Biological Docking and BSA Binding Studies of 1,4-Disubstituted Piperdine Containing 1,2,4-Triazoles: Comparative Synthesis Leveraging Microwave-Assisted and Conventional Protocols †
Abstract
:1. Introduction
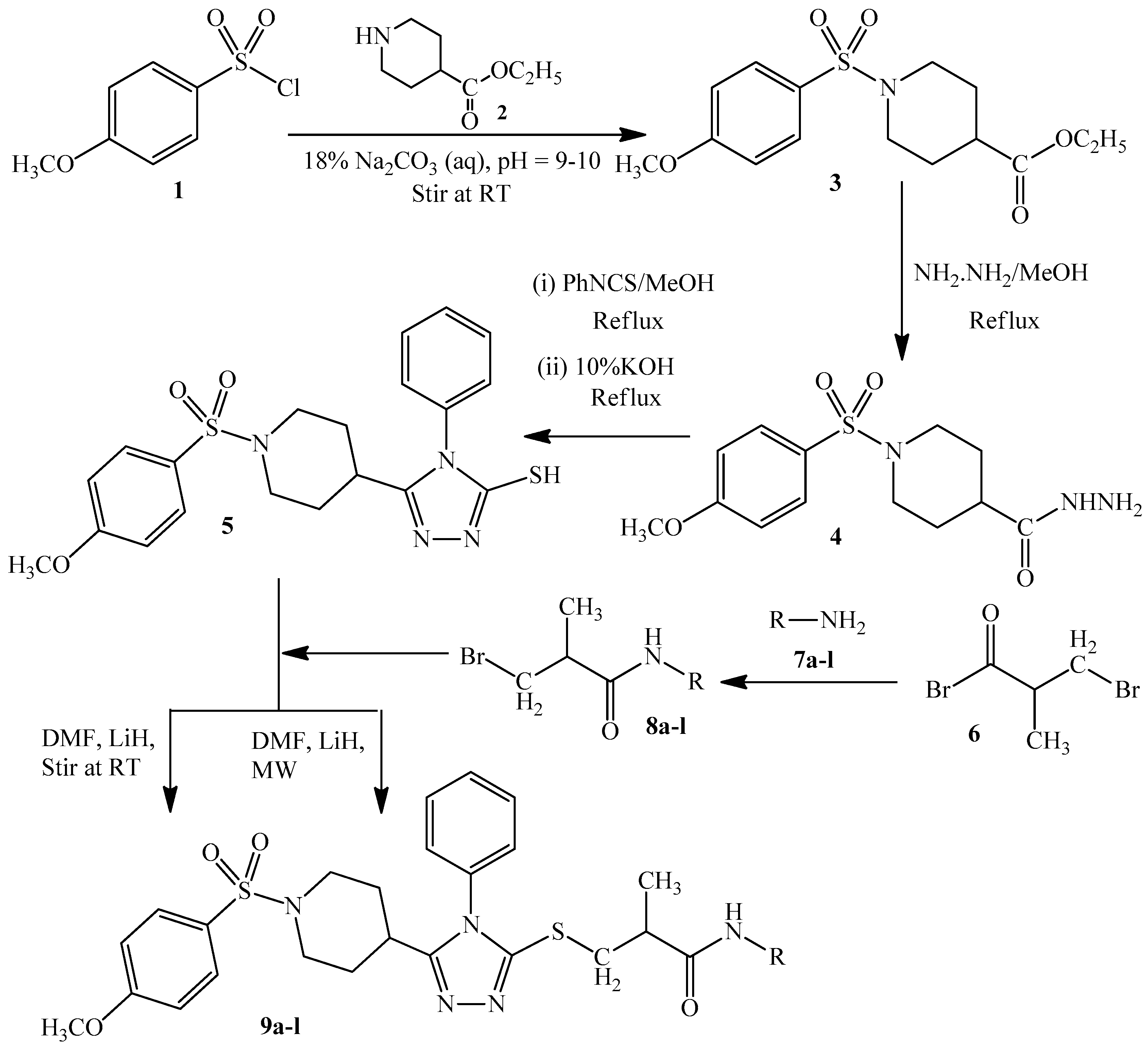
2. Experimental Section
2.1. Synthesis of Ethyl 1-[(4-Methoxyphenyl)sulfonyl]-4-piperidincarboxylate (3)
2.2. Synthesis of 1-(4-Methoxyphenylsulfonyl)piperidin-4-carbohydrazide (4)
2.3. Synthesis of 5-{1-[(4-Methoxyphenyl)sulfonyl]-4-piperidinyl}-4-methyl-4H-1,2,4-triazole-3-thiol (5)
2.4. Synthesis of N-(Substituted)-2-bromopropanamides (8a–8l)
2.5. General Procedure for the Synthesis of N-(Substituted)-2-[(5-{1-[(4-methoxyphenyl) sulfonyl]-4-piperidinyl}-4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl] Propanamide (9a–9l)
2.5.1. N-(2,5-Dimethylphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9a)
2.5.2. N-(2,6-Dimethylphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9b)
2.5.3. N-(3,5-Dimethylphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9c)
