in hospital expenditures of over $1.5 billion [20].
Emergency and inpatient doctors need to select appropriate antibiotic therapy based on the specific pathogen type and drug resistance. Therapy needs to be determined within hours of the first sign of sepsis; however, blood cultures take three to five days. When the patient is in septic shock, every hour delay in administering the appropriate antibiotic therapy has a 6% drop in patient survival [21,22]. Alternative approaches to determine the pathogen type and drug resistance enzymes can employ a 12-h blood culture followed by multiplex PCR using sophisticated instruments. These approaches have a 13–17 h sample-to-result turnaround time, are too expensive for many hospitals, are too large for many emergency departments, and can incur culture errors that cause false-positive and false-negative outcomes.
The electrochemically detectable oligonucleotide tag platform can be configured to operate in a mobile test cartridge and detect up to 32 low concentration targets. Targets can include a suite of sepsis pathogens and drug resistance enzymes. A rapid mobile test has the potential to allow emergency room and inpatient doctors to select the right antibiotics in about 1 hour instead of 3 to 5 days with blood cultures. The mobile electrochemical reader can fit in an emergency room’s limited available space. In addition, it can potentially be used by telehealth practitioners and physician-at-home specialists to test patients with sepsis-related vital signs for blood infections before they are sent to the hospital which can save valuable time.
Quantification of the signal peak amplitude is proportional to the target concentration, which can also be used to determine the effectiveness of a specific antibiotic by measuring the change in signal strength from a second sample incubated a few hours in different antibiotic inoculums.
Author Contributions
Conceptualization, N.G., G.P. and R.B.; methodology, G.P., N.G. and R.B.; software, N.G.; validation, R.B. and N.G.; formal analysis, N.G., G.P. and R.B.; investigation, N.G., G.P. and R.B.; resources, N.G. and R.B.; data curation, N.G., G.P. and R.B.; writing—original draft preparation, N.G.; writing—review and editing, R.B. and G.P.; visualization, N.G. and G.P.; supervision, N.G.; project administration, R.B.; funding acquisition, N.G. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for this research was provided by the Centers for Disease Control and Prevention as SBIR 1R43CK000522-01.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest. Neil Gordon is the founder and president of Guanine, Inc., and has a financial interest in the company. Raj Bawa, PhD, MD, is vice president and chief IP officer of Guanine, Inc., and he also serves as scientific advisor to Teva Pharmaceutical Industries, Ltd., Israel.
References
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Diculescu, A.C.; Chiorcea-Paquim, A.M.; Oliveira-Brett, A.M. Applications of a DNA-electrochemical biosensor. Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Labuda, J.; Oliveira-Brett, A.M.; Evtugynm, G.; Fojta, M.; Mascini, M.; Ozsoz, M.; Palchetti, I.; Palecek, E.; Wang, J. Electrochemical nucleic-acid-based biosensors: Concepts, terns, and methodology (IUPAC Technical Report). Pure Appl. Chem. 2010, 82, 1161–1187. [Google Scholar] [CrossRef]
- Palecek, E.; Jelen, F. Towards Electrochemical Sensors for Genomics and Proteomics. In Electrochemistry of Nucleic acids and Proteins; Palecek, E., Scheller, F., Wang, J., Eds.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 74–174. [Google Scholar]
- Gordon, N. Bioanalyte Signal Amplification and Detection with Artificial Intelligence Diagnosis. U.S. Patent No. 11105801, 31 August 2021. [Google Scholar]
- Gordon, N.; Bawa, R.; Palmateer, G.; Rajabi, M.; Gordon, J.B.; Kotb, N.M.; Balasubramaniyam, R.; Gordon, B.R. Carbapenem-Resistant Enterobacteriaceae Testing in 45 Minutes Using Oligonucleotide Detection Tags. In Advances in Medical Imaging, Detection, and Diagnosis; Bawa, R., Audette, G.F., Patel, B., Bawa, S.R., Johnson, B.D., Eds.; Jenny Stanford Publishing Pte. Ltd.: Singapore, 2023; Volume 4, pp. 813–828. ISBN 978-981-4877-46-6. [Google Scholar]
- Gordon, N. Ultra-Sensitive Bioanalyte Quantification from Self-Assembled Quadruplex Tags. U.S. Patent No. 11175285, 16 November 2021. [Google Scholar]
- Jayamohan, H.; Gale, B.K.; Minson, B.J.; Lambert, C.J.; Gordon, N.; Sant, H.J. Highly sensitive bacteria quantification using immunomagnetic separation and electrochemical detection of guanine-labeled secondary beads. Sensors 2015, 15, 12034–12052. [Google Scholar] [CrossRef] [PubMed]
- Gordon, N. Ultra-Sensitive Detection of Extremely Low Level Biological Analytes Using Electrochemical Signal Amplification and Biosensor. U.S. Patent No. 9624532, 18 April 2017. [Google Scholar]
- Centers for Disease Control and Prevention. What Is Sepsis? Available online: https://www.cdc.gov/sepsis/what-is-sepsis.html (accessed on 1 May 2023).
