In silico Evaluation of Antimicrobial Activity of Some Thiadiazoles Using Molecular Docking Approach †
Abstract
1. Introduction
2. Computational Methodology—Docking Protocol
3. Results and Discussion
4. Conclusions
References
- Oruç, E.E.; Rollas, S.; Kandemirli, F.; Shvets, N.; Dimoglo, A.S. 1,3,4-Thiadiazole derivatives. synthesis, structure elucidation, and structure−antituberculosis activity relationship investigation. J. Med. Chem. 2004, 47, 6760–6767. [Google Scholar] [CrossRef] [PubMed]
- Serban, G.; Stanasel, O.; Serban, E.; Bota, S. 2-Amino-1,3,4-thiadiazole as a potential scaffold for promising antimicrobial agents. Drug Des. Devel. Ther. 2018, 12, 1545–1566. [Google Scholar] [CrossRef] [PubMed]
- Razus, A.; Birzan, L.; Cristea, M.; Tecuceanu, V.; Draghici, C.; Haganu, A.; Maganu, M.; Pintilie, L.; Ungureanu, E.-M. New (azulen-1-yldiazenyl)-heteroaromatic compounds containing 1,2,5-thiadiazol-3-yl moieties. Rev. Chim. 2019, 70, 1518–1529. [Google Scholar] [CrossRef]
- 3M4I Piton, J.; Petrella, S.; Delarue, M.; Andre-Leroux, G.; Jarlier, V.; Aubry, A.; Mayer, C. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS ONE 2010, 5, e12245. [Google Scholar]
- Grillot, A.L.; Tiran, A.L.; Shannon, D.; Krueger, E.; Liao, Y.; O’Dowd, H.; Li, P. Second-generation antibacterial benzimidazole ureas: Discovery of a preclinical candidate with reduced metabolic liability. J. Med. Chem. 2014, 57, 8792–8816. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.R.; Zhang, X.; Blumenthal, R.M.; Cheng, X. Structures of Escherichia coli DNA adenine methyltransferase (DAM) in complex with a non-GATC sequence: Potential implications for methylation-independent transcriptional repression. Nucleic Acids Res. 2015, 43, 4296–4308. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shao, Y.; Molnar, L.F.; Jung, Y.; Kussmann, J.; Ochsenfeld, C.; Brown, S.T.; DiStasio, R.A., Jr. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [Google Scholar] [CrossRef] [PubMed]
- Hehre, W.J. A Guide to Molecular Mechanics and Quantum Chemical Calculations; Wavefunction, Inc.: Irvine, CA, USA, 2003. [Google Scholar]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Li, C.Y.; Wang, X.M.; Yang, Y.H.; Zhu, H.L. 1,3,4-Thiadiazole: Synthesis, reactions, and applications in medicinal, agricultural, and materials chemistry. Chem. Rev. 2014, 114, 5572–5610. [Google Scholar] [CrossRef] [PubMed]
- Matysiak, J. Biological and pharmacological activities of 1,3,4-Thiadiazole based compounds. Mini Reviews Med. Chem. 2012, 15, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Thomasco, M.; Gadwood, R.C.; Weaver, E.A.; Ochoada, J.M.; Ford, C.W.; Zurenko, G.E.; Hamel, J.C.; Stapert, D.; Moerman, J.K.; Schaadt, R.D.; et al. The synthesis and antibacterial activity of 1,3,4-thiadiazole phenyl oxazolidinone analogues. Bioorg. Med. Chem. Lett. 2003, 13, 4196. [Google Scholar] [CrossRef] [PubMed]
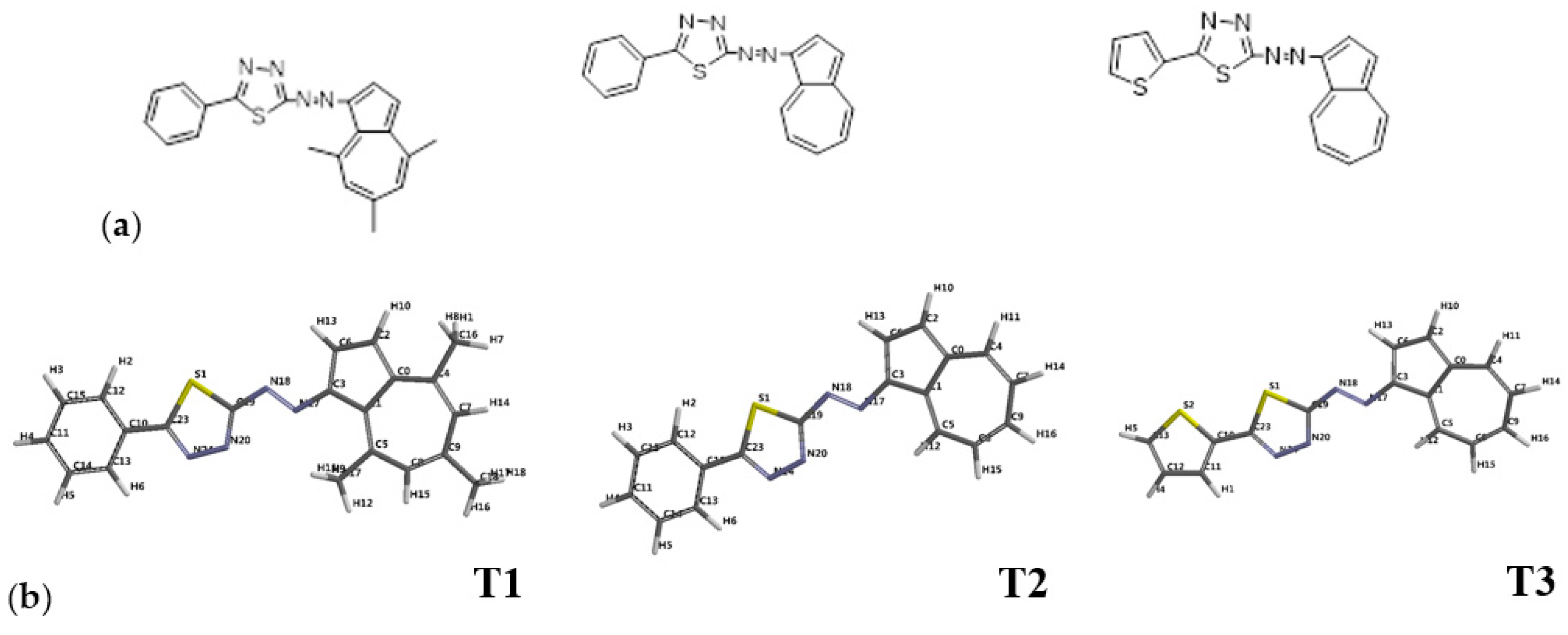
| Ligand/ Protein Fragment Source | MW (g·mol−1) | HBD | HBA | LogP | Flexible Bonds | Lipinski’s Violations |
|---|---|---|---|---|---|---|