2.5.4. 2-[(5-{1-[(4-Methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]-N-phenylpropanamide (9d)
2.5.5. 2-[(5-{1-[(4-Methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]-N-(2-methylphenyl)propanamide (9e)
2.5.6. 2-[(5-{1-[(4-Methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]-N-(3-methylphenyl)propanamide (9f)
2.5.7. N-(2-Ethylphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9g)
2.5.8. N-(4-Ethoxyphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9h)
2.5.9. N-(2-Ethyl-6-methylphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9i)
2.5.10. N-Cyclohexyl-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9j)
2.5.11. Methyl 2-({2-[(5-{1-[(4-Methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanoyl}amino)benzoate (9k)
2.5.12. N-(3,4-Dimethylphenyl)-2-[(5-{1-[(4-methoxyphenyl)sulfonyl]-4-piperidinyl}-4-phenyl-4H-1,2,4-triazol-3-yl)sulfanyl]propanamide (9l)
2.6. Antioxidant Activity by DPPH Method
2.7. Butyryl Cholinesterase Inhibition Assay
2.8. Urease Inhibition Assay
2.9. Statistical Analysis
2.10. Molecular Docking Studies
3. Results and Discussion
3.1. Chemistry
3.2. Acetyl Cholinesterase Inhibition Potential
3.3. Antioxidant Activity Studies
3.4. Urease Inhibition Studies
3.5. Butyryl Cholinesterase (BChE) Studies
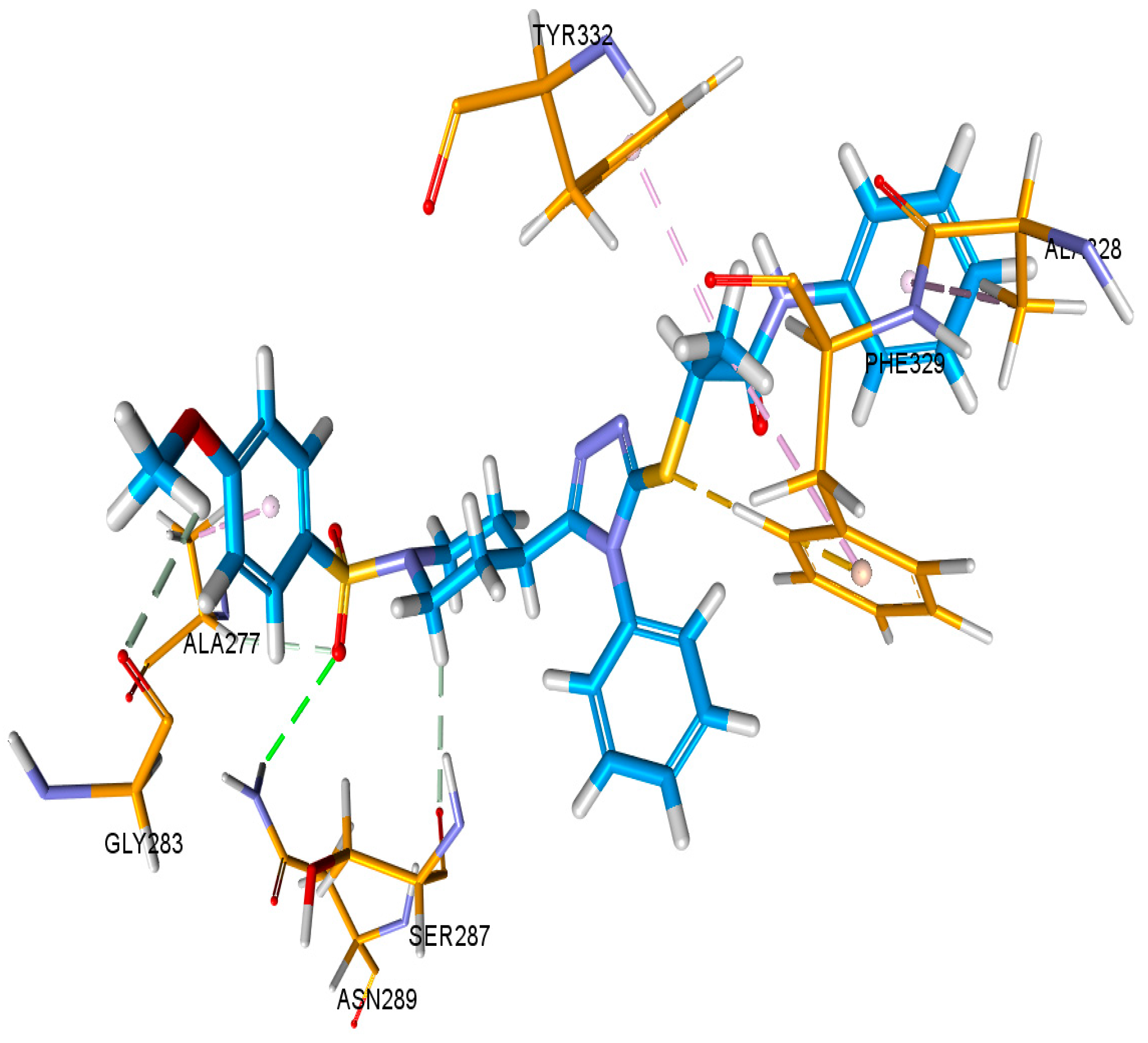
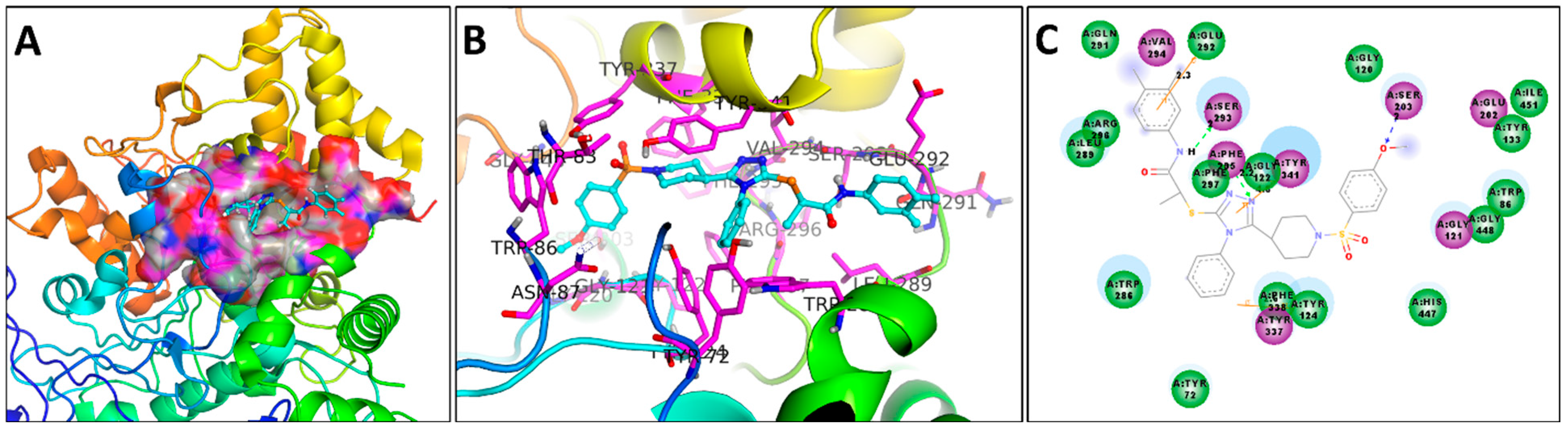
3.6. Docking Studies
3.6.1. Docking Studies against Urease and BChE Enzymes
3.6.2. Docking Studies against AChE Enzyme
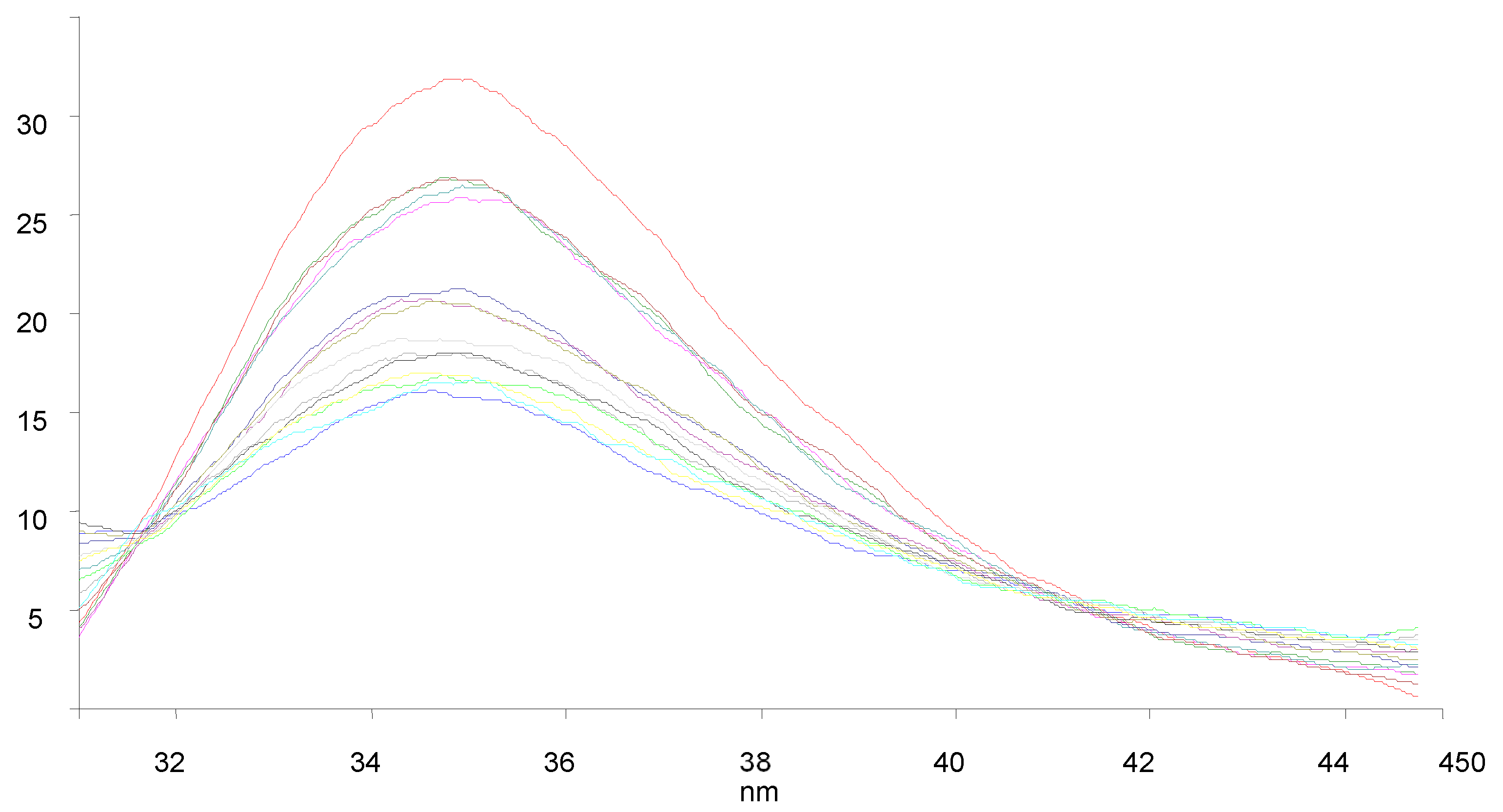
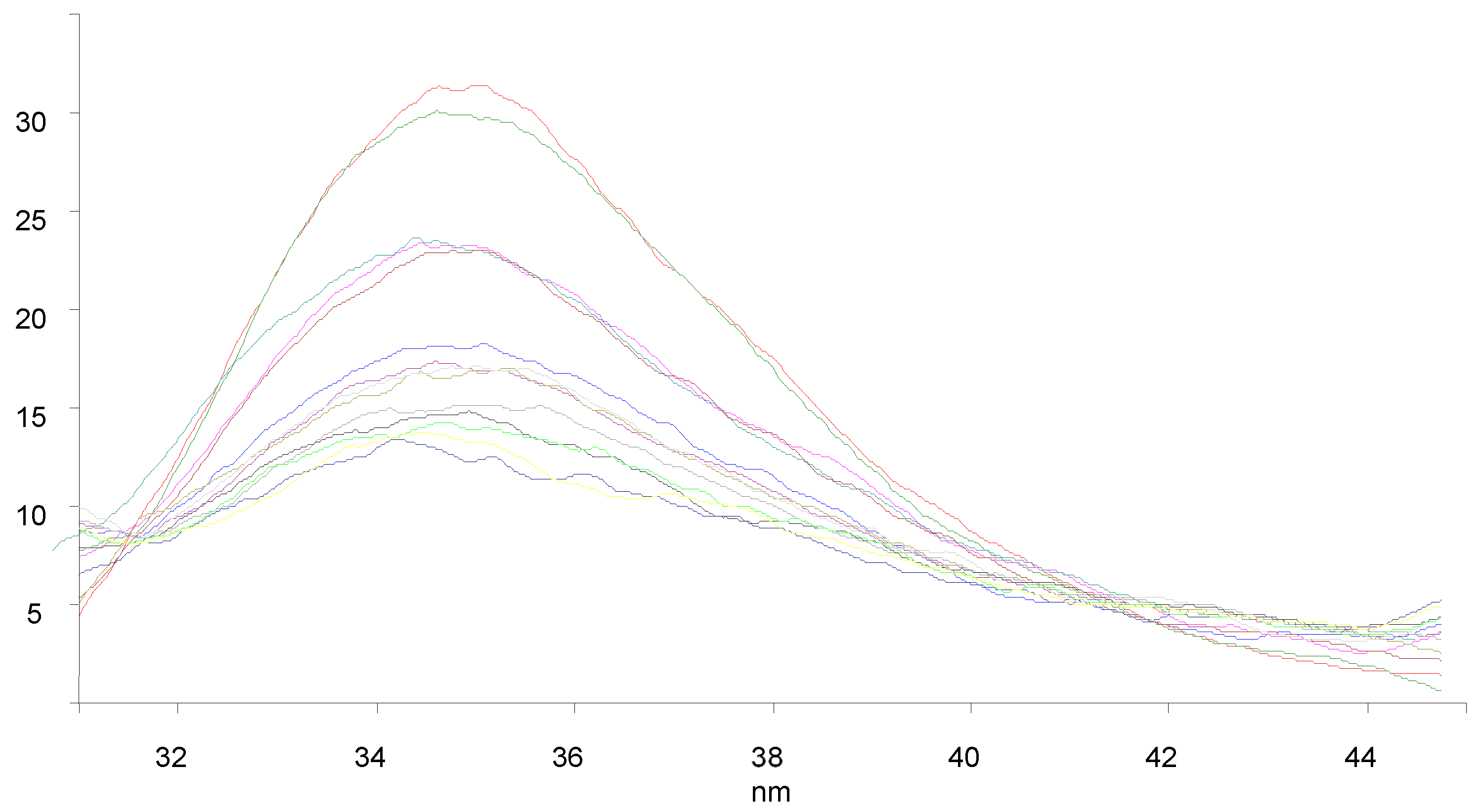
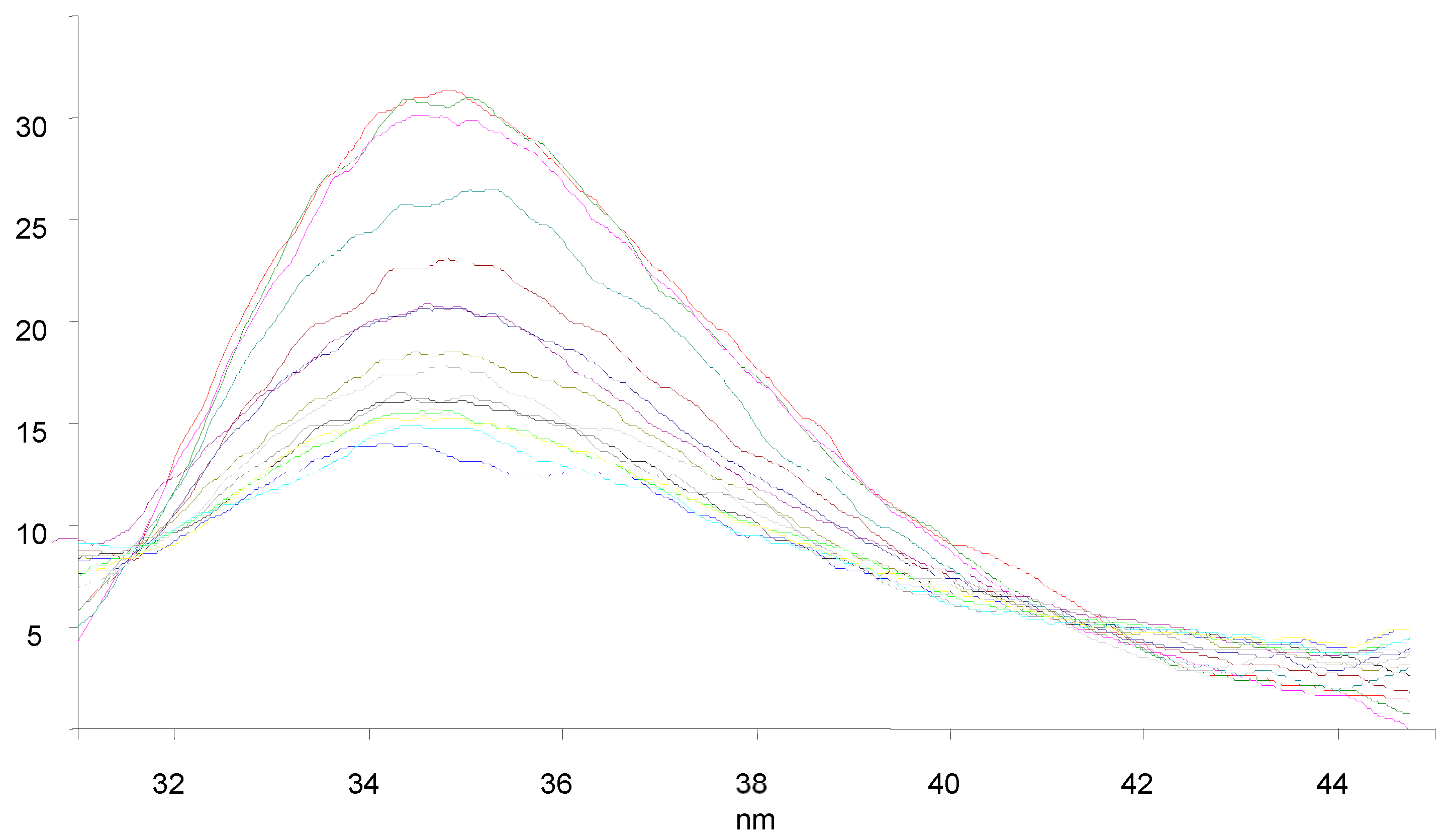
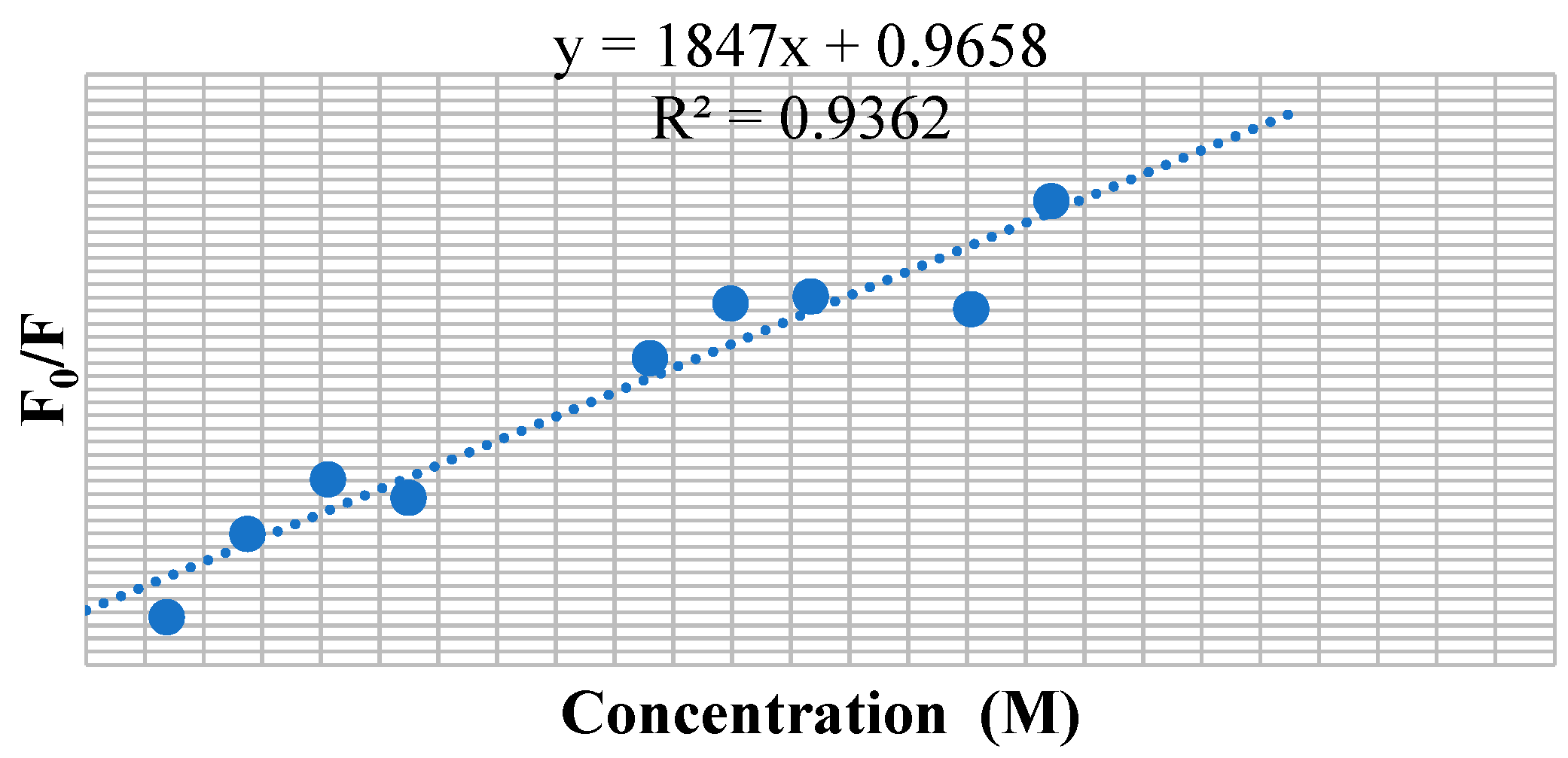
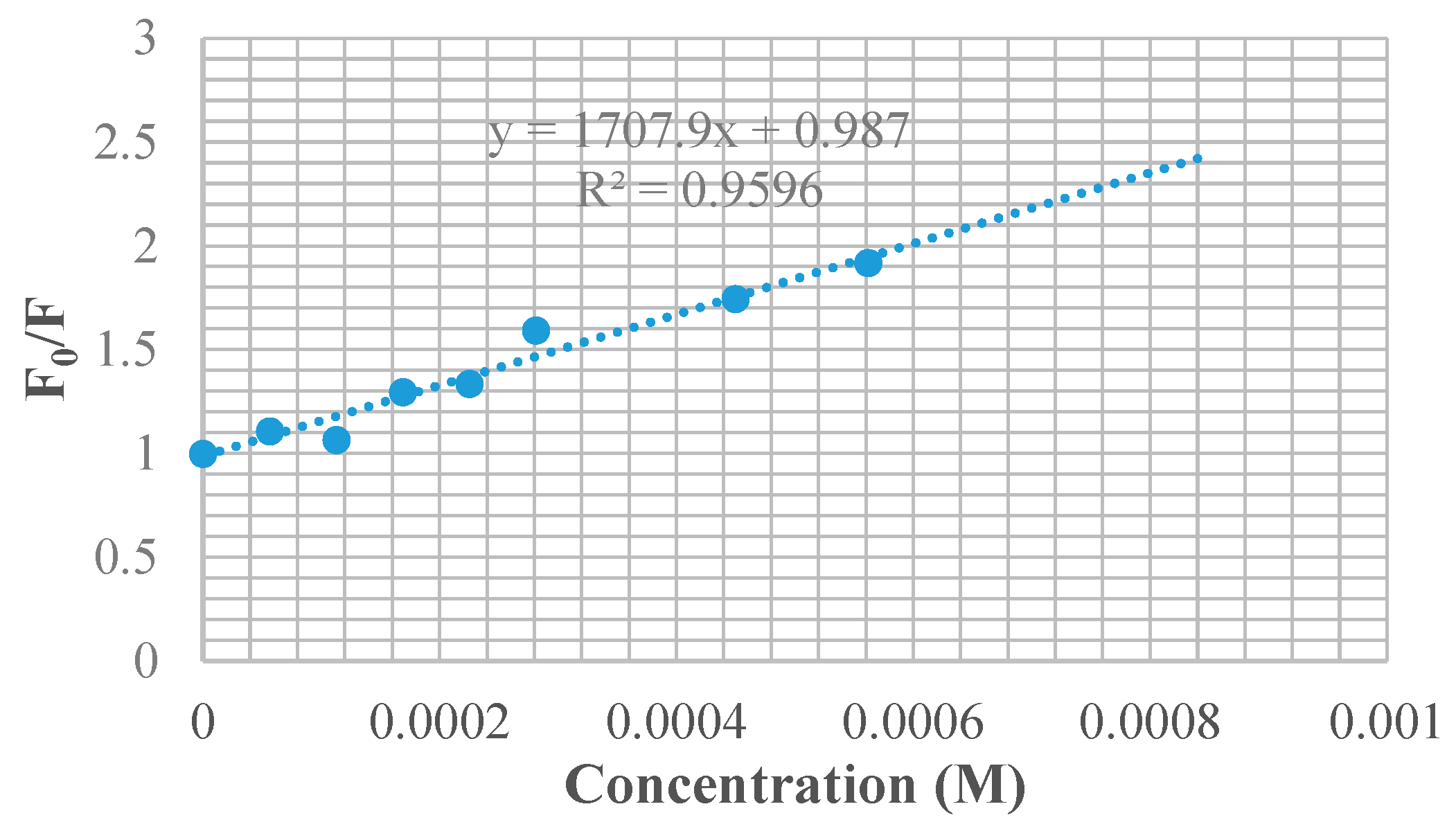
3.7. BSA Binding Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jhuang, H.S.; Liu, Y.W.; Reddy, D.M.; Tzeng, Y.Z.; Lin, W.Y.; Lee, C.F. Microwave assisted synthesis of thioesters from aldehydes and thiols in water. J. Chin. Chem. Soc. 2018, 65, 24–27. [Google Scholar] [CrossRef]
