Evaluation of the Ellman’s Reagent Protocol for Free Sulfhydryls Under Protein Denaturing Conditions
Abstract
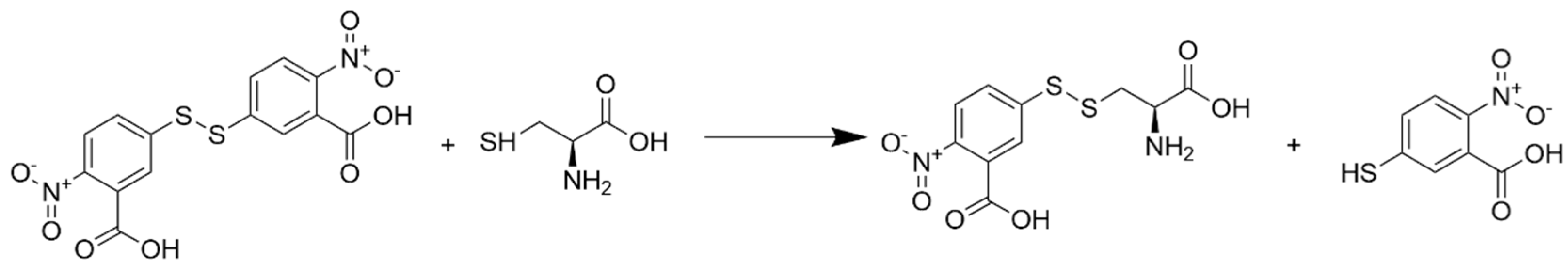
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Absorbance Spectroscopy
3. Results
3.1. Reagent Stability and Assay Conditions
3.2. Denaturants and Buffer Modifications
3.3. Application to Peptide Model Systems
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Balachandra, S.; Kusin, S.B.; Lee, R.; Blackwell, J.M.; Tiro, J.A.; Cowell, L.G.; Chiang, C.M.; Wu, S.Y.; Varma, S.; Rivera, E.L.; et al. Blood-based biomarkers of human papillomavirus-associated cancers: A systematic review and meta-analysis. Cancer 2021, 127, 850–864. [Google Scholar] [CrossRef] [PubMed]
- Lyman, D.F.; Bell, A.; Black, A.; Dingerdissen, H.; Cauley, E.; Gogate, N.; Liu, D.; Joseph, A.; Kahsay, R.; Crichton, D.J.; et al. Modeling and integration of N-glycan biomarkers in a comprehensive biomarker data model. Glycobiology 2022, 32, 855–870. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Martinez, F.; Wojciechowska, G.; Szczerbinski, L.; Kretowski, A. Circulating Nucleic Acid-Based Biomarkers of Type 2 Diabetes. Int. J. Mol. Sci. 2021, 23, 295. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef]
- Varesi, A.; Carrara, A.; Pires, V.G.; Floris, V.; Pierella, E.; Savioli, G.; Prasad, S.; Esposito, C.; Ricevuti, G.; Chirumbolo, S.; et al. Blood-Based Biomarkers for Alzheimer’s Disease Diagnosis and Progression: An Overview. Cells 2022, 11, 1367. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fournier, M.; Lopez-Molina, M.; Torres Iglesias, G.; Botella, L.; Chamorro, B.; Laso-Garcia, F.; Puertas, I.; Tallon Barranco, A.; Otero-Ortega, L.; Frank-Garcia, A.; et al. Antibody Content against Epstein-Barr Virus in Blood Extracellular Vesicles Correlates with Disease Activity and Brain Volume in Patients with Relapsing-Remitting Multiple Sclerosis. Int. J. Mol. Sci. 2023, 24, 4192. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P.; et al. Biomarkers in Stress Related Diseases/Disorders: Diagnostic, Prognostic, and Therapeutic Values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef]
- Hristova, V.A.; Chan, D.W. Cancer biomarker discovery and translation: Proteomics and beyond. Expert. Rev. Proteom. 2019, 16, 93–103. [Google Scholar] [CrossRef]
- Minutentag, I.W.; Seneda, A.L.; Barros-Filhos, M.C.; de Carvalho, M.; Souza, V.G.P.; Hasimoto, C.N.; Moraes, M.P.T.; Marchi, F.A.; Lam, W.L.; Reis, P.P.; et al. Discovery of Novel miRNAs in Colorectal Cancer: Potential Biological Roles and Clinical Utility. Non-Coding RNA 2023, 9, 65. [Google Scholar] [CrossRef]
- Pessoa, L.S.; Heringer, M.; Ferrer, V.P. ctDNA as a cancer biomarker: A broad overview. Crit. Rev. Oncol. Hematol. 2020, 155, 103109. [Google Scholar] [CrossRef]
- Fritschi, N.; Vaezipour, N.; Buettcher, M.; Portevin, D.; Naranbhai, V.; Ritz, N. Ratios from full blood count as markers for TB diagnosis, treatment, prognosis: A systematic review. Int. J. Tuberc. Lung Dis. 2023, 27, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jeong, J.Y.; Seo, A.N. Biomarkers for Predicting Response to Personalized Immunotherapy in Gastric Cancer. Diagnostics 2023, 13, 2782. [Google Scholar] [CrossRef]
- Xie, Q.Y.; Oh, S.; Wong, A.; Yau, C.; Herold, K.C.; Danska, J.S. Immune responses to gut bacteria associated with time to diagnosis and clinical response to T cell-directed therapy for type 1 diabetes prevