Molecular Docking and Dynamics of a Series of Aza-Heterocyclic Compounds Against Effector Protein NleL †
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Docking Procedure
2.2. Molecular Dynamics Details
3. Results
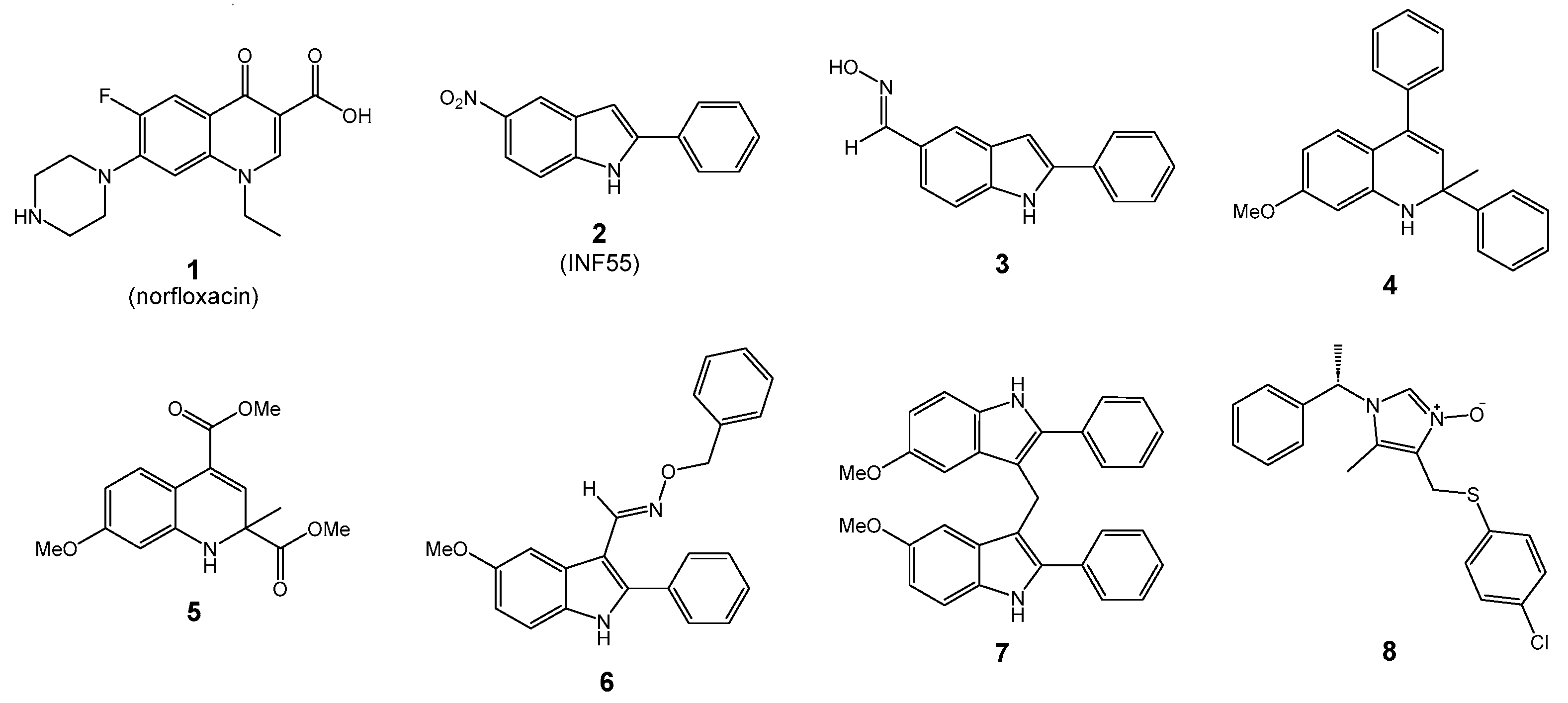
3.1. Molecular Docking Results
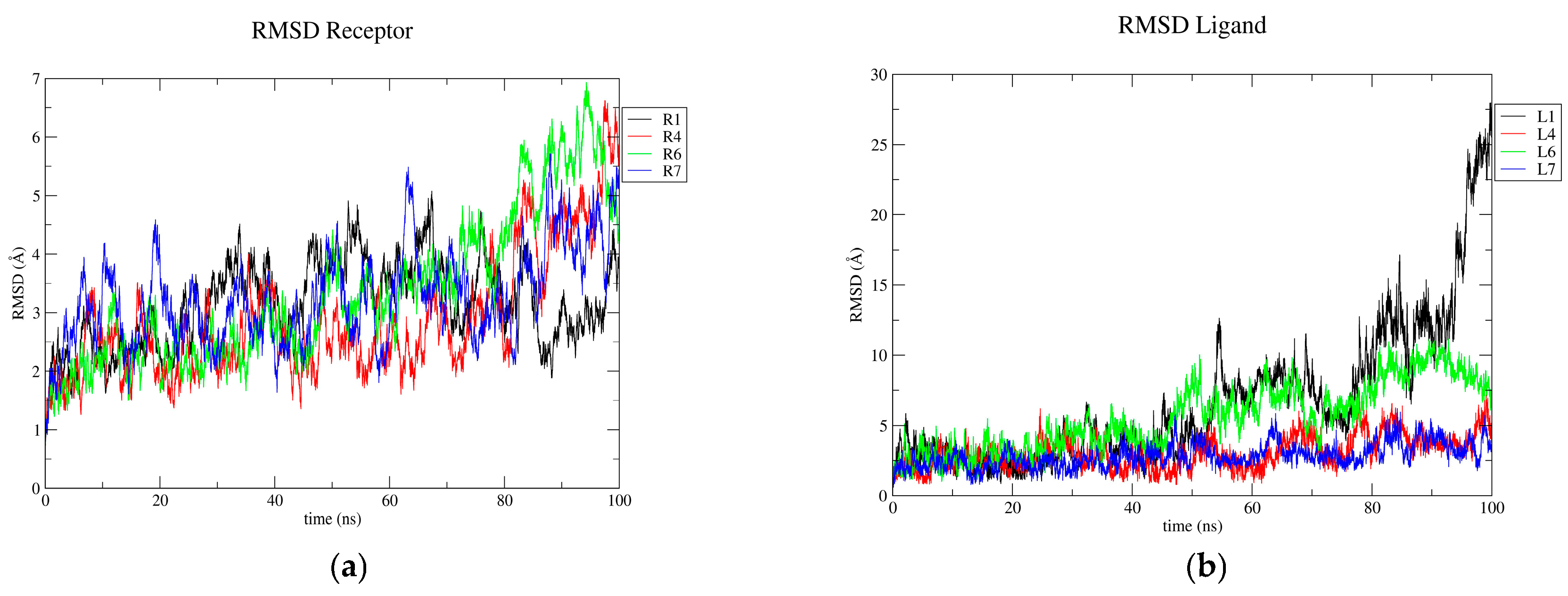
3.2. Molecular Dynamics Results
4. Discussion
4.1. Molecular Docking Analysis
4.2. Molecular Dynamics Simulations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bush, L.M. Infecciones por Escherichia coli. Available online: https://www.msdmanuals.com/es/hogar/infecciones/infecciones-bacterianas-bacterias-gramnegativas/infecciones-por-escherichia-coli (accessed on 10 September 2025).
- World Health Organization. E. coli. Available online: https://www.who.int/es/news-room/fact-sheets/detail/e-coli (accessed on 10 September 2025).
- Lin, D.Y.W.; Diao, J.; Zhou, D.; Chen, J. Biochemical and Structural Studies of a HECT-like Ubiquitin Ligase from Escherichia coli O157:H7. J. Biol. Chem. 2011, 286, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.Y.W.; Diao, J.; Chen, J. Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions. Proc. Natl. Acad. Sci. USA 2012, 109, 1925–1930. [Google Scholar] [CrossRef]