- Lidström, P.; Tierney, J.; Watheyb, B.; Westmana, J. Microwave assisted organic synthesis—A review. Tetrahedron 2001, 57, 9225–9283. [Google Scholar] [CrossRef]
- Lee, J.; Hong, M.; Jung, Y.; Cho, E.J.; Rhee, H. Synthesis of 1,3,5-trisubstituted-1,2,4-triazoles by microwave-assisted N-acylation of amide derivatives and the consecutive reaction with hydrazine hydrochlorides. Tetrahedron 2012, 68, 2045–2051. [Google Scholar] [CrossRef]
- Sebeka, A.A.H.; Osman, A.M.A.; ElSayed, I.E.T.; ElBahanasawy, M.; Tantawy, M.A. Synthesis and antiproliferative activity of novel neocryptolepine-hydrazides hybrids. J. Appl. Pharm. Sci. 2017, 7, 9–15. [Google Scholar]
- Potewar, T.M.; Petrova, K.T.; Barros, M.T. Efficient microwave assisted synthesis of novel 1,2,3-triazole–sucrose derivatives by cycloaddition reaction of sucrose azides and terminal alkynes. Carbohydr. Res. 2013, 379, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Sangshetti, J.N.; Chabukswar, A.R.; Shinde, D.B. Microwave assisted one pot synthesis of some novel 2,5-disubstituted1,3,4-oxadiazoles as antifungal agents. Bioorg. Med. Chem. Lett. 2011, 21, 444–448. [Google Scholar] [CrossRef] [PubMed]
