Abstract
Here, we report the optimization of a methodology using docking and virtual screening to identify novel clinical uses for already approved drugs. The molecular targets selected were MvfR and PqsD due to their crucial role in quorum sensing and biofilm formation and development. The FDA-approved subset of the ZINC database was screened after careful validation of the virtual screening protocol, and molecules obtained in the top 1% for each target were further analyzed. Presented here are the top five molecules selected for each target.
1. Introduction
Pseudomonas aeruginosa is an opportunistic Gram-negative bacterium, responsible for acute and chronic infections. It is a highly adaptable pathogen, and it is becoming extremely difficult to eradicate due to acquired resistance and tolerance to drugs [1,2]. This bacterium can be found in planktonic state or in an association called biofilm, the ultimate method of protection in adverse conditions [3]. Biofilms are an association of microorganisms organized within a self-produced extracellular polymeric substance matrix. This matrix confers stability and works like a protective armor against antimicrobial compounds as well as providing increased virulence that often leads to chronic infections [4,5,6,7].
Like many other bacterial species, P. aeruginosa can control the expression of genes, population density and biofilm formation through a process called quorum sensing (QS). Quorum sensing is a cell–cell communication mechanism controlled by the release, detection, and response of signaling molecules called autoinducers. It controls, among other aspects, biofilm formation and the transcription of several virulence genes [8].
Quorum sensing in P. aeruginosa is rather complex and hierarchical. It uses four types of signaling systems, two of which are based on acyl homoserine lactones (LasR, RhlR); one that uses quinolone as signaling molecules (PQS) and one whose mechanism and targets are still unknown (IQS) [9,10]. The LasR system is at the top of the hierarchy, but integration with the RhIR and PQS systems is fundamental as a regulatory link to control the direct and indirect expression of several virulence genes [11] Targeting the QS system will not kill the bacteria, but it will hamper its pathogenicity and the possibility of resistance is diminished as there is less selective pressure on the bacteria [12].
The focus of this work is the PQS system, more specifically the proteins PqsR (also known as Multiple Virulence Factor Regulator—MvfR) and PqsD. PqsD is a Anthraniloyl-CoA anthraniloyltransferase required for the biosynthesis of several signaling molecules such as HHQ. It catalyzes the transfer of the anthraniloyl moiety from antraniloyl-CoA to malonyl-CoA to form 2-aminobenzoylacetyl-CoA (2-ABACoA). This involves the formation of a covalent bond between Cys112 and antraniloyl-CoA [13]. MvfR is a transcriptional regulator responsible for the transcription of virulence genes). It interacts with two native ligands: 2-Heptyl-3-hydroxy-4(1H)-quinolone (also called the Pseudomonas Quinolone Signal, PQS), and its precursor 2-heptyl-4- hydroxyquinoline (HHQ). It controls its own activity by upregulating the expression of genes in the pqsABCDE and phnAB operons which encode other enzymes [1]. Studies have shown that interfering with PqsR and PqsD leads to a more efficient attenuation of pathogenicity than single target approaches [14].
In this work, a docking and virtual screening (VS) protocol was applied to discover new inhibitors for MvfR and PqsD proteins, using the ZINC FDA-approved database as starting point. Drug repurposing is becoming an attractive approach to the drug discovery process since the repurposed drugs are, safe, already in use, and well characterized, reducing the drug-development time and cost [15].
2. Materials and Methods
2.1. Docking Protocol Validation
The Protein Databank [16] and the Biofilms Structural database [17] were explored to find molecular structures of MvfR and PqsD. A total of 12 strucutres for MvfR and 3 for PqsD were found. All fifteen X-ray structures were prepared for docking using Pymol, with the extraction of water molecules and crystallographic ligands (these were saved in separate files to be used as reference in the following steps). For MvfR, there is a variety of X-ray structures and X-ray ligands; however, that is not the case for PqsD, as there are only 3 protein structures and 2 ligands.
For this work, the docking software GOLD [18] was used (with all its scoring functions (SFs): CHEMPLP, GoldScore, ChemScore and ASP). The purpose of testing all the different scoring functions was to evaluate which one is the best for these specific hydrophobic targets, as it has been demonstrated that docking results can vary significantly depending on the type of protein target and ligand [19,20]. The docking conditions were the same for every SF and every target to ensure consistency and reproducibility. The optimized conditions consisted of binding site coordinates and radius, number of runs and search efficiency. The protocol described was applied separately for MvfR and PqsD.
