1. Introduction
Aptamers are short, single-stranded nucleic acid sequences selected in vitro via the process of the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) to bind to specified targets [
1]. They present a substantial opportunity for the development of diagnostic sensors, serving to either “capture” targets or report on successful aptamer-target binding. For their latter application in sensors, aptamers are frequently modified by conjugation to different reporting agents [
2,
3,
4,
5,
6]. Conventional SELEX generates a pool of candidate aptamer sequences capable of binding to the desired target under the experimental conditions employed in the SELEX approach followed. However, this does not always result in aptamers capable of binding the same target with high affinities. Once sequences are modified for use in biosensors, conjugation of specific reporting agents to aptamers results in decreased binding affinities to their targets [
4,
5], requiring laborious screening of individual sequences for their compatibility with selected reporting agents when conjugated to different molecules.
The Polymerase Chain Reaction (PCR) is readily used to introduce modifications into aptamer pools, with forward primers conjugated with specific reporter agent groups at their 5′ sites, e.g., [
7]. For this study, a 5′-biotin modification commonly used for aptamer sensor generation [
4,
5,
6] was selected. However, the PCR is not free of biases, preventing specific sequences from amplifying with high fidelity [
8], which may further be influenced by the presence of labels such as 5′-biotin, as has been reported for other moieties [
9]. This may also affect downstream sequencing and ligation reactions, where the 5′-phosphate group found in unmodified DNA sequences is required to be accessible.
Here, we propose an amended SELEX process to source potential aptamers that can simultaneously bind to the target and report on its presence effectively. This study proposes revisiting pools of aptamer candidates that had already been enriched for a specific target and adding in a new step in which the desired reporting agent is included as a modification at the 5′ end of the aptamer, via PCR. Modified aptamers would then be re-selected for suitable binding to the target. Ultimately, this study will test whether alterations in the enrichment of specific sequences in the aptamer pool occur due to the presence of the modification, creating a new pool of aptamer candidates that are compatible with the modification used for subsequent reporting.
As the first stage in this amendment, we present a study that seeks to address a fundamental concern: whether integration of reporting agents via PCR during amplification of the aptamer candidates affects the composition of the aptamer pool that will subsequently undergo re-selection and enrichment towards the target. We report on the results of PCR-amplification using unmodified and 5′-biotin-C6-phosphoramidite modified forward primers on the same pool and the subsequent bioinformatic analysis of the sequenced pools.
2. Materials and Methods
Briefly, the aptamer pool used in this study (5′-TCGCACATTCCGCTTCTACC(N
40)CGTAAGTCCGTGTGTGCGAA-3′) was prepared in a previous study, where 9 rounds of selection were conducted against human chorionic gonadotropin (hCG), via a novel-selection SELEX strategy [
10]. Two fractions of the 9th round of selection were used as samples. PCR was carried out using unmodified and biotinylated (i.e., containing a biotin-(CH
2)
6)-phosphoramidite modification at the 5′ end) forward primers, using GoTaq Hotstart Polymerase, and the cycling parameters followed as in
Table S1. Following PCR, the amplified pools were separately purified (NucleoSpin
® Gel and PCR clean-up kit; MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany) and concentrated (ethanol-precipitated). A commercial NGS sequencing kit (Oxford Nanopore Technologies (ONT); Oxford, United Kingdom) was used to generate the NGS library, and sequencing using the ONT MinKNOW software v24.02.8 was performed. Bioinformatic and statistical analyses using various types of software were conducted using the retrieved and processed data. Initial filtering of data was conducted using AptaSUITE v0.9.8. Distance computing and tree analyses were conducted using UGENE Unipro v50. Other statistical analyses were conducted using BlueSky Statistics v10.4.3.
3. Results and Discussion
To investigate whether primer modification affected the sequences amplified in the enriched aptamer pools, the raw sequencing data obtained from two aptamer pools (presence and absence of modified (5′-biotin-(CH
2)
6) forward primers) was analysed using AptaSUITE, including a cluster analysis conducted using the AptaCluster built-in tool [
11,
12]. Supplementary results are detailed in
S2 of the Supplementary Material.
3.1. AptaSUITE Analysis
Tabulated results from AptaSUITE analysis, detailed in
Supplementary Material S2;
Table S2(a), shows the sequencing output results obtained from the ONT MinKNOW software, with the total number of reads generated and base called.
