Design, Synthesis and Studies of Novel Imidazoles †
Abstract
:1. Introduction
2. Materials and Methods
- Docking Strategies:
- Molecular Modelling Studies:
- i.
- Protein Preparation:The X-ray-co-crystallized structures of all of the protein molecules (PDB ID: 1RT2, 2VF5) used in the study were retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) [26]. From every protein molecule, co-crystallized water molecules were deleted and polar hydrogens were added as well as Gasteiger charges assigned, and it was saved in PDBQT format using AutoDock 4.2.6 software.
- ii.
- Ligand Preparation:All of the ligands were prepared by minimizing their energies using PRODRG server [27]. PDBQT formats of all of the ligands were saved.
- iii.
- Receptor grid Generation:Autogrid was used to generate specific grid maps for each and every ligand. The generation of the grid box was carried out by taking the dimensions of the three coordinates (X, Y, and Z) at 24 × 24 × 24, with grid spacing of 0.100 Å. The values of X, Y, and Z centers were taken according to the crystallographic positions of the native ligand of each receptor.
- iv.
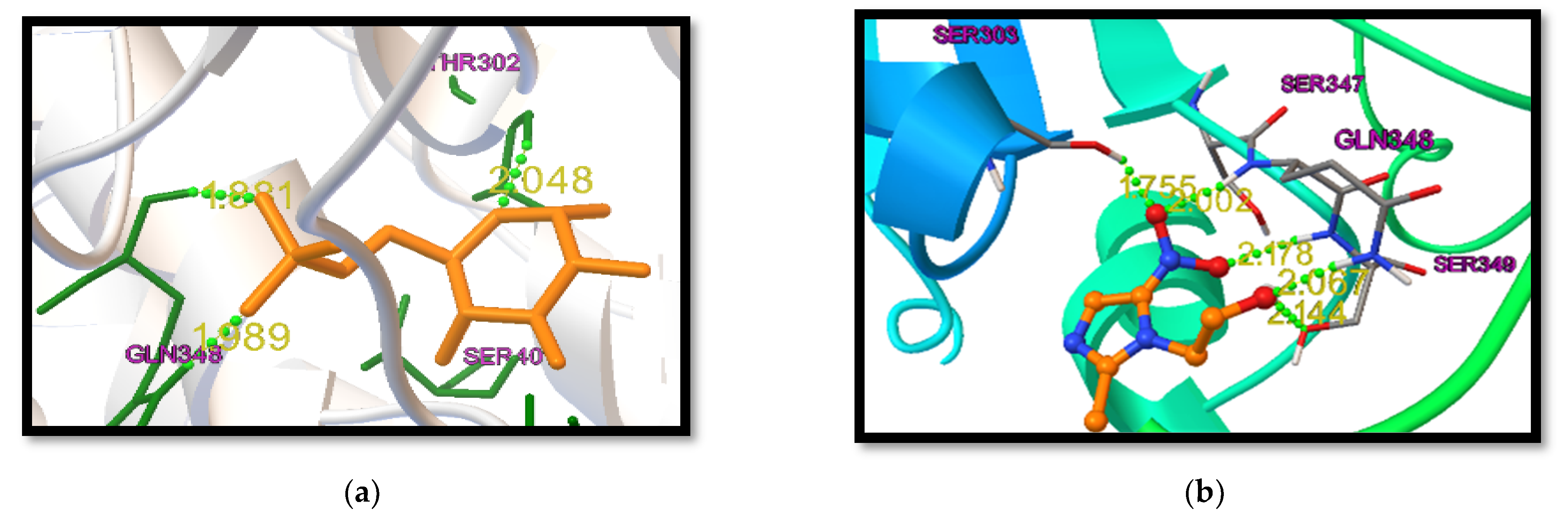
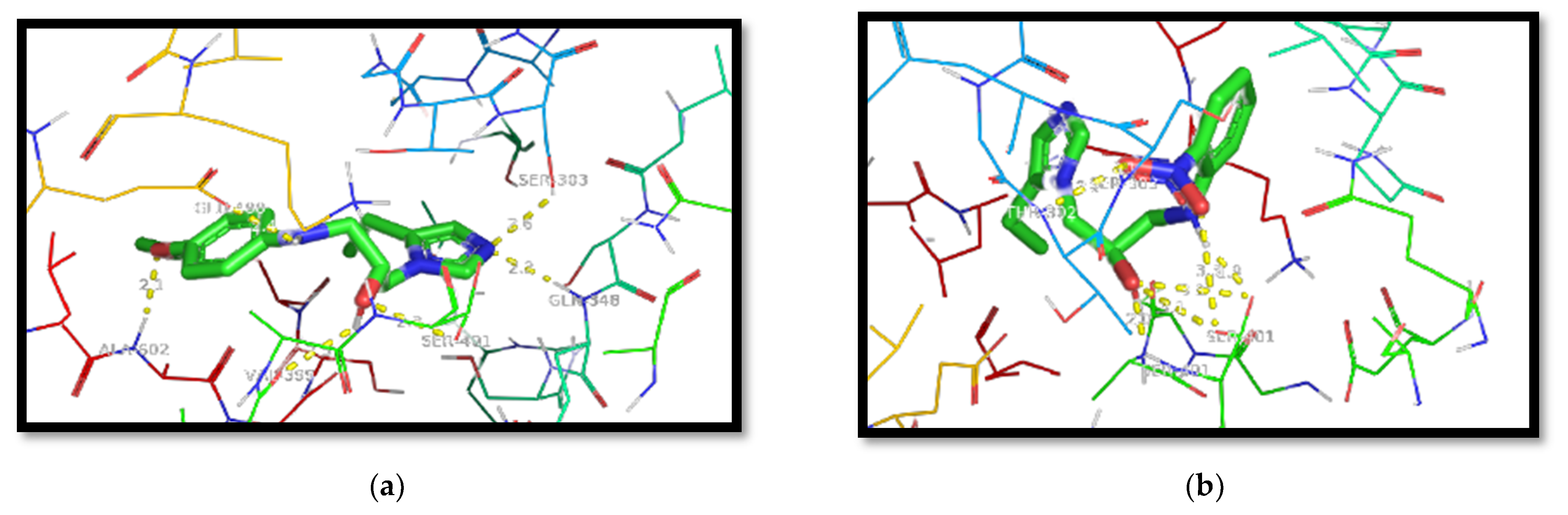
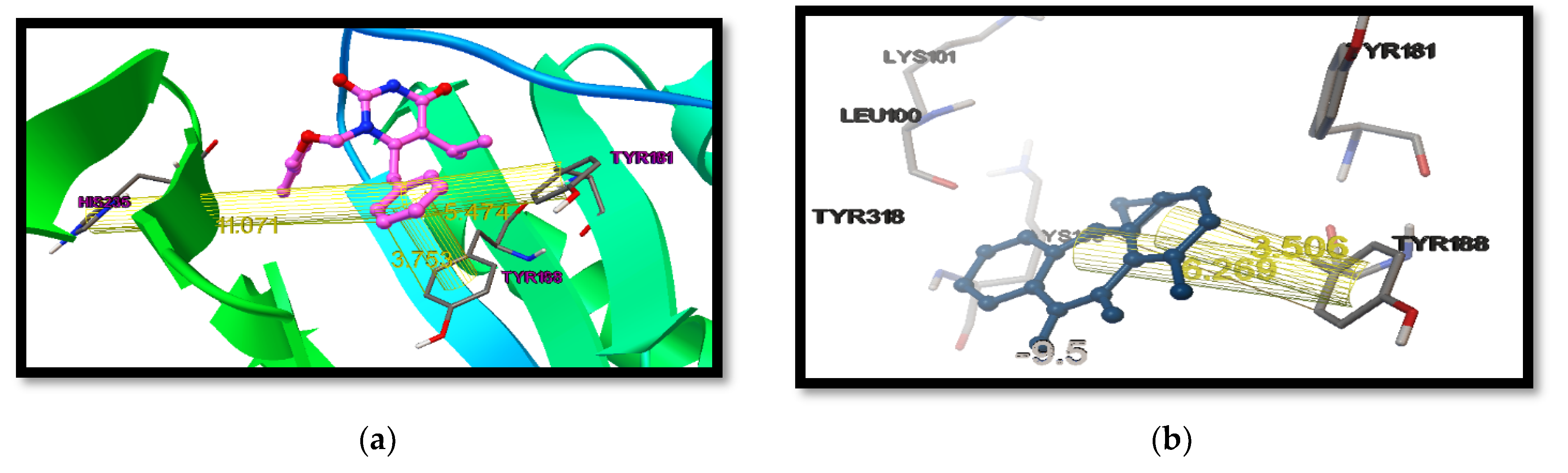
- Docking Protocol Validation:For computational studies, AutoDock 4.2.6 was used. This software was used to predict the different binding mode of co-crystallized ligands as well as test molecules with all of the receptors taken to carry out the study. To carry out the docking procedure, the method was validated to check the robustness of the software. The extracted ligand (previously mentioned) was corrected and then it was redocked using the same protein. The standard drugs were docked into the active site of the respective receptors along with the other test molecules using the same procedure; thereafter, the different conformations were compared. The generated docking scores and conformations of the co-crystallized ligand and the standard drugs were compared with the docking scores of other test molecules to choose the best molecule.