- Zhang, R.; Kennedy, M.A. Current Understanding of the Structure and Function of Pentapeptide Repeat Proteins. Biomolecules 2021, 11, 638. [Google Scholar] [CrossRef]
- Gutiérrez, R.U.; Correa, H.C.; Bautista, R.; Vargas, J.L.; Jerezano, A.V.; Delgado, F.; Tamariz, J. Regioselective Synthesis of 1,2-Dihydroquinolines by a Solvent-Free MgBr2-Catalyzed Multicomponent Reaction. J. Org. Chem. 2013, 78, 9614–9626. [Google Scholar] [CrossRef]
- Gutiérrez, R.U.; Rebollar, A.; Bautista, R.; Pelayo, V.; Várgas, J.L.; Montenegro, M.M.; Espinoza-Hicks, C.; Ayala, F.; Bernal, P.M.; Carrasco, C.; et al. Functionalized α-oximinoketones as building blocks for the construction of imidazoline-based potential chiral auxiliaries. Tetrahedron Asymmetry 2015, 26, 230–246. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.; Thiessen, P.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Wavefunction_Inc. Spartan’20 (Version 20.1.3); Q-CHEM: Pleasanton, CA, USA, 2020. [Google Scholar]
- BIOVIA. Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, 2019; Dassault Systèmes: San Diego, CA, USA, 2019. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F. AutoDockFR Home Page. Available online: https://ccsb.scripps.edu/adfr/ (accessed on 10 September 2025).