A similar number of raw reads were generated for both pools, with similar mean quality score and average read lengths (
Table S2(b), Supplementary S2), indicating a broadly similar sequencing behaviour from both pools. However, following quality control and filtering, differences between the two pools were evident. A significant proportion of sequence reads were removed from the original. FASTQ input file retrieved from sequencing, largely due to filtering due to primer mismatches, i.e., the presence of expected 5′ and 3′ primers is checked, and the subsequent sequence is removed as a result of primer mismatches (dependent on the set primer tolerance). Thus, a primer tolerance of 5 resulted in 98.3% of sequences being removed from the biotinylated pool, while 90.9% of the unmodified pool was subsequently removed. This was mainly attributed to sequences containing a 5′ primer error, which were significant in both pools, but were higher in the biotinylated pool. This may indicate possible primer-dependent bias, mismatches during amplification, or sequencing errors.
After filtering, 563 suitable sequences were present in the biotinylated pool and 2842 in the unmodified pool. While a slightly lower amount of the biotinylated pool was inputted for sequencing compared to the unmodified pool (9.8 ng vs. 12.8 ng,
Supplementary S1), this input difference should not result in a nearly four-fold difference in filtered reads between these two pools. A potential reason for this disparity may be attributed more to the presence of the 5′-phosphoamaridite biotin group which may inhibit the T4 DNA ligase (used in conjunction with the sourced NGS kit) from accessing the phosphate group of the double-stranded DNA (dsDNA) for adapter and barcode ligation during the library preparation step of NGS. A similar occurrence during enzymatic reactions has been reported with a 5′-DMT group conjugated onto dsDNA [
9]. The 5′-phosphoamaridite biotin group used in this study may cause steric hindrance, preventing the transfer of the adenylyl group and thus the formation of phosphodiester bonds between the relevant adapter and/or barcode and the dsDNA of interest [
13]. Thus, this AptaSUITE-filtered data was subjected to further analysis.
The nucleotide distribution for each of the four bases (A,C,G,T) in the variable regions of both aptamer pools (
Table S2(b), Supplementary S2) showed no significant difference in overall abundance: χ
2(3) = 0.09;
p = 0.993), indicating that the sequences within each filtered pool had largely similar abundances.
3.2. Cluster Sequence Analysis
AptaCluster resolved the two pools into a total of 306 clusters for the biotinylated pool and 1559 individual clusters for the unmodified pool. This is reasonable, given the small edit distance used for clustering (d = 2), used to identify very closely related sequences. Subsequent cluster data was subjected to analysis of nucleotide prevalence, phylogenetic relationships, as well as a principal component analysis (PCA) to identify if primer modification introduced any bias to the sequences enriched within the pool.
Of the clusters resolved for these two pools, only 24 cluster sequences co-occurred in both pools, possibly due to the stringent clustering parameters used. Co-occurrent sequences accounted for ~4.6% of the sequences read within the unmodified pool and ~9.5% of the biotinylated pool. Enriched clusters (i.e., with multiple sequences within each pool) comprise ~9.3% of the unmodified pool’s reads (contained within 60 clusters) and ~8.6% of the biotinylated pool (16 clusters). The disparity in enriched clusters is likely due to the significantly smaller number of filtered reads present in the biotinylated pools.
Co-occurrence within the two pools themselves represents evidence of enrichment of these sequences, as separate fractions of the same aptamer pool were used to create the unmodified and biotinylated sequence pools in this study. Enriched sequences that co-occur comprise a large proportion of the enriched clusters of both pools: approximately 39% of the unmodified pool’s enriched sequences and 56% of the biotin pools. However, the indication of large numbers of significantly enriched sequences that do not co-occur may indicate that selective enrichment caused by amplification with the two different forward primers might have occurred.
Table S2(c) (Supplementary S2) presents the AptaCluster-calculated diversity and counts per million (CPM) values for the top 10 most-abundant clusters common and enriched in both pools, and
Table S2(d) (
Supplementary S2) compares these parameters between pools, using Spearman coefficient correlations for the diversity and prevalence (CPM) values of both pools’ top 10 most-common cluster sequences. A moderate correlation can be observed between the pools’ prevalence (R
2 = 0.502), whilst a weak correlation exists between the diversities (R
2 = 0.129). This indicates a difference in the enrichment of clusters within each pool, which may be largely owing to the disparity in the number of filtered reads between the biotinylated and unmodified pools. This provides evidence of the effect of the 5′ modification employed during PCR described above.