3. Predictive ADME Studies
4. Results and Discussion
Predictive ADME Studies-
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Block, J.H., Jr.; Beale, J.H. Wilson and Gisvold’s Textbook of Organic Medicinal and Pharmaceutical Chemistry, 11th ed.; Lippincotts Williams and Wilkins: Philadelphia, PA, USA, 2004; p. 504. [Google Scholar]
- Butler, K.; Howes, H.L.; Lynch, J.E.; Pirie, D.K. Nitroimidazole Derivatives. Relationship between Structure and Antitrichomonal Activity. J. Med. Chem. 1967, 10, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Tweit, R.C.; Muir, R.D.; Ziecina, S. Nitroimidazoles with Antibacterial Activity against Neisseria Gonorrhoeae. J. Med. Chem. 1977, 20, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Mital, A. Synthetic Nitroimidazoles: Biological Activities and Mutagenicity Relationships. Sci. Pharm. 2009, 77, 497–520. [Google Scholar] [CrossRef] [Green Version]
- Edwards, D. Nitroimidazole drugs-action and resistance mechanisms: Mechanism of action. J. Antimicrob. Chemother. 1993, 31, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Raether, W.; Hänel, H.; Ha, H. Nitroheterocyclic Drugs with Broad Spectrum Activity. Parasitol. Res. 2003, 90 (Suppl. 1), S19–S39. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.S.; Wilkinson, S.R. Activation of benznidazole by trypanosomal type I nitroreductases results in glyoxal formation. Antimicrob. Agents Chemother. 2012, 56, 115–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finar, I.L. Stereochemistry and chemistry of natural products. Org. Chem. 2002, 2, 622–629. [Google Scholar]
- Veraldi, S. Isoconazole nitrate: A unique broad-spectrum antimicrobial azole effective in the treatment of dermatomycoses, both as monotherapy and in combination with corticosteroids. Mycoses 2013, 56, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Ganellin, C.R. Analogue-Based Drug Discovery; John Wiley & Sons: New York, NY, USA, 2006; p. 509. ISBN 9783527607495. [Google Scholar]
- Seidman, L.S.; Skokos, C.K. An evaluation of Butoconazole nitrate 2% Site Release® vaginal cream (Gynazole-1®) compared to fluconazole 150 mg tablets (Diflucan®) in the time to relief of symptoms in patients with vulvovaginal candidiasis. Infect. Dis. Obstet. Gynecol. 2005, 13, 197–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigger-Alberti, W.; Kluge, K.; Elsner, P. Clinical effectiveness and tolerance of climbazole containing dandruff shampoo in patients with seborrheic scalp eczema. Praxis 2001, 90, 346–1349. [Google Scholar]