As the first step in the protocol validation, re-docking was performed to evaluate the ability of the docking software to reproduce the geometry and orientation of the crystallographic pose. The root mean square deviation (RMSD) between the heavy atoms of the crystallographic and docked poses was calculated, and the resulting scores were evaluated. The docking conditions were optimized with the goal of obtaining the lowest RMSD possible. The next step toward protocol validation was to perform cross-docking. This strategy, as a measure of robustness of target structures and methodology, is quite simple to perform. All the crystallographic ligand structures isolated from the protein structures of both targets were “docked” into the different X-ray structures. This test aimed to evaluate the ability of individual X-ray structures in enabling the correct docking of different X-ray ligands, co-crystallized in other X-ray structures. The RMSDs in both cases was calculated using DockRMSD [21]. A good result is the one that presents a high positive score and a RMSD below 2 Å.
2.2. Virtual Screening Protocol Validation
For this stage, all the structures that presented mutations were removed. Only the best structures obtained in the docking protocol validation stage were selected to move on to the VS protocol validation (4JVI and 6B8A for MvfR and 3H76 and 3H77 for PqsD). The VS protocol was validated with a benchmark dataset to ensure that it provides reliable results. For MvfR and PqsD, a specific virtual screening training library was prepared to evaluate and optimize the ability of the protocol in discriminating between binders and non-binders. After an initial query in the ChEMBL [22] and BindingDB [23] databases and a brief literature review, 40 molecules with experimental activity against MvfR were found; this was the active pool of the test set. Using the DUD-E [24] database, a set of 50 decoys for each ligand was created. Decoys are molecules that resemble the ligands in their physical properties but are chemically and topologically different, so they are most likely non-binders. The total number of decoys generated was 2000. The final test set for MvfR was composed of 2040 compounds. The same protocol was followed for PqsD and the final dataset was composed of 59 active molecules and 2950 decoys.
The discriminatory ability of the five scoring functions was assessed and the evaluation metrics were calculated using a web-based application, Screening Explorer [25], as well as Excel. The metrics used for the evaluation of the VS results were the enrichment factor at 1% (EF 1%), receiver operating characteristic (ROC) curves, respective area under the curve (AUC), and total gain (TG). TG quantifies the discrimination of actives over decoys attributable to score variations. TG values over 0.25 combined with an AUC over 0.5 indicate a good performance and reproducibility from the VS protocol [25].
2.3. Virtual Screening of ZINC FDA Approved Compounds
At this stage, only the best SFs and the X-ray structures that yielded a better actives/decoys discrimination in the validation stage was selected.
FDA-approved drugs are a subset of ZINC [26] a free database of commercially available compounds for virtual screening. ZINC contains over 230 million purchasable compounds. At the time of the VS experiments, the FDA-approved drugs dataset had 3207 compounds that were all docked against the target. The top five compounds for each of the protein targets were selected to move on to further studies.
3. Results and Discussion
The two X-ray structures from each target, which provided the highest scores and lowest RMSD values (data not shown) in the re-docking and cross-docking stage, were selected to move on to the VS stage.
Table 1 summarizes the results obtained for these two chosen structures for all the SFs tested, for MvfR. CHEMPLP, ChemScore and ASP provided good discriminatory ability between binders and non-binders for both structures with an EF1% of 10.40. However, CHEMPLP did not provide a good TG value for both structures. The TG value for ASP and 4JVI structure was also not satisfactory. Ultimately, the best TG value was obtained for structure 6B8A and ASP SF, and that was the combination that was used in VS of the FDA approved compounds.

Table 1.
Evaluation metrics for virtual screening results for X-ray structures for MvfR (4JVI and 6B8A).
The same analysis was performed for PqsD and the results are presented in Table 2. In this case, the SFs that provided the best results across all the metrics were CHEMPLP and GoldScore in structure 3H76. Because the AUC of CHEMPLP is slightly higher, that was the SF selected to move on to the next stage.

Table 2.
Evaluation metrics for virtual screening results for x-ray structures for PqsD (3H76 and 3H77).
