Abstract
The 2019 coronavirus (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has significantly impacted human lives, overburdened the healthcare system, and weakened global economies. The lack of specific drugs against SARS-CoV-2 is a significant hurdle toward the successful treatment of COVID-19. The SARS-CoV-2 Main protease (Mpro) is considered an appealing target because of its role in replication in host cells. Plant-derived natural compounds are being largely tested for their efficacy against COVID-19 targets to combat SARS-CoV-2 infection. To discover hit compounds that can be used alone or in combination with repositioned drugs, we curated a set of 224,205 natural product structures from the ZINC database and virtually screened it against COVID-19 Mpro. Sequential docking protocols involving different levels of exhaustiveness were performed to screen a library of natural compounds. The final 88 compounds were selected and post-processed using the MM-GBSA analysis for the generation of binding free energies. The top four compounds (ZINC000085626103, ZINC000085569275, ZINC000085625768, and ZINC000085488571) showed higher affinity against the COVID-19 Mpro enzyme selected for MD simulation studies. The RMSD, RMSF, and RoG analysis of all four compound–protein complexes indicated absolute stability during a 100 ns MD run. Furthermore, the post-MD simulation binding free energies were calculated for all four compounds and were found to be in the range of −38.29 to −18.07 kcal/mol. The in silico virtual screening results suggested that the selected natural compounds have the potential to be developed as a COVID-19 Mpro inhibitor and can be explored further for experimental research to evaluate the in vitro and in vivo efficacy of these compounds for the treatment of COVID-19.
1. Introduction
Novel coronavirus SARS-CoV-2 caused a worldwide pandemic and remains a severe threat to the entire human population due to the lack of specific therapeutic agents to control the sudden outbreak of SARS-CoV-2 [1]. The SARS-CoV-2 main protease (Mpro) protein is a vital target for drug discovery studies against the recent coronavirus pandemic [2]. In silico screening of phytochemical databases has gained increasing interest in drug discovery research for the identification of new drug leads or drug molecules [3,4]. Virtual screening based on molecular docking emerges as an important tool for obtaining new antiviral molecules, where researchers can use this tool as a complementary approach so that the synthesis of new compounds or the repositioning of drugs can be assigned. The objective of the study is to perform a Virtual Screening of Natural Compounds from the ZINC database to discover possible antiviral agents with protease inhibitory potential against SARS-CoV-2.
2. Result and Discussion
2.1. Docking Studies
In the pursuit of identifying the potential drug candidates targeting the COVID-19 Main Protease (Mpro) enzyme, we conducted a rigorous virtual screening process using docking studies employing the Smina molecular docking software. The sequential docking protocols, involving varying levels of exhaustiveness, were carried out to effectively screen the extensive library of approximately 224,205 natural product structures sourced from the ZINC database. Initially, all compounds were docked on the Mpro enzyme with a default exhaustiveness setting of 8, and subsequently, the top 10% of compounds with the best docking scores were selected for further screening. The obtained subset of approximately 10,000 compounds was further docked on the Mpro enzyme with an exhaustiveness setting of 24. Once again, the top 10% of compounds with the highest docking scores were retained, and in the final phase of screening, approximately 1000 compounds were subjected to rigorous docking simulations, employing an exhaustiveness setting of 48. Ultimately, we selected only the highest-scoring compound from this final set, ensuring the most stringent selection criteria. To calculate binding free energies, shed light on the thermodynamic aspects of the ligand–receptor interactions, and to gain deeper insights into the interactions between the selected 88 compounds and the Mpro enzyme, post-docking MM-GBSA analysis was performed. Based on all the above results, the final four compounds, namely ZINC000085626103, ZINC000085625768, ZINC000085488571, and ZINC000085569275, were selected with the highest docking and MM-GBSA scores for further ligand–enzyme interaction analysis and MD simulation studies (Table 1, Figure 1).

Table 1.
Docking results of selected compounds against Mpro enzyme.
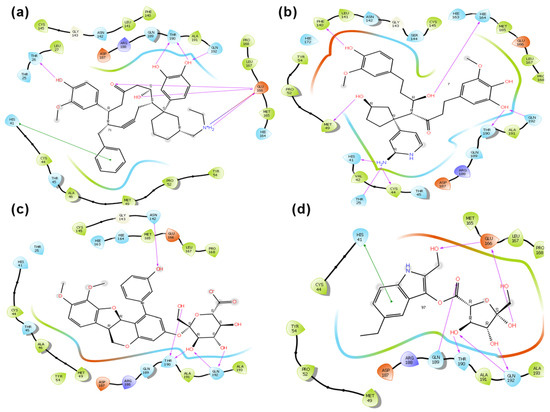
Figure 1.
Two-Dimensional images of (a) ZINC000085626103, (b) ZINC000085625768, (c) ZINC000085488571, and (d) ZINC000085569275 at active site of SARS-CoV-2 main protease (Mpro).
2.2. MD Simulation Studies
Comprehensive molecular dynamics (MD) simulations for a duration of 100 ns were carried out to validate the stability and dynamic behavior of the Mpro enzyme upon binding to all four selected compounds. The MD simulation study was also performed for the apo protein structure to further support the analysis. The key parameters, such as the Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of Gyration (RoG), and ligand–protein interactions, were used to evaluate the molecular stability of complex systems, which helps to provide insight into the conformational changes that occur during compound-Mpro enzyme interaction (Figure 2 and Figure 3).
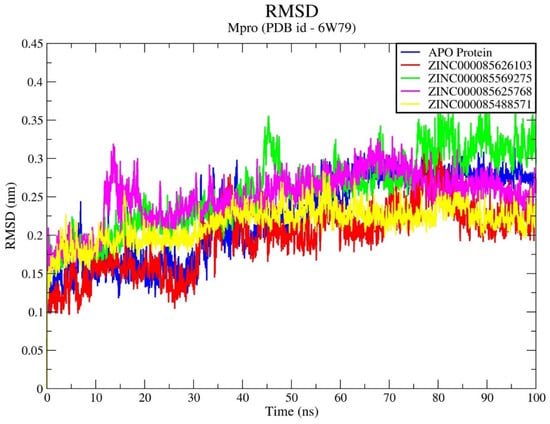
Figure 2.
Time-dependent review of RMSD for Mpro enzyme upon binding all compounds.
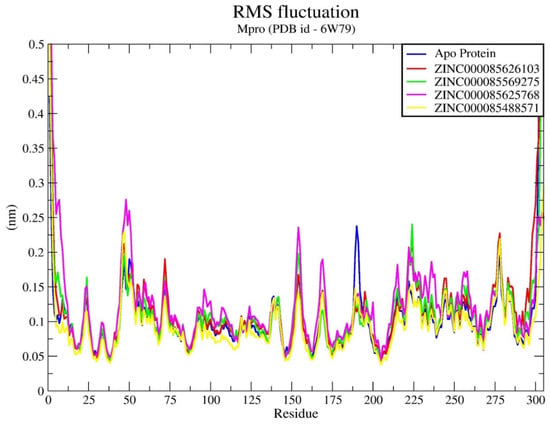
Figure 3.
Time-dependent evaluation of RMSF Rg for Mpro enzyme upon binding to all compounds.
The analysis of RMSD values for the Mpro enzyme–compound complexes reveals that all systems exhibit remarkable stability over a timeframe of 100 ns (Figure 2). Like the apo structure of the Mpro enzyme, all four Mpro enzyme–compound complexes were shown to stabilize below 0.30–0.35 nm of RSMD values. While RMSD values within the Mpro enzyme–ZINC000085625768 system and Mpro enzyme–ZINC000085569275 exhibit some degree of fluctuation, these fluctuations tend to stabilize within the narrow range of 0.225 to 0.325 and 0.3 to 0.375 nm. Similarly, Mpro enzyme–ZINC000085626103 showed minimal fluctuation as denoted by its RMSD values during an entire simulation run, while Mpro enzyme–ZINC000085488571 showed highly stable compounds throughout the MD simulation cycle with RMSD values tending to stabilize within the range of 0.15 to 0.275 nm. The analysis of RMSD indicated that the incorporation of all compounds into the active site of the Mpro enzyme leads to a consistent and steady behavioral pattern across these systems. The stability observed throughout the MD simulations emphasizes the potential therapeutic importance of these compounds in modulating the activity of the Mpro enzyme.