- Becker, K.L. Principle and Practice of Endrocrinology and Metabolism; Lippincotts Williams and Wilkins; Wiley: Philadelphia, PA, USA, 2001; p. 1197. ISBN 9780781717502. [Google Scholar]
- Croxtall, J.D.; Plosker, G.L. Sertaconazole. Drugs 2009, 69, 339–359. [Google Scholar] [CrossRef] [PubMed]
- Alomar, A.; Videla, S.; Delgadillo, J.; Gich, I.; Izquierdo, I.; Forn, J.; Muntañola, A.A.; Masoliver, A.A.; Corominas, A.B.; Montesinos, E.B.; et al. Flutrimazole 1% dermal cream in the treatment of dermatomycoses: A multicentre, double-blind, randomized, comparative clinical trial with bifonazole 1% cream. Dermatology 1995, 190, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Rovati, R.A.; Mestres, R.C. Dichloro-Substituted Imidazole Derivatives as Antifungal Agents. U.S. Patent 07887103, 19 May 1993. [Google Scholar]
- Hiroyoshi, K.; Yoshiki, N.; Masanori, Y. Antifungal Agent, Its Compound, Production Thereof and Its Usage. Japan Patent 1997100279A, 15 April 1997. [Google Scholar]
- Turner, B.G.; Summers, M.F. Structural biology of HIV. J. Mol. Biol. 1999, 285, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Gupta, S.; Singh, J.; Arsi, T. Azoles as non-nucleoside reverse transcriptase inhibitors (NNRTIs): Mini review. Int. J. Pharm. Sci. Res. 2017, 8, 29. [Google Scholar]
- De Clercq, E. New approaches toward anti-HIV chemotherapy. J. Med. Chem. 2005, 48, 1297–1313. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. The design of drugs for HIV and HCV. Nat. Rev. Drug Discov. 2007, 6, 1001–1018. [Google Scholar] [CrossRef]
- Miller, M.W.; Howes, H.L.; Kasubick, R.V.; English, A.R. Alkylation of 2-methyl-5- nitroimidazole. Some potent antiprotozoal agents. J. Med. Chem. 1970, 13, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Mouilleron, S.; Badet-Denisot, M.A.; Golinelli-Pimpaneau, B. Ordering of C-terminal loop and glutaminase domains of glucosamine-6-phosphate synthase promotes sugar ring opening and formation of the ammonia channel. J. Mol. Biol. 2008, 377, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, W.; Morris, G.M.; Weber, C.; Huey, R. AutoDock 4. The Scripps Research Institute; Molecular Graphics Laboratory: La Jolla, CA, USA, 2008. [Google Scholar]
- Hopkins, A.L.; Ren, J.; Esnouf, R.M.; Willcox, B.E.; Jones, E.Y.; Ross, C.; Miyasaka, T.; Walker, R.T.; Tanaka, H.; Stammers, D.K.; et al. Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J. Med. Chem. 1996, 39, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- RCSB PDB. Available online: http://www.rcsb.org/pdb (accessed on 10 February 2021).