- Carly, J.; Paoli, C.J.; Reynolds, M.A.; Sinha, M.; Gitlin, M.; Crouser, E. Epidemiology and Costs of Sepsis in the United States—An Analysis Based on Timing of Diagnosis and Severity Level. Crit. Care Med. 2018, 46, 1889–1897. [Google Scholar] [CrossRef]
- Liu, V.; Escobar, G.J.; Greene, J.D.; Soule, J.; Whippy, A.; Angus, D.C.; Iwashyna, T.J. Hospital deaths in patients with sepsis from 2 independent cohorts. JAMA 2014, 312, 90–92. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.S. Sepsis, severe sepsis and septic shock: Changes in incidence, pathogens and outcomes. Expert Rev. Anti-Infect. Ther. 2012, 10, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.J.; Williams, S.N.; DeFrances, C.J.; Golosinskiy, A. Inpatient Care for Septicemia or Sepsis: A Challenge for Patients and Hospitals, 2000–2008. National Center for Health Sta-tistics. Data Brief No. 62; June 2011. Available online: http://www.cdc.gov/nchs/data/databriefs/db62.pdf (accessed on 1 May 2023).
- HCUP National Inpatient Sample (NIS). Healthcare Cost and Utilization Project (HCUP); Agency for Healthcare Re-search and Quality: Rockville, MD, USA, 2013. Available online: www.hcup-us.ahrq.gov/nisoverview.jsp (accessed on 1 May 2023).
- Torio, C.M.; Moore, B.J. National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2013; HCUP Statistical Brief #204 [Internet]; Agency for Healthcare Research and Quality: Rockville, MD, USA, May 2016. Available online: https://www.hcup-us.ahrq.gov/reports/statbriefs/sb204-Most-Expensive-Hospital-Conditions.jsp (accessed on 1 May 2023).
- Shankar-Hari, M.; Rubenfeld, G.D. Understanding Long-Term Outcomes Following Sepsis: Implications and Challenges. Curr. Infect. Dis. Rep. 2016, 18, 37. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.L.; Ashton, C.M.; Kiehne, L.; Gigliotti, E.; Bell-Gordon, C.; Pinn, T.T.; Tran, S.K.; Nicolas, J.C.; Rose, A.L.; Shirkey, B.A.; et al. The Sepsis Early Recognition and Response Initiative (SERRI). Jt. Comm. J. Qual. Patient Saf. 2016, 42, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Popp, P.L. Hiding in plain sight: Why are we worried about Ebola and not sepsis? J. Healthc. Risk Manag. 2016, 35, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Sepsis Facts. Available online: http://www.world-sepsis-day.org/ (accessed on 1 May 2023).
- Dellinger, R.P.; Carlet, J.M.; Masur, H.; Gerlach, H.; Calandra, T.; Cohen, J.; Gea-Banacloche, J.; Keh, D.; Marshall, J.C.; Parker, M.M.; et al. Surviving sepsis: Campaign guidelines for management of severe sepsis and septic shock. Crit. Care Med. 2004, 32, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.L.; Marshall, J.C.; Ñamendys-Silva, S.A.; François, B.; Martin-Loeches, I.; Lipman, J.; Reinhart, K.; Antonelli, M.; Pickkers, P.; Njimi, H.; et al. Assessment of the worldwide burden of critical illness: The intensive care over nations (ICON) audit. Lancet Respir. Med. 2014, 2, 380–386. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).