- Medvedeva, A.S.; Demina, M.M.; Kon, T.V.; Nguyen, T.L.H.; Afonin, A.V.; Ushakov, I.A. Microwave assisted solvent-and catalyst-free three-component synthesis of NH-1,2,3-triazoloimines. Tetrahedron 2017, 73, 3979–3985. [Google Scholar] [CrossRef]
- Kharb, R.; Sharma, P.C.; Yar, M.S. Pharmacological significance of triazole scaffold. J. Enzyme Inhib. Med. Chem. 2011, 26, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Upmanyu, N.; Mehta, A. In vitro anti-microbial activity of some 5-aryl-4-(substituted amino)-3-mercapto-1,2,4-triazoles. Indian J. Microbiol. 2006, 46, 401–404. [Google Scholar]
- Sahu, S.K.; Dubey, B.K.; Tripathi, A.C.; Koshy, M.; Saraf, S.K. Piperidin-4-one: The potential pharmacophore. Mini Rev. Med. Chem. 2013, 13, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, M.; Thanusu, J.; Kanagarajan, V.; Govindaraju, R. Unexpected formation of 3-chloro-1-hydroxy-2,6-diarylpiperidin-4-ones: Synthesis, antibacterial and antifungal activities. J. Enzyme Inhib. Med. Chem. 2009, 24, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Koleva, I.I.; VanBeek, T.A.; Linssen, J.P.H.; Groot, A.D.; Evstatieva, L.N. Screening of plant extracts for antioxidant activity: A comparative study on three testing methods. Phytochem. Anal. 2002, 13, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Cygler, M.; Schrag, J.D.; Sussman, J.L.; Harel, M.; Silman, I.; Gentry, M.K.; Doctor, B.P. Relationship between sequence conservation and three-dimensional structure in a large family of esterases, lipases, and related proteins. Protein Sci. 1993, 2, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Tougu, V. Acetyl cholinesterase: Mechanism of catalysis and inhibition. Curr. Med. Chem. 2001, 1, 155–170. [Google Scholar]
- Mobley, H.L.; Cortesia, M.J.; Rosenthal, L.E.; Jones, B.D. Characterization of urease from Campylobacterpylori. J. Clin. Microbiol. 1988, 26, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, A.; Ponnuraj, K. Crystal structure of the first plant urease from jackbean: 83 years of journey from itsfirst crystal to molecular structure. J. Mol. Biol. 2010, 400, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Nachon, F.; Carletti, E.; Ronco, C.; Trovaslet, M.; Nicolet, Y.; Jean, L.; Renard, P.Y. Crystal structures of human cholinesterases in complex with huprine Wandtacrine: Elements of specificity for anti-Alzheimer’s drugs targeting acetyl-and butyryl-cholinesterase. Biochem. J. 2013, 453, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Elmer, P. ChemBioDraw Professional, version (15.0.0.106); Cambridge Soft Waltham: Waltham, MA, USA, 2017.
- BIOVIA Discovery Studio Client, v16.1.0.15350; Dassault Systemes: SanDiego, CA, USA, 2015.
- FRED, version 3.0.0; Open Eye Scientific Software: SantaFe, NM, USA, 2013. Available online: www.eyesopen.com(accessed on 12 April 2023).
- OMEGA, version 2.4.6; Open Eye Scientific Software: SantaFe, NM, USA, 2013. Available online: www.eyesopen.com(accessed on 12 April 2023).