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLoS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.; Jo, S.; Brooks III, C.L.; Lee, H.S.; Im, W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017, 38, 1879–1886. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Fletcher, R.; Reeves, C.M. Function minimization by conjugate gradients. Comput. J. 1964, 7, 149–154. [Google Scholar] [CrossRef]
- Martyna, G.J.; Tobias, D.J.; Klein, M.L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994, 101, 4177–4189. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef]
| Ligand | Affinity Energies |
|---|---|
| 1 | −7.2 |
| 2 | −7.4 |
| 3 | −8.2 |
| 4 | −9.3 |
| 5 | −7.7 |
| 6 | −9.0 |
| 7 | −9.1 |
| 8 | −8.3 |
| Ligand | Donor | Acceptor | % of Occupancy |
|---|---|---|---|
| 1 | GLN605 | LIG | 8.60 |
| 4 | LIG | SER604 | 54.46 |
| 6 | LIG | LEU775 | 43.08 |
| LIG | SER604 | 43.14 | |
| VAL779 | LIG | 6.06 | |
| 7 | VAL608 | LIG | 29.60 |
| LIG | LEU775 | 38.58 | |
| LIG | ASN630 | 37.40 |
| Ligand | van der Waals | Binding Free Energies | |
|---|---|---|---|
| 1 | −16.09 | −7.48 | −8.20 |
| 4 | −28.44 | −7.14 | −20.58 |
| 6 | −41.18 | −20.79 | −32.57 |
| 7 | −37.26 | −13.81 | −23.80 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortiz-Vargas, K.A.; Palomares-Báez, J.P.; Aviña-Verduzco, J.A.; García-Gutiérrez, H.A.; Herrera-Bucio, R.; Navarro-Santos, P. Molecular Docking and Dynamics of a Series of Aza-Heterocyclic Compounds Against Effector Protein NleL. Chem. Proc. 2025, 18, 16. https://doi.org/10.3390/ecsoc-29-26894
Ortiz-Vargas KA, Palomares-Báez JP, Aviña-Verduzco JA, García-Gutiérrez HA, Herrera-Bucio R, Navarro-Santos P. Molecular Docking and Dynamics of a Series of Aza-Heterocyclic Compounds Against Effector Protein NleL. Chemistry Proceedings. 2025; 18(1):16. https://doi.org/10.3390/ecsoc-29-26894
Chicago/Turabian StyleOrtiz-Vargas, Karen Astrid, Juan Pedro Palomares-Báez, Judit A. Aviña-Verduzco, Hugo A. García-Gutiérrez, Rafael Herrera-Bucio, and Pedro Navarro-Santos. 2025. "Molecular Docking and Dynamics of a Series of Aza-Heterocyclic Compounds Against Effector Protein NleL" Chemistry Proceedings 18, no. 1: 16. https://doi.org/10.3390/ecsoc-29-26894
APA StyleOrtiz-Vargas, K. A., Palomares-Báez, J. P., Aviña-Verduzco, J. A., García-Gutiérrez, H. A., Herrera-Bucio, R., & Navarro-Santos, P. (2025). Molecular Docking and Dynamics of a Series of Aza-Heterocyclic Compounds Against Effector Protein NleL. Chemistry Proceedings, 18(1), 16. https://doi.org/10.3390/ecsoc-29-26894