3.2.1. Nucleotide Prevalence
Figure 1 shows the nucleotide prevalence for each base (A,C,G,T) in the variable region of the total sequence clusters generated for both pools.
Figure 1 compares the nucleotide prevalence from each position of the variable region, between the pools. An overall correlation of the nucleotides’ positional prevalence between the pools is evident, consistent with the overall nucleotide compositional similarities (
Section 3.1, (
Table S2(b), Supplementary S2)). Overall, there appears to be a general nucleotide consensus within the variable region sequences between the two pools, with all
Figure 1a–d producing an R
2 value of ~0.99.
3.2.2. Phylogenetic Distance Between Sequences in Both Pools
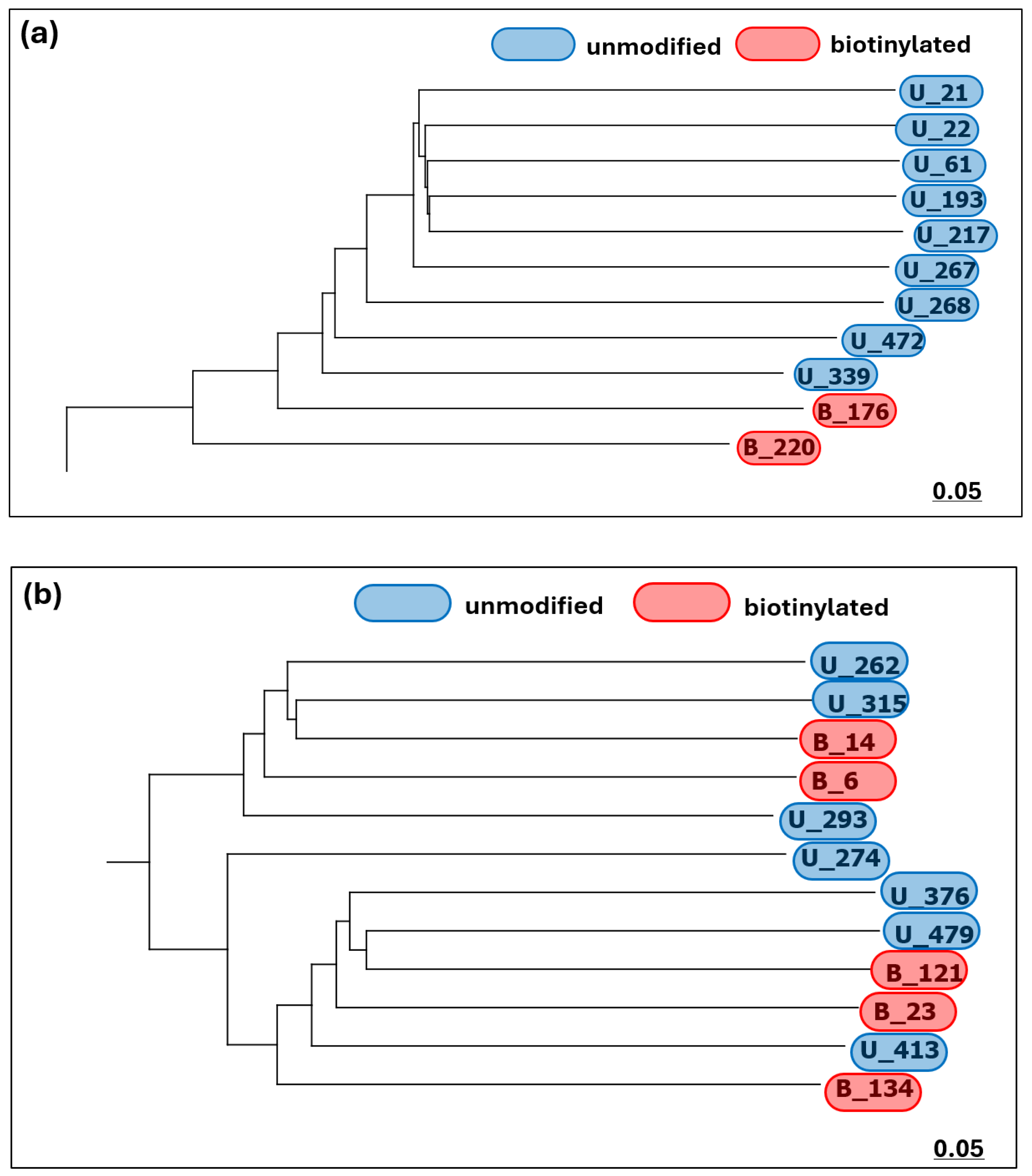
Two different segments of the phylogenetic tree generated from the total sequence cluster data for both pools can be seen in
Figure 2.