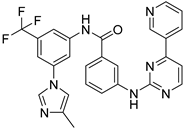
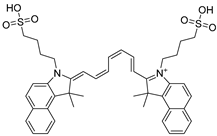
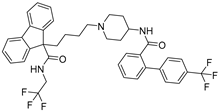
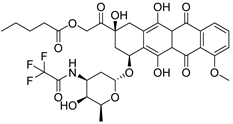
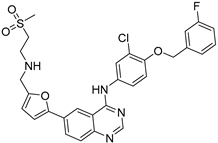
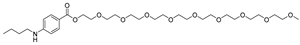
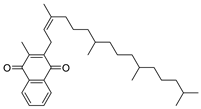
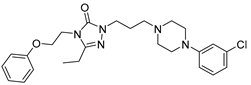
After performing the VS protocol for the ZINC FDA-approved database, only the molecules present in the top 1% were analyzed, corresponding to a total of 30 compounds for each protein target. Table 3 lists the top five results obtained for MvfR, and Table 4 lists the top five compounds for PqsD. A brief description of the pharmaceutical use of each compound is provided, along with the score that was obtained in the VS. Different SFs use different metrics and scales, hence the difference between the ASP and CHEMPLP scores.

Table 3.
Top 5 hits of the FDA-approved drugs database for MvfR.

Table 4.
Top 7 hits of the FDA-approved drugs database for PqsD.
The top five molecules selected for each target are different structurally, with the molecules for PqsD presenting, in general, a higher molecular weight and long aliphatic tails. This is to be expected, as the characteristics of each binding pocket are different. However, two compounds also stood out. Lapatinib, one of the top five results for PqsR, was also present in the top 10 molecules for PqsD. The same occurred with Salmeterol (a top-five result for PqsD) which was also one of the top 20 molecules for PqsR. These are two strong candidates for dual inhibition.
4. Conclusions
A docking protocol was optimized using the crystallographic ligand as a validation tool for the reproducibility of the pose generated by the docking software. The virtual screening protocol was adjusted to obtain the best discriminatory ability between known binders and non-binders, and it was applied to a database of 3207 FDA-approved compounds for both MvfR and PqsD targets.
The top five compounds of each database obtained using the optimized VS protocol were presented and described. Further computational studies will be performed for all these compounds, using molecular dynamics simulation and free energy calculations, to confirm the docking binding predictions and stability of protein–ligand complexes. Additionally, experimental testing must be performed to confirm the quality and predictability of this in silico protocol. This optimized protocol can also be used in the future to screen additional chemical libraries in the search for novel drug candidates targeting MvfR and PqsD.
Author Contributions
Conceptualization, S.F.S.; methodology, S.F.S.; software, T.V.; validation, T.V., R.M. and S.F.S.; formal analysis, S.F.S.; investigation, T.V. and R.M.; resources, S.F.S.; writing—original draft preparation, T.V.; writing—review and editing, S.F.S.; visualization, S.F.S.; supervision, S.F.S.; project administration, S.F.S.; funding acquisition, S.F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by national funds from Fundação para a Ciência e a Tecnologia [grant numbers UIDP/04378/2020 and UIDB/04378/2020, SFRH/BD/137844/2018, 2020.01423.CEECIND]. Some of the calculations were produced with the support of INCD funded by FCT and FEDER under project 01/SAICT/2016 number 022153 and projects CPCA/A00/7140/2020 and CPCA/A00/7145/2020.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The compounds are available at the ZINC database of compound cited in the publication, available at https://zinc.docking.org/ accessed on 28 January 2021.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Allegretta, G.; Maurer, C.K.; Eberhard, J.; Maura, D.; Hartmann, R.W.; Rahme, L.; Empting, M. In-depth Profiling of MvfR-Regulated Small Molecules in Pseudomonas aeruginosa after Quorum Sensing Inhibitor Treatment. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Camara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Kamaruzzaman, N.F.; Tan, L.P.; Yazid, K.A.M.; Saeed, S.I.; Hamdan, R.H.; Choong, S.S.; Wong, W.K.; Chivu, A.; Gibson, A.J. Targeting the Bacterial Protective Armour; Challenges and Novel Strategies in the Treatment of Microbial Biofilm. Materials 2018, 11, 1705. [Google Scholar] [CrossRef] [Green Version]
- Jamal, M.; Tasneem, U.; Hussain, T.; Andleeb, S. Bacterial Biofilm: Its Composition, Formation and Role in Human Infections. Res. Rev. J. Microbiol. Biotechnol. 2015, 4, 1–14. [Google Scholar]
