Study of the Stability, Solubility and Geometry of the Complex of Inclusion β-CD with Nimesulide by Computer Chemistry Methods †
Abstract
1. Introduction
2. Computational Chemistry Methods Used
3. Results and Discussion
3.1. Optimization of the Geometry of the Carrier Molecule β-CD
3.2. RMSD Trajectory Analysis
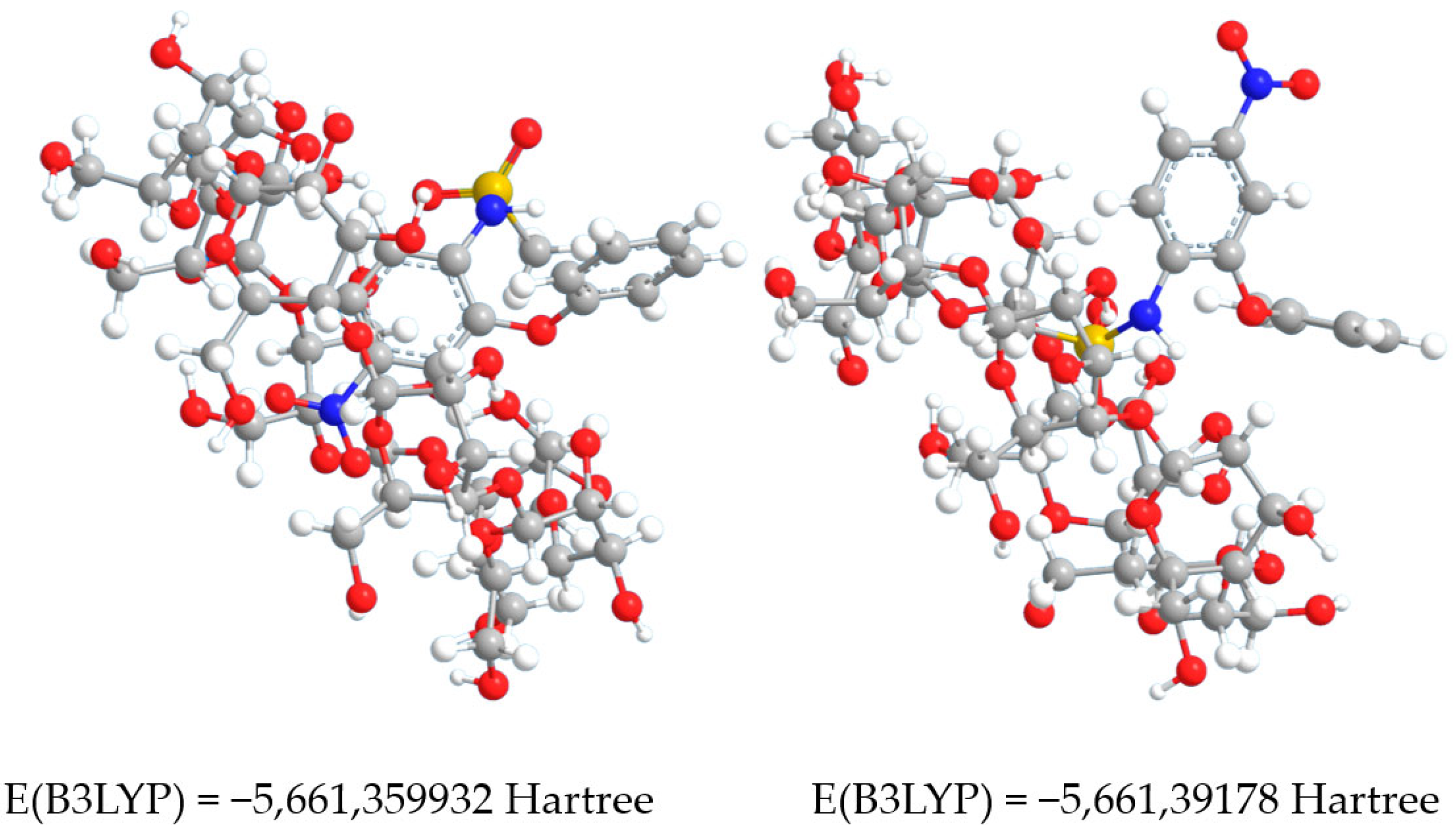
3.3. Taking into Account the Effects of the Concatenation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chan, S.L.; Labute, P. Training a Scoring Function for theAlignment of Small Molecules. J. Chem. Inf. Model. 2010, 50, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Hönig, S.M.N.; Lemmen, C.; Rarey, M. Small molecule superposition: A comprehensive overview on pose scoring of the latest methods. WIREs Comput. Mol. Sci. 2023, 13, e1640. [Google Scholar] [CrossRef]
- Oliveira, T.A.d.; Silva, M.P.d.; Maia, E.H.B.; Silva, A.M.d.; Taranto, A.G. Virtual Screening Algorithms in Drug Development: A Review of Machine and Deep Learning Methods. Drugs Drug Candidates 2023, 2, 311–334. [Google Scholar] [CrossRef]
- Melanie, F.; Gratteri, P.; Adamo, M.; Boachchini, K. Field interaction and geometric overlap: A new computational procedure based on simplex and experimental design for the superimposition of small molecules of ligands. J. Med. Chem. 2003, 46, 1359–1371. [Google Scholar] [CrossRef] [PubMed]
- Cleve, A.E.; Jane, A.N. 3D-ForceGen Conformants Structure and Generation: From small molecules, like lead, to macrocyclic preparations. J. Comput. Aided. Mol. Des. 2017, 31, 419–439. [Google Scholar] [CrossRef] [PubMed]
- Steffen, A.; Apostolakis, J. On the simplicity of predicting thermodynamic properties of complexes involving beta-cyclodextrin. Cent. Chem. J. 2007, 1, 29. [Google Scholar] [CrossRef] [PubMed]