Figure 2 represents two different segments belonging to the same generated phylogram, generated with a distance scale of 0.05.
Figure 2a illustrates a set of diverging clades predominantly belonging to unmodified pool sequences, whilst
Figure 2b illustrates a mixture of biotinylated and unmodified pool sequences. While there appears to be some formation of diverging clades among the overall sequences (indicating the presence of genetically diverse sequences within the group, especially observed with the unmodified sequences), there is no distinct clustering based on the modifications of the pools: the above example (
Figure 2b) of the expanded phylogram shows that (given the difference in the numbers of sequences generated by the two pools), a proportionate mixture of unmodified and biotinylated sequences is generally obtained. For a more detailed investigation into the extent of sequence diversity within each sequenced pool, Principal Component Analysis (PCA) was conducted, based on the Hamming distances between each cluster’s main sequence.
3.2.3. Principal Component Analysis (PCA)
Figure 3 exhibits the principal component analysis plots comparing the top 3 components based on the sequence cluster data for both pools. For the unmodified pool, the top 500 clusters were used, due to computational limitations.
Figure 3 exhibits the PCA conducted to further analyse the clustering discussed in
Figure 2. Very few individual clusters appear to resolve by PCA, with the majority of the sequences forming a large central group, when viewed on either Component 1 vs. 2 (
Figure 3a) or Component 2 vs. 3 (
Figure 3b): the distant cluster in the top-right of each PCA potentially indicates a disparate grouping of sequences. However, enriched clusters (discussed in
Section 3.2) found in both groups tended to co-occur; these are visible as the numerous darker large circles (indicating co-occurrence) appearing in both of the plots. Despite the co-occurrence, numerous enriched sequences in the unmodified pools do not appear to have a closely related biotinylated counterpart, and many of the non-enriched sequences show no co-occurrence, indicating that subtle differences in the clustered sequences occur between these two different pools. Due to the differences in the number of cluster sequences, further studies are required to validate this.
4. Conclusions
This study was conducted to determine the effect of a 5′ primer biotin modification on an aptamer pool that was previously enriched towards hCG via 9 rounds of SELEX. Overall, the findings from this work indicate that there does not appear to be a significant difference in the sequence profiles, clustering patterns or phylogenetics between the two primer pools. However, PCA suggested subtle cluster formation that differed due to pool modifications. Given the disparity in the number of sequences filtered through AptaSUITE, which followed through to cluster analysis, there may be an amplification or sequencing bias based on the biotin primer modification which may interfere with subsequent sequencing and analysis and thus requires further evaluation.
Future research objectives include refining the aptamer selection based on prior SELEX pools and assessing the effect of primer modification upon binding to hCG. Aptamer candidates that meet the requirements of the refined selection process will be considered for their end application in the development of commercialisable diagnostics.
Supplementary Materials
The following supporting information can be downloaded at
https://www.mdpi.com/article/10.3390/engproc2025109007/s1, Table S1 Cycling parameters used in PCR, Table S2(a): Sequencing output results from the Oxford Nanopore Technologies MinKNOW software, Table S2(b): General statistics of initial reads compared to AptaSUITE analysis, Table S2(c): Prevalence statistics of the top 10 most-abundant cluster sequences common to both pools (biotinylated and unmodified), Table S2(d): Spearman’s rank correlation coefficients correlating diversity or CPM measurements of the prevalent clusters and comparing them between both biotinylated and unmodified pools. References [
10,
14,
15,
16] are cited in the supplementary materials.