Furthermore, an in-depth analysis of RMSF of the Cα atoms of amino acid residues was performed in all systems, as illustrated in Figure 3. The investigation unveiled that the Cα atoms of amino acids located in the loop region of the enzyme noticed the most significant atomic fluctuations. Significantly, the most noteworthy fluctuations were primarily observed in the region encompassing amino acid residues 45 to 50 and 150 to 200, which corresponds to a domain associated with the loop located away from the active site. It is worth mentioning that similar levels of fluctuation were observed in all Mpro enzyme–compound systems. Here, the analysis of RMSF presented the additional evidence supporting the overall stability of the Mpro enzyme in complex with all four compounds. During the simulations, we continuously monitored the interactions between the Mpro enzyme and ligands, encompassing hydrogen bonds, hydrophobic interactions, ionic interactions, and water bridges. Figure 4, which presents stacked bar charts illustrating the protein–ligand interactions, revealed that both compounds engaged in a greater number of interactions aimed at stabilizing the complex with the Mpro enzyme throughout the simulations.
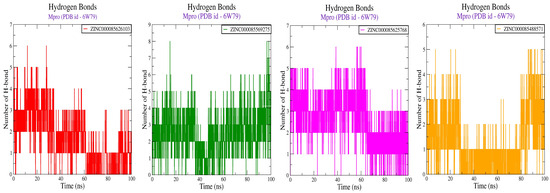
Figure 4.
Bar diagram showing protein–ligand H-bonds obtained after 100 ns MD simulation studies for compound-Mpro complexes.
These overall observations further served as confirmation of the prevailing belief that both drugs form stable complexes within the active site of the Mpro enzyme with minimal structural changes.
2.3. Binding Free Energy Calculations of the Complexes Using MM-GBSA Analysis
To assess the reliability of the binding affinity of all compounds with the Mpro enzyme, we conducted post-MD simulation MM-GBSA calculations. The MM-PBSA ∆G bind values were determined by assessing the energy difference between the bound and unbound states of the complexes. The average ∆G binding free energy values for ZINC000085626103, ZINC000085625768, ZINC000085488571, and ZINC000085569275 against the Mpro enzyme were found to be −19.17 ± 17.54, −38.29 ± 5.84, −25.84 ± 5.74, and −20.56 ± 5.53 kcal/mol, respectively (Table 2). These results indicate that all compounds have a significant affinity for binding to the enzyme. Notably, ZINC000085625768 exhibited a higher affinity for the Mpro enzyme, suggesting that complexes formed with it may be more stable.

Table 2.
MM-GBSA ΔG binding free energy of docked compounds in complex with MAO-A enzyme.
3. Conclusions
The four compounds, namely ZINC000085626103, ZINC000085625768, ZINC000085488571, and ZINC000085569275, were found to exhibit remarkable binding affinities for the Mpro of SARS-CoV-2, after the screening of natural compound from the ZINC database against the SARS-CoV-2 Mpro enzyme. All four compounds were found to be highly effective against the Mpro enzyme based on docking score, MMGBSA free binding energy, and post-processing ∆G binding energy. These compounds could therefore serve as a starting point for the development of potent and successful antiviral drugs against the deadly COVID-19.
4. Methodology
4.1. Docking Methodology
The molecular docking investigation was conducted using the Smina molecular docking tool [5], following a procedure in alignment with the methodologies previously outlined by our research group [6,7,8]. Initially, the 3D structures of both compounds were obtained from the PubChem database and were minimized utilizing the steepest descent method via the Open Babel chemical toolbox. For all enzymes, X-ray crystal structures were acquired from the Protein Data Bank, and their preparation for docking was performed via Dock Prep, an integrated tool within UCSF Chimera Software. The binding site was chosen by employing the coordinates of the co-crystal ligand of enzymes, with an additional 4 Å extension in each dimension. The lower energy conformers of both ligands were then subsequently docked within the selected active site of the enzyme, using the default scoring function of Smina. The academic version of Maestro software (Schrodinger, New York, NY, USA) was used to visualize scores and poses and save images of the docking results.
4.2. Molecular Dynamics (MD) Simulation
MD simulation studies of the compound-Mpro enzyme complexes were carried out using GROMACS 2021, following a methodology detailed in our prior publication [6]. In summary, we employed the CHARMM36 force field to establish the protein’s topological structure. The topology and parameters for the ligands were generated using the AnteChamber Python Parser interface (ACPYPE) [9]. Subsequently, the system was placed within a cubic box with periodic boundary conditions (PBC), with the TIP3P water model utilized for solvation. Counter ions were introduced to achieve a neutralized system. To minimize the system’s energy, we applied the steepest descent algorithm with a tolerance value set at 1000 kJ mol−1 nm−1. Following energy minimization, the system underwent equilibration under both the NVT and NPT ensembles, each lasting 1000 ps. The Berendsen algorithm was employed to control the thermostat and barostat during the equilibration process. Subsequently, the system was subjected to a production MD simulation spanning 100 nanoseconds (ns), with trajectory snapshots saved at 50 ps intervals, resulting in approximately 2000 frames for subsequent analysis. Throughout the MD simulations, the temperature and pressure were held constant at 300 K and 1.01325 bar, respectively. We employed standard analysis techniques to compute parameters such as RMSD, RMSF, Rg, and the formation of hydrogen bonds over the simulation duration. For post-simulation molecular mechanics with generalized born and surface area (MM-GBSA) analysis, we utilized the gmx_MMPBSA tool [10].