- Rekharsky Mikhail, V.; Inouet, Y. Thermodynamics of cyclodextrins complex formation. Chem. Rev. 1998, 98, 1875–1918. [Google Scholar] [CrossRef] [PubMed]
- Grekhneva, E.V.; Barteneva, E.S.; Efanov, K.S. Peculiarities of Obtaining and Modeling the Structure of Nimesulide Clathrate Complexes with β- and γ-Cyclodextrins. Chem. Proc. 2022, 12, 53. [Google Scholar] [CrossRef]
- Ekaterina, B.; Elena, G.; Kirill, E. Substantiation of the Possibility of Obtaining Complex Including Nimesulide with γ-CD by Computer Modeling Methods. Polym. Sci. Peer. Rev. J. PSPRJ 2023, 4, 000596. [Google Scholar] [CrossRef]
- Andreev, P.Y.; Barteneva, E.S.; Grekhneva, E.V.; Efanov, K.S.; Breskin, K.A. A New Approach to the Preparation of Inclusion Complexes with Cyclodextrins: Studying Their Stability Using Molecular Dynamics Methods. Eng. Proc. 2023, 56, 245. [Google Scholar] [CrossRef]
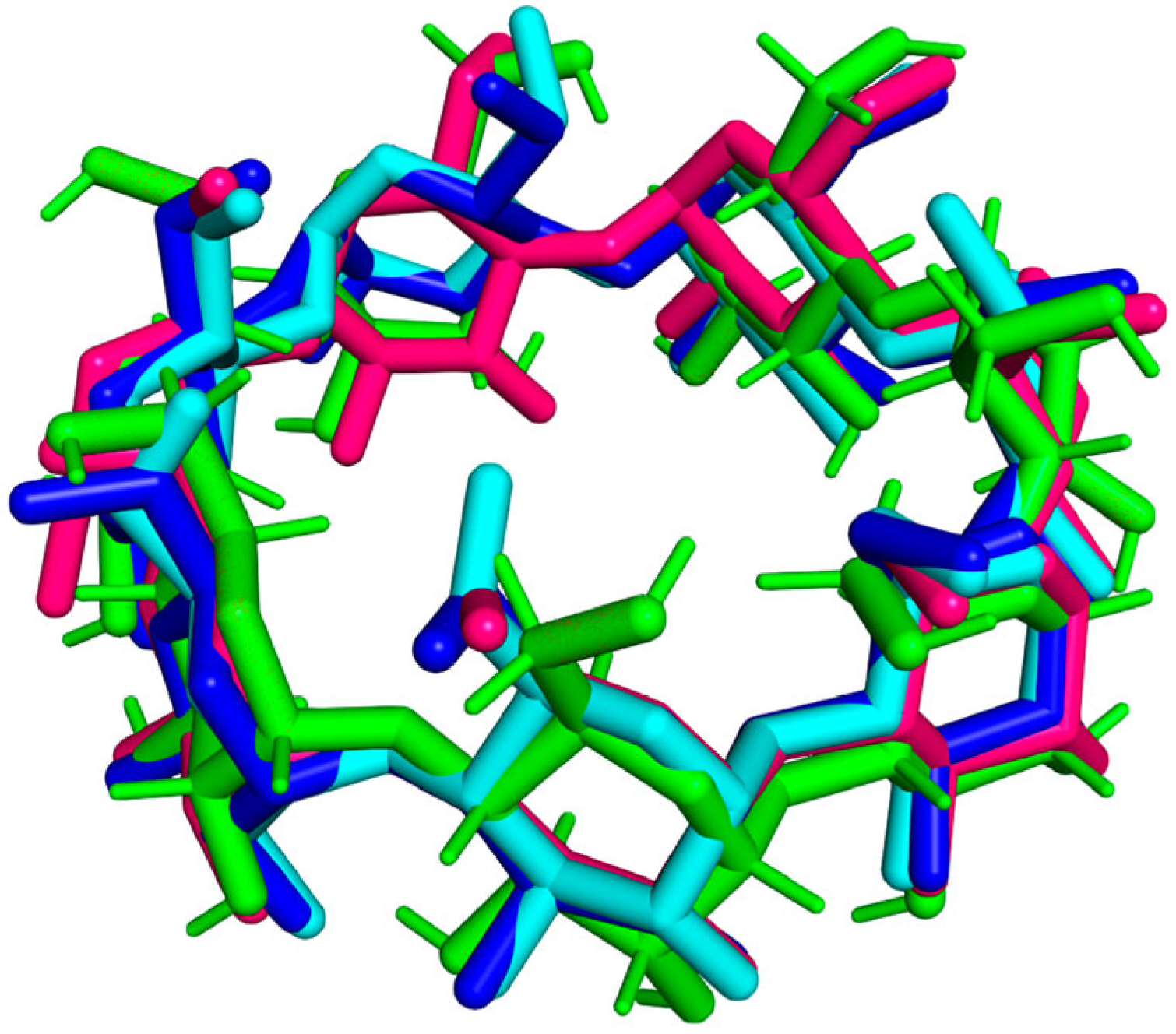
| № | PDB ID | Method | Resolution, Å | RMSD, Å (Atoms of 77) | Atoms Aligned of 77 |
|---|---|---|---|---|---|
| 1 | 1VFO | X-ray | 2.81 | 0.768 | 73 |
| 2 | 2V8L | X-ray | 1.8 | 0.767 | 73 |
| 3 | 2Z1K | X-ray | 2.3 | 0.630 | 74 |
| 4 | 3CGT | X-ray | 2.40 | 0.000 | 77 |
| 5 | 3JUV | X-ray | 3.12 | 0.879 | 73 |
| 6 | 5E6Z | X-ray | 1.878 | 0.759 | 73 |
| 7 | 6JEQ | X-ray | 1.802 | 0.846 | 73 |
| Element Name | Solvation Accounting | S, kcal/mol × K | Sum of Electronic and Thermal Free Energies, Hartree/Particle | Sum of Electronic and Thermal Enthalpies, Hartree/Particle | E(B3LYP), Hartree | Solvation Energy, Hartree |
|---|---|---|---|---|---|---|
| β-CD | - | 383.432 | −4273.99089 | −4273.80871 | −4275.10458 | 0.0696683 |