| co-crystalized MPDA-1 /3M4I (M. tuberculosis) | 118.17 | 2 | 2 | 0.27 | 2 | 0 |
| co-crystalized 883B 301 /4P8O (S. aureus) | 376.37 | 2 | 8 | 1.61 | 4 | 0 |
| co-crystalized SFG /4RTO (E. coli) | 382.39 | 10 | 12 | −3.22 | 7 | 2 |
| T1 | 358.46 | 0 | 4 | 5.24 | 3 | 1 |
| T2 | 326.46 | 0 | 4 | 5.49 | 3 | 1 |
| T3 | 322.41 | 0 | 4 | 5.21 | 3 | 1 |
| Target/Ligand | Interacting Group | Hydrogen Bonds/Length(Å) | Docking Score/RMSD |
|---|---|---|---|
| 3M4I/co-crystallized MPDA-1 | ARG451, HIS525, PRO450, TYR524, HIS560, GLY520, ILE519, LEU522, ARG523 | O4(sp3)—O(sp2) LEU522/3.302 | −25.91/0.86 |
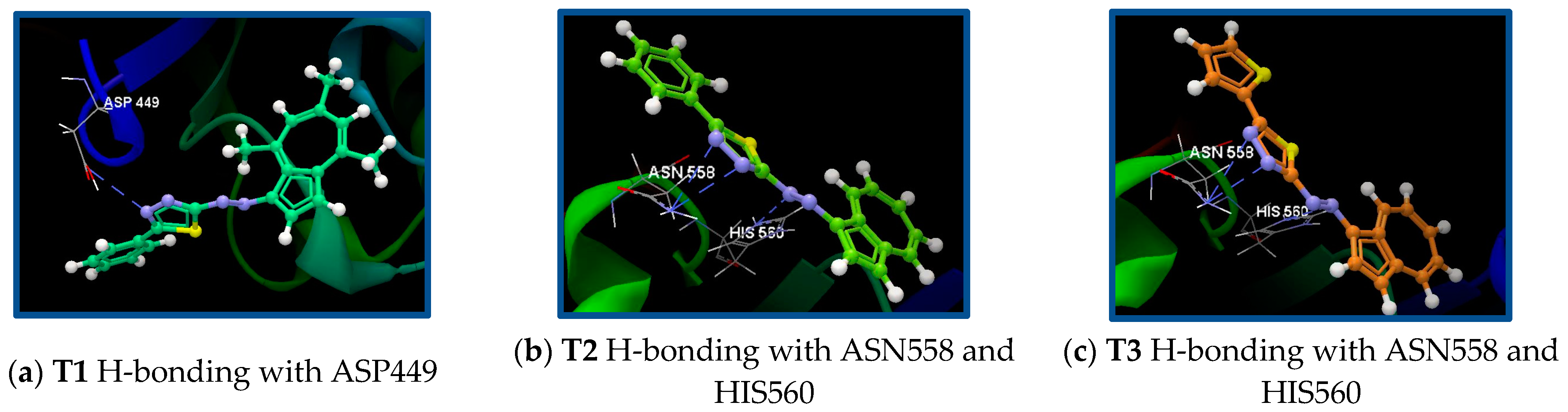
| 3M4I/T1 | ASP449, ARG451, PRO450, TYR524, ARG523, LEU522, LYS521, GLY520, ALA508, LEU509, GLY510, THR507 | N24(sp2)—O(sp3) ASP449/3.247 | −38.19/0.06 |
| 3M4I/T2 | ASN558, HIS560, ILE519, HIS525, ARG451, PRO450, GLY520, TYR524, LEU522, ARG523, LYS521 | N18(sp2)—N(sp2) HIS560/3.057 N20(sp2)—N(sp2) ASN558/3.126 N24(sp2)—N(sp2) ASN558/3.103 | −43.19/0.69 |
| 3M4I/T3 | GLU557, ASN558, HIS560, ILE519, HIS525, ARG451, LYS452, ASP449, PRO450, TYR524, GLY520, LEU522, ARG523, LYS521 | N17 (sp2)—N (sp2) HIS560/3.187 N20 (sp2)—N(sp2) ASN558/2.914 N24 (sp2)—N (sp2) ASN558/3.135 | −40.95/0.72 |
| Target/Ligand | Interacting Group | Hydrogen Bonds/Length (Å) | Docking Score/RMSD |
|---|---|---|---|
| 4P8O/ co-crystallized | ASN54, VAL52, ILE51, ILE102, VAL79, ILE175, VAL174, THR80, THR173, PRO87, GLY85, ASP81, ARG144, ARG84, GLY83, GLU58, SER55, ILE86 | N25(sp2)—N(sp2) ARG144/2.769 N6(sp2)—O(sp2) ASP81/2.797 N3(sp2)—O(sp2) ASP81/2.914 N3(sp2)—O(sp3) SER55/3.081 | −70.22/0.08 |