Author Contributions
Conceptualization, D.K.L. and P.M.N.; Methodology, D.K.L. and S.R.C.; software, D.K.L. and S.R.C.; validation, A.P.S. and V.R.U.; formal analysis, V.R.U.; investigation, D.K.L.; resources, P.M.N. and S.P.J.; data curation, S.R.C.; writing—original draft preparation, S.R.C. and A.P.S.; writing—review and editing, D.K.L.; visualization, D.K.L.; supervision, D.K.L. and P.M.N.; project administration, P.M.N. and S.P.J.; funding acquisition, P.M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kumari, M.; Lu, R.-M.; Li, M.-C.; Huang, J.-L.; Hsu, F.-F.; Ko, S.-H.; Ke, F.-Y.; Su, S.-C.; Liang, K.-H.; Yuan, J.P.-Y.; et al. A Critical Overview of Current Progress for COVID-19: Development of Vaccines, Antiviral Drugs, and Therapeutic Antibodies. J. Biomed. Sci. 2022, 29, 68. [Google Scholar] [CrossRef] [PubMed]
- Kronenberger, T.; Laufer, S.A.; Pillaiyar, T. COVID-19 Therapeutics: Small-Molecule Drug Development Targeting SARS-CoV-2 Main Protease. Drug Discov. Today 2023, 28, 103579. [Google Scholar] [CrossRef] [PubMed]
- Rani, I.; Kalsi, A.; Kaur, G.; Sharma, P.; Gupta, S.; Gautam, R.K.; Chopra, H.; Bibi, S.; Ahmad, S.U.; Singh, I.; et al. Modern Drug Discovery Applications for the Identification of Novel Candidates for COVID-19 Infections. Ann. Med. Surg. 2022, 80, 104125. [Google Scholar] [CrossRef] [PubMed]
- Das, A.P.; Agarwal, S.M. Recent Advances in the Area of Plant-Based Anti-Cancer Drug Discovery Using Computational Approaches. Mol. Divers. 2023, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Koes, D.R.; Baumgartner, M.P.; Camacho, C.J. Lessons Learned in Empirical Scoring with Smina from the CSAR 2011 Benchmarking Exercise. J. Chem. Inf. Model. 2013, 53, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Kardile, R.A.; Sarkate, A.P.; Lokwani, D.K.; Tiwari, S.V.; Azad, R.; Thopate, S.R. Design, Synthesis, and Biological Evaluation of Novel Quinoline Derivatives as Small Molecule Mutant EGFR Inhibitors Targeting Resistance in NSCLC: In Vitro Screening and ADME Predictions. Eur. J. Med. Chem. 2023, 245, 114889. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.V.; Sarkate, A.P.; Lokwani, D.K.; Pansare, D.N.; Gattani, S.G.; Sheaikh, S.S.; Jain, S.P.; Bhandari, S.V. Explorations of Novel Pyridine-Pyrimidine Hybrid Phosphonate Derivatives as Aurora Kinase Inhibitors. Bioorganic Med. Chem. Lett. 2022, 67, 128747. [Google Scholar] [CrossRef] [PubMed]
- Thorat, N.M.; Khodade, V.S.; Ingale, A.P.; Lokwani, D.K.; Sarkate, A.P.; Thopate, S.R. Molecular Docking Studies and Application of 6-(1-Arylmethanamino)-2-Phenyl-4 H-Chromen-4-Ones as Potent Antibacterial Agents. Polycycl. Aromat. Compd. 2022, 1–14. [Google Scholar] [CrossRef]
- Sousa da Silva, A.W.; Vranken, W.F. ACPYPE—AnteChamber PYthon Parser interfacE. BMC Res. Notes 2012, 5, 367. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).