- Dufour, D.; Leung, V.; Lévesque, C.M. Bacterial biofilm: Structure, function, and antimicrobial resistance. Endod. Top. 2010, 22, 2–16. [Google Scholar] [CrossRef]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, S.; Sen, S.K.; Pattanaik, S.; Raut, S. Review on bacterial biofilm: An universal cause of contamination. Biocatal. Agric. Biotechnol. 2016, 7, 56–66. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576. [Google Scholar] [CrossRef]
- Li, S.; Chen, S.; Fan, J.; Cao, Z.; Ouyang, W.; Tong, N.; Hu, X.; Hu, J.; Li, P.; Feng, Z.; et al. Anti-biofilm effect of novel thiazole acid analogs against Pseudomonas aeruginosa through IQS pathways. Eur. J. Med. Chem. 2018, 145, 64–73. [Google Scholar] [CrossRef]
- Abelyan, N.; Grabski, H.; Tiratsuyan, S. Identification of flavone and its derivatives as potential inhibitors of transcriptional regulator LasR of Pseudomonas aeruginosa using virtual screening. bioRxiv 2019, 523381. [Google Scholar] [CrossRef] [Green Version]
- Lazdunski, A.M.; Ventre, I.; Sturgis, J.N. Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2004, 2, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Reuter, K.; Steinbach, A.; Helms, V. Interfering with Bacterial Quorum Sensing. Perspect. Med. Chem. 2016, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storz, M.P.; Maurer, C.K.; Zimmer, C.; Wagner, N.; Brengel, C.; De Jong, J.C.; Lucas, S.; Müsken, M.; Häussler, S.; Steinbach, A.; et al. Validation of PqsD as an Anti-biofilm Target in Pseudomonas aeruginosa by Development of Small-Molecule Inhibitors. J. Am. Chem. Soc. 2012, 134, 16143–16146. [Google Scholar] [CrossRef] [PubMed]
- Thomann, A.; Brengel, C.; Börger, C.; Kail, D.; Steinbach, A.; Empting, M.; Hartmann, R.W. Structure-Activity Relationships of 2-Sufonylpyrimidines as Quorum-Sensing Inhibitors to Tackle Biofilm Formation and eDNA Release ofPseudomonas aeruginosa. ChemMedChem 2016, 11, 2522–2533. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Berman, H.M.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Magalhães, R.P.; Vieira, T.F.; Fernandes, H.S.; Melo, A.; Simões, M.; Sousa, S.F. The Biofilms Structural Database. Trends Biotechnol. 2020, 38, 937–940. [Google Scholar] [CrossRef]
- Jones, G.H.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef] [Green Version]
- Vieira, T.F.; Sousa, S.F. Comparing AutoDock and Vina in Ligand/Decoy Discrimination for Virtual Screening. Appl. Sci. 2019, 9, 4538. [Google Scholar] [CrossRef] [Green Version]
- Tf, V.; Rp, M.; Sf, S. Tailoring Specialized Scoring Functions For More Efficient Virtual Screening. Front. Drug Chem. Clin. Res. 2019, 2, 1–4. [Google Scholar] [CrossRef]
- Bell, E.W.; Zhang, Y. DockRMSD: An open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J. Cheminform. 2019, 11, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaulton, A.; Hersey, A.; Nowotka, M.; Bento, A.P.; Chambers, J.; Mendez, D.; Mutowo, P.; Atkinson, F.; Bellis, L.J.; Cibrián-Uhalte, E.; et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016, 44, D1045–D1053. [Google Scholar] [CrossRef] [PubMed]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Empereur-Mot, C.; Zagury, J.-F.; Montes, M. Screening Explorer–An Interactive Tool for the Analysis of Screening Results. J. Chem. Inf. Model. 2016, 56, 2281–2286. [Google Scholar] [CrossRef]
- Sterling, T.; Irwin, J.J. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Choulis, N.H. Miscellaneous drugs, materials, medical devices, and techniques. Side Eff. Drugs Annu. 2009, 757–769. [Google Scholar] [CrossRef]
- Alonso, R.; Cuevas, A.; Mata, P. Lomitapide: A review of its clinical use, efficacy, and tolerability. Core Évid. 2019, 14, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Opdam, F.L.; Guchelaar, H.; Beijnen, J.H.; Schellens, J.H.M. Lapatinib for Advanced or Metastatic Breast Cancer. Oncologist 2012, 17, 536–542. [Google Scholar] [CrossRef] [Green Version]
- Davis, R.; Whittington, R.; Bryson, H.M. Nefazodone. Drugs 1997, 53, 608–636. [Google Scholar] [CrossRef]
- Anwar, M.; El-Haggar, R.S.; Zaghary, W.A. Salmeterol Xinafoate. Profiles Drug Subst. Excip. Relat. Methodol. 2015, 40, 321–369. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, J.; Huang, M.; Zhang, X. Efficacy and safety of polidocanol in the treatment of varicose veins of lower extremities. Medicine 2021, 100, e24500. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).