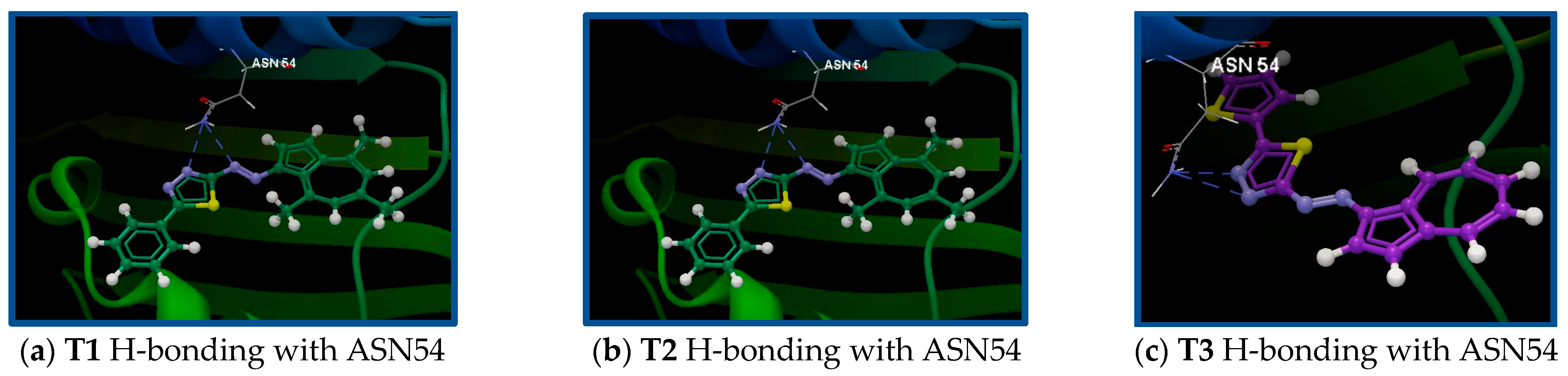
| 4P8O/T1 | SER55, ASN54, GLU58, ASP81, GLY83, GLY172, ARG84, GLY85, ILE86, PRO87, ARG144, ILE102, SER128, THR173 | N20(sp2)—N(sp2) ASN54/3.062 N18(sp2)—N(sp2) ASN54/3.135 | −58.08/0.10 |
| 4P8O/T2 | VAL52, VAL79, ASN54, ILE51, GLU50, SER55, THR80, ASP81, GLU88, GLY83, ARG84, THR173, VAL174, ILE175, GLY85, ARG144, ILE86, PRO87, ILE102 | N24(sp2)—N(sp2) ASN54/2.790 N20(sp2)—N(sp2) ASN54/2.944 | −56.49/0.18 |
| 4P8O/T3 | ASP81, GLU58, GLY83, THR80, SER55, VAL79, ASN54, ILE51, ILE175, VAL174, THR173, ARG84, GLY85, ARG144, ILE86, PRO87, ILE102 | N24(sp2)—N(sp2) ASN54/2.954 N20(sp2)—N(sp2) ASN54/2.903 | −53.61/0.19 |
| Target/Ligand | Interacting Group | Hydrogen Bonds/Length (Å) | Docking Score/RMSD |
|---|---|---|---|
| 4RTO/co-crystalized SFG | ASN56, ILE55, PHE201, GLU163, SER164, GLN205, TYR165, SER168, LEU59, ASP54, PRO183, PHE35, ALA53, PRO182, PRO34, ASP181, GLU33, TYR179, VAL36, TYR184, VAL41, LYS14, SER40, GLY39, GLY13, GLY12, GLY37, ALA38, ALA11, TRP10 | N1 (sp2)—N (sp2) TYR165/3.129 O2′ (sp3)—O (sp2) ASP54/2.654 O3′ (sp3)—O (sp3) ASP54/2.567 O3′ (sp3)—N(sp2) TRP10/3.128 O (sp2)—N (sp2) ALA38/2.834 OXT (sp2)—O (sp3) SER40/2.980 N (sp3)—O s(sp3) ASP181/2.426 | −67.74/0.79 |
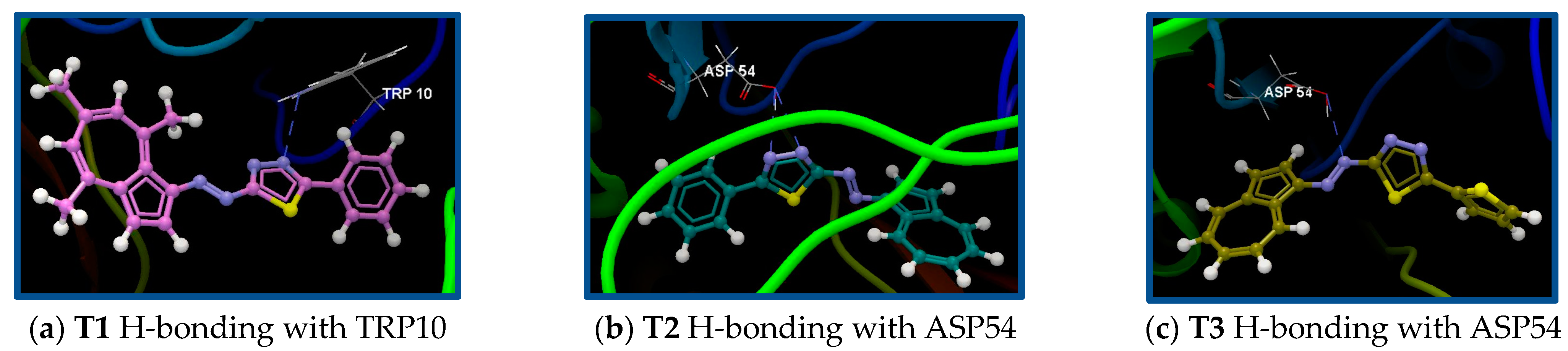
| 4RTO/T1 | ALA53, VAL36, GLU163, PRO34, ASP54, PHE35, ILE55, SER164, TYR165, ALA166, GLN205, PHE201, SER200, PRO183, ASN120, LEU122, CYS123, ALA11, TRP10, LYS59, ASN115, GLY121 | N24 (sp2)—N(sp2) TRP10/3.101 | −72.21/0.07 |