| Compound | R | Compound | R |
|---|---|---|---|
| 9a | 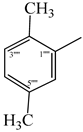 | 9g | 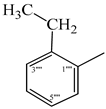 |
| 9b |  | 9h | 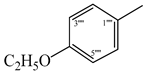 |
| 9c | 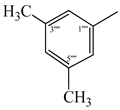 | 9i |  |
| 9d | 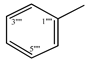 | 9j | 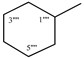 |
| 9e | 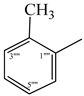 | 9k | 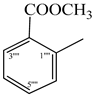 |
| 9f | 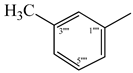 | 9l |  |
| Compounds | Reaction Time | Reaction Yield (%) | ||
|---|---|---|---|---|
| Conventional (hours) | Microwave (sec) | Conventional | Microwave | |
| 9a | 10 | 32 | 62 | 94 |
| 9b | 13 | 36 | 58 | 92 |
| 9c | 14 | 43 | 66 | 93 |
| 9d | 11 | 33 | 61 | 88 |
| 9e | 8 | 59 | 70 | 86 |
| 9f | 11 | 39 | 67 | 89 |
| 9g | 12 | 40 | 72 | 67 |
| 9h | 14 | 48 | 54 | 92 |
| 9i | 16 | 47 | 68 | 90 |
| 9j | 13 | 52 | 76 | 88 |
| 9k | 9 | 38 | 49 | 85 |
| 9l | 8 | 49 | 58 | 82 |
| Compounds | IC50 Values | |||
|---|---|---|---|---|
| AChE | Antioxidant | Urease Inhibition | BChE | |
| 9a | 270.83 ± 1.21 | 65.2 ± 0.15 | 21.1 ± 0.11 | 66.1 ± 0.62 |
| 9b | 269.25 ± 1.13 | 52.1 ± 0.26 | 36.2 ± 0.21 | 79.2 ± 0.22 |
| 9c | 134.63 ± 1.12 | 45.2 ± 0.15 | 39.4 ± 0.45 | 77.7 ± 0.36 |
| 9d | 215.91 ± 1.13 | 49.5 ± 0.21 | >500 | 15.5 ± 0.39 |
| 9e | 63.27 ± 1.21 | 72.1 ± 0.69 | >500 | 15.9 ± 0.67 |
| 9f | 169.83 ± 1.12 | 77.7 ± 0.29 | >500 | 19.2 ± 0.67 |
| 9g | 63.45 ± 1.21 | 75.5 ± 0.32 | 20.2 ± 0.21 | 29.2 ± 0.26 |
| 9h | 87.32 ± 1.18 | 85.5 ± 0.55 | 45.5 ± 0.52 | 26.1 ± 0.14 |
| 9i | 182.73 ± 1.15 | 82.2 ± 0.56 | 49.2 ± 0.26 | 39.5 ± 0.21 |
| 9j | - | 55.5 ± 0.20 | 55.2 ± 0.11 | 79.8 ± 0.29 |
| 9k | 133.91 ± 1.12 | 52.1 ± 0.09 | 19.2 ± 0.09 | 72.1 ± 0.35 |
| 9l | 402.83 ± 1.12 | 47.7 ± 0.29 | - | 29.2 ± 0.37 |
| BHA | 44.2 ± 0.41 | - | - | |
| Thiourea | - | 21.3 ± 0.24 | - | |
| Eserine | 0.19 ± 0.05 | - | - | 7.8 ± 0.05 |
| Compound | ChemGauss4 Score against Urease | ChemGauss4 Score against BChE |
|---|---|---|
| 9a | −6.57 | −9.80 |
| 9b | −6.06 | −9.92 |
| 9c | −7.49 | −8.98 |
| 9d | −6.23 | −8.82 |
| 9e | −6.56 | −9.47 |
| 9f | −4.66 | −8.94 |
| 9g | −4.17 | −8.95 |
| 9h | −5.37 | −8.21 |
| 9i | −6.17 | −9.32 |
| 9j | −4.27 | −9.02 |
| 9k | −6.75 | −8.68 |
| 9l | −4.21 | −9.12 |
| Docking Complex = AChE- 9l | |||||||
|---|---|---|---|---|---|---|---|
| C Score a | Crash Score b | Polar Score c | D Score d | PMF Score e | G Score f | Chem Score g | Amino Acid Interaction |
| 6.84 | −3.89 | 0.09 | −215.074 | 15.098 | −345.68 | −32.964 | Glu292, Ser203, Phe295, Arg296, Phe338 |
| Compounds | KSV × 101 (M−1) | kq × 1011 (M−1 s−1) | Ka (M−1) | n |
|---|---|---|---|---|
| 9a | 1.238 | 1.238 | 61.2 | 0.58 |
| 9c | 1.847 | 1.847 | 2.33 × 103 | 1.03 |
| 9f | 1.708 | 1.708 | 4.11 × 103 | 1.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iqbal, J.; Virk, N.A.; Rehman, A.U.; Kuznetsov, A.; Rasool, S.; Yasir, M.; Shah, S.A.A. Biological Docking and BSA Binding Studies of 1,4-Disubstituted Piperdine Containing 1,2,4-Triazoles: Comparative Synthesis Leveraging Microwave-Assisted and Conventional Protocols. Chem. Proc. 2023, 14, 106. https://doi.org/10.3390/ecsoc-27-16281
Iqbal J, Virk NA, Rehman AU, Kuznetsov A, Rasool S, Yasir M, Shah SAA. Biological Docking and BSA Binding Studies of 1,4-Disubstituted Piperdine Containing 1,2,4-Triazoles: Comparative Synthesis Leveraging Microwave-Assisted and Conventional Protocols. Chemistry Proceedings. 2023; 14(1):106. https://doi.org/10.3390/ecsoc-27-16281
Chicago/Turabian StyleIqbal, Javed, Naeem Akhtar Virk, Aziz Ur Rehman, Aleksey Kuznetsov, Shahid Rasool, Muhammad Yasir, and Syed Adnan Ali Shah. 2023. "Biological Docking and BSA Binding Studies of 1,4-Disubstituted Piperdine Containing 1,2,4-Triazoles: Comparative Synthesis Leveraging Microwave-Assisted and Conventional Protocols" Chemistry Proceedings 14, no. 1: 106. https://doi.org/10.3390/ecsoc-27-16281
APA StyleIqbal, J., Virk, N. A., Rehman, A. U., Kuznetsov, A., Rasool, S., Yasir, M., & Shah, S. A. A. (2023). Biological Docking and BSA Binding Studies of 1,4-Disubstituted Piperdine Containing 1,2,4-Triazoles: Comparative Synthesis Leveraging Microwave-Assisted and Conventional Protocols. Chemistry Proceedings, 14(1), 106. https://doi.org/10.3390/ecsoc-27-16281