| PCM | 386.579 | −4274.06354 | −4273.87986 | −4275.17425 | ||
| Nim | - | 143.736 | −1386.05638 | −1385.98809 | −1386.2509 | 0.01683559 |
| PCM | 144.997 | −1386.07425 | −1386.00536 | −1386.26774 | ||
| Complex No.1 | - | 491.886 | −5660.034603 | −5659.800892 | −5661.359932 | 0.08395511 |
| PCM | 498.4 | −5660.12313 | −5659.88632 | −5661.44389 | ||
| Complex No.2 | - | 488.694 | −5660.06363 | −5659.83143 | −5661.39178 | 0.08010679 |
| PCM | 501.576 | −5660.15194 | −5659.91363 | −5661.47189 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barteneva, E.S.; Andreev, P.Y.; Grekhneva, E.V.; Efanov, K.S. Study of the Stability, Solubility and Geometry of the Complex of Inclusion β-CD with Nimesulide by Computer Chemistry Methods. Chem. Proc. 2024, 16, 80. https://doi.org/10.3390/ecsoc-28-20197
Barteneva ES, Andreev PY, Grekhneva EV, Efanov KS. Study of the Stability, Solubility and Geometry of the Complex of Inclusion β-CD with Nimesulide by Computer Chemistry Methods. Chemistry Proceedings. 2024; 16(1):80. https://doi.org/10.3390/ecsoc-28-20197
Chicago/Turabian StyleBarteneva, Ekaterina S., Pavel Y. Andreev, Elena V. Grekhneva, and Kirill S. Efanov. 2024. "Study of the Stability, Solubility and Geometry of the Complex of Inclusion β-CD with Nimesulide by Computer Chemistry Methods" Chemistry Proceedings 16, no. 1: 80. https://doi.org/10.3390/ecsoc-28-20197
APA StyleBarteneva, E. S., Andreev, P. Y., Grekhneva, E. V., & Efanov, K. S. (2024). Study of the Stability, Solubility and Geometry of the Complex of Inclusion β-CD with Nimesulide by Computer Chemistry Methods. Chemistry Proceedings, 16(1), 80. https://doi.org/10.3390/ecsoc-28-20197