| 4RTO/T2 | ALA53, ASP54, GLU163, PRO34, PHE35, ILE55, SER164, TYR165, GLN205, PHE201, TYR184, PRO183, PRO182, ASP181, ALA11, GLY12, TRP10, GLY13 | N24 (sp2)—O (sp3) ASP54/3.000 N20 (sp2)—O (sp3) ASP54/2.982 | −71.15/0.07 |
| 4RTO/T3 | TRP10, ALA11, GLY12, GLY37, VAL36, ASP54, ALA53, ILE55, GLU163, SER164, PHE35, PRO34, TYR165, ASP181, PRO183, PRO182, SER200, TYR184, PHE201, GLN205 | N18 (sp2)—O (sp3) ASP54/3.304 | −66.42/0.23 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stefaniu, A.; Pintilie, L.; Anastasoaie, V.; Ungureanu, E.-M. In silico Evaluation of Antimicrobial Activity of Some Thiadiazoles Using Molecular Docking Approach. Chem. Proc. 2021, 3, 116. https://doi.org/10.3390/ecsoc-24-08319
Stefaniu A, Pintilie L, Anastasoaie V, Ungureanu E-M. In silico Evaluation of Antimicrobial Activity of Some Thiadiazoles Using Molecular Docking Approach. Chemistry Proceedings. 2021; 3(1):116. https://doi.org/10.3390/ecsoc-24-08319
Chicago/Turabian StyleStefaniu, Amalia, Lucia Pintilie, Veronica Anastasoaie, and Eleonora-Mihaela Ungureanu. 2021. "In silico Evaluation of Antimicrobial Activity of Some Thiadiazoles Using Molecular Docking Approach" Chemistry Proceedings 3, no. 1: 116. https://doi.org/10.3390/ecsoc-24-08319
APA StyleStefaniu, A., Pintilie, L., Anastasoaie, V., & Ungureanu, E.-M. (2021). In silico Evaluation of Antimicrobial Activity of Some Thiadiazoles Using Molecular Docking Approach. Chemistry Proceedings, 3(1), 116. https://doi.org/10.3390/ecsoc-24-08319