Author Contributions
Conceptualization, J.L., R.F. and T.S.; methodology, T.S. and R.F.; validation, T.S., R.F. and J.L.; formal analysis, R.F. and T.S.; investigation, T.S.; resources, J.L.; data curation, T.S. and R.F.; writing—original draft preparation, T.S.; writing—review and editing, R.F., T.S. and J.L.; visualisation, T.S. and R.F.; supervision, R.F. and J.L.; project administration, J.L. funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the DSI/NRF South African Research Chair in Biotechnology Innovation & Engagement, grant number 95319. TS acknowledges the National Research Foundation (NRF) for postgraduate funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is available upon reasonable request.
Acknowledgments
This research was supported by access to the Nano-Micro Manufacturing Facility, funded by the Department of Science and Innovation (South African Research Infrastructure Roadmap).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; nor in the decision to publish the results.
References
- Bayat, P.; Nosrati, R.; Alibolandi, M.; Rafatpanah, H.; Abnous, K.; Khedri, M.; Ramezani, M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie 2018, 154, 132–155. [Google Scholar] [CrossRef] [PubMed]
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef] [PubMed]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Amaya-González, S.; López-López, L.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Affinity of aptamers binding 33-mer gliadin peptide and gluten proteins: Influence of immobilization and labeling tags. Anal. Chim. Acta 2015, 873, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Challier, L.; Miranda-Castro, R.; Barbe, B.; Fave, C.; Limoges, B.; Peyrin, E.; Ravelet, C.; Fiore, E.; Labbé, P.; Coche-Guérente, L.; et al. Multianalytical Study of the Binding between a Small Chiral Molecule and a DNA Aptamer: Evidence for Asymmetric Steric Effect upon 3′-versus 5′-End Sequence Modification. Anal. Chem. 2016, 88, 11963–11971. [Google Scholar] [CrossRef] [PubMed]
- Klose, A.M.; Miller, B.L. A Stable Biotin-Streptavidin Surface Enables Multiplex, Label-Free Protein Detection by Aptamer and Aptamer-Protein Arrays Using Arrayed Imaging Reflectometry. Sensors 2020, 20, 5745. [Google Scholar] [CrossRef] [PubMed]
- Amaya-González, S.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Aptamer Binding to Celiac Disease-Triggering Hydrophobic Proteins: A Sensitive Gluten Detection Approach. Anal. Chem. 2014, 86, 2733–2739. [Google Scholar] [CrossRef] [PubMed]
- Aird, D.; Ross, M.G.; Chen, W.S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011, 12, R18. [Google Scholar] [CrossRef] [PubMed]
- Shchur, V.V.; Burankova, Y.P.; Zhauniarovich, A.I.; Dzichenka, Y.V.; Usanov, S.A.; Yantsevich, A.V. 5′-DMT-protected double-stranded DNA: Synthesis and competence to enzymatic reactions. Anal. Biochem. 2021, 617, 114115. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.; Flanagan, S.P.; Fogel, R.; Limson, J.L. Generation of epitope-specific hCG aptamers through a novel targeted selection approach. PLoS ONE 2024, 19, e0295673. [Google Scholar] [CrossRef] [PubMed]
- Hoinka, J.; Backofen, R.; Przytycka, T.M. AptaSUITE: A Full-Featured Bioinformatics Framework for the Comprehensive Analysis of Aptamers from HT-SELEX Experiments. Mol. Ther.-Nucleic Acids 2018, 11, 515–517. [Google Scholar] [CrossRef] [PubMed]
- Hoinka, J.; Berezhnoy, A.; Sauna, Z.E.; Gilboa, E.; Przytycka, T.M. AptaCluster—A method to cluster HT-SELEX aptamer pools and lessons from its application. In Research in Computational Molecular Biology; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2014; Volume 8394, pp. 115–128. [Google Scholar] [CrossRef]
- Lehman, I.R. DNA Ligase: Structure, Mechanism, and Function: The joining of DNA chains by DNA ligase is an essential component of DNA repair. replication, and recombination. Science 1974, 186, 790–797. [Google Scholar] [CrossRef] [PubMed]
- ThermoFisher Scientific. Sodium Acetate Precipitation of Small Nucleic Acids. Available online: https://www.thermofisher.com/za/en/home/references/protocols/nucleic-acid-purification-and-analysis/dna-protocol/sodium-acetate-precipitation-of-small-nucleic-acids.html (accessed on 9 September 2025).
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP-Phylogeny Inference Package (Ver. 3.2). Cladistics 1989, 5, 164–166. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).