- Prodrg 2 Server. Available online: http://prodrg1.dyndns.org/ (accessed on 15 April 2021).
- SwissADME. Available online: http://www.swissadme.ch (accessed on 17 May 2021).
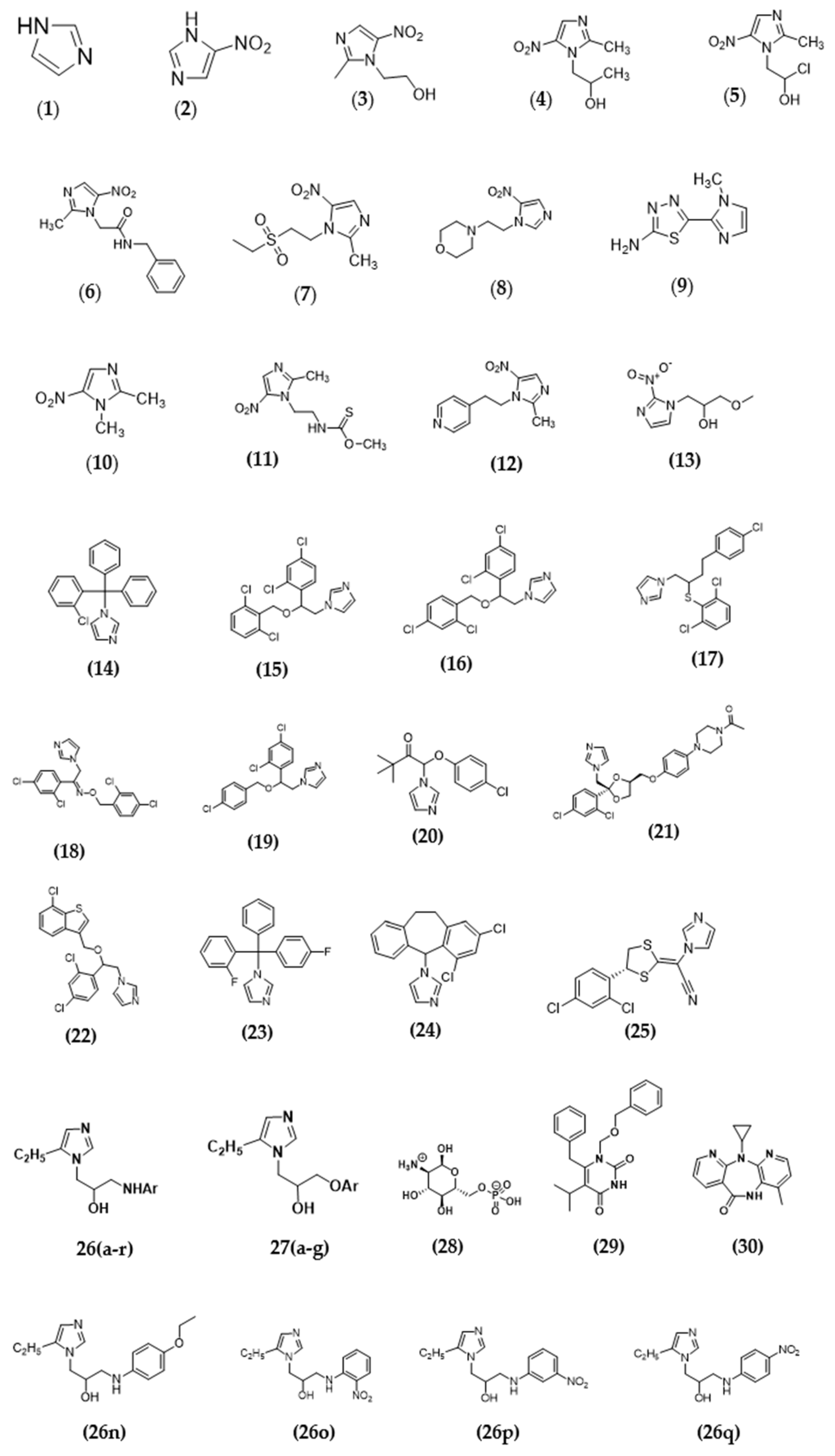

| Compound | Ar | Compound | Ar | Compound | Ar |
|---|---|---|---|---|---|
| 26a | 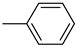 | 26g | 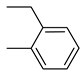 | 26m | 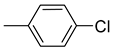 |
| 26b | 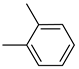 | 26h | 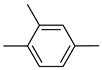 | 26n | 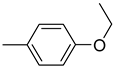 |
| 26c | 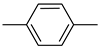 | 26i | 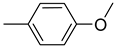 | 26o | 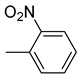 |
| 26d | 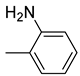 | 26j |  | 26p | 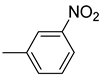 |
| 26e | 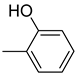 | 26k | 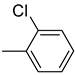 | 26q | 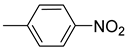 |
| 26f | 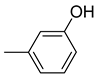 | 26l | 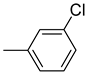 | 26r | 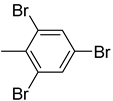 |
| Compound | 27a | 27b | 27c | 27d | 27e | 27f | 27g |
|---|---|---|---|---|---|---|---|
| Ar | 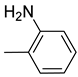 | 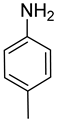 |  | 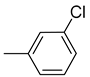 | 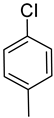 | 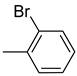 | 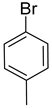 |
| Compound | Docking Scores on | |
|---|---|---|
| 1RT2 | 2VF5 | |
| Native Ligand | −11.9 | −7.9 |
| Standard Drug | −9.5 | −7.5 |
| 26a | −8.3 | −6.3 |
| 26b | −8.6 | −6.2 |
| 26c | −8.3 | −6.4 |
| 26d | −7.9 | −6.8 |
| 26e | −7.9 | −6.8 |
| 26f | −7.9 | −6.6 |
| 26g | −8.6 | −6.6 |
| 26h | −8.4 | −6.4 |
| 26i | −7.8 | −6.1 |
| 26j | −7.9 | −6.1 |
| 26k | −8.3 | −6.3 |
| 26l | −8.6 | −6.7 |
| 26m | −7.9 | −6.7 |
| 26n | −7.9 | −6.7 |
| 26o | −8.1 | −7.4 |
| 26p | −8.2 | −7.1 |
| 26q | −8.3 | −7.4 |
| 26r | −5.2 | −6.6 |
| 27a | −8.3 | −7.2 |
| 27b | −8.2 | −7.2 |
| 27c | −8.7 | −7.0 |
| 27d | −8.7 | −7.4 |
| 27e | −8.1 | −7.1 |
| 27f | −8.3 | −7.0 |
| 27g | −8.0 | −7.0 |
| Compound | Mol. Wt. | HBA | HBD | MR | TPSA | Log P O/W | Solubility (mg/)mL | Lipinski | Veber’s | Leadlikeness |
|---|---|---|---|---|---|---|---|---|---|---|
| 26a | 245.32 | 4 | 2 | 72.28 | 35.5 | 1.30 | 1.30 | Yes | Yes | No |
| 26b | 259.35 | 4 | 2 | 77.63 | 35.5 | 1.56 | 1.55 | Yes | Yes | Yes |
| 26c | 259.35 | 4 | 2 | 77.63 | 35.5 | 1.56 | 2.01 | Yes | Yes | Yes |
| 26d | 260.33 | 5 | 3 | 75.02 | 61.52 | 0.47 | 2.24 | Yes | Yes | Yes |
| 26e | 261.32 | 5 | 5 | 73.48 | 55.73 | 0.47 | 1.52 | Yes | Yes | Yes |
| 26f | 261.32 | 5 | 3 | 73.48 | 55.73 | 0.47 | 1.52 | Yes | Yes | Yes |
| 26g | 273.37 | 4 | 2 | 82.44 | 35.5 | 1.81 | 9.23 | Yes | Yes | Yes |
| 26h | 273.37 | 4 | 2 | 82.99 | 35.5 | 1.81 | 9.17 | Yes | Yes | Yes |
| 26i | 275.35 | 5 | 2 | 78.21 | 44.73 | 0.73 | 7.09 | Yes | Yes | Yes |
| 26j | 277.39 | 4 | 2 | 80.24 | 74.3 | 1.3 | 3.15 | Yes | Yes | Yes |
| 26k | 279.77 | 4 | 2 | 77.11 | 35.5 | 1.56 | 1.99 | Yes | Yes | Yes |
| 26l | 279.77 | 4 | 2 | 77.11 | 35.5 | 1.56 | 1.99 | Yes | Yes | Yes |
| 26m | 279.77 | 4 | 2 | 77.11 | 35.5 | 1.56 | 1.99 | Yes | Yes | Yes |
| 26n | 289.37 | 5 | 2 | 83.01 | 44.73 | 0.98 | 4.15 | Yes | Yes | No |
| 26o | 290.32 | 7 | 2 | 74.47 | 38.74 | 0.15 | 8.55 | Yes | Yes | Yes |
| 26p | 290.32 | 7 | 2 | 74.47 | 38.74 | 0.15 | 8.55 | Yes | Yes | Yes |
| 26q | 290.32 | 7 | 2 | 74.47 | 38.74 | 0.15 | 8.55 | Yes | Yes | Yes |
| 26r | 482.21 | 4 | 2 | 96 | 35.5 | 2.41 | 3.81 | Yes | Yes | No |
| 27a | 261.32 | 3 | 2 | 74.46 | 73.3 | 0.44 | 1.59 | Yes | Yes | Yes |
| 27b | 280.75 | 3 | 1 | 75.06 | 47.28 | 1.53 | 1.93 | Yes | Yes | Yes |
| 27c | 280.75 | 3 | 1 | 75.06 | 47.28 | 1.53 | 1.93 | Yes | Yes | Yes |
| 27d | 280.75 | 3 | 1 | 75.06 | 47.28 | 1.53 | 1.93 | Yes | Yes | Yes |
| 27e | 325.2 | 3 | 1 | 77.75 | 47.28 | 1.65 | 1.09 | Yes | Yes | Yes |
| 27f | 325.2 | 3 | 1 | 77.75 | 47.28 | 1.65 | 1.09 | Yes | Yes | Yes |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandra, P.; Ganguly, S.; Ghosh, M. Design, Synthesis and Studies of Novel Imidazoles. Chem. Proc. 2022, 8, 78. https://doi.org/10.3390/ecsoc-25-11628
Chandra P, Ganguly S, Ghosh M. Design, Synthesis and Studies of Novel Imidazoles. Chemistry Proceedings. 2022; 8(1):78. https://doi.org/10.3390/ecsoc-25-11628
Chicago/Turabian StyleChandra, Priyanka, Swastika Ganguly, and Manik Ghosh. 2022. "Design, Synthesis and Studies of Novel Imidazoles" Chemistry Proceedings 8, no. 1: 78. https://doi.org/10.3390/ecsoc-25-11628
APA StyleChandra, P., Ganguly, S., & Ghosh, M. (2022). Design, Synthesis and Studies of Novel Imidazoles. Chemistry Proceedings, 8(1), 78. https://doi.org/10.3390/ecsoc-25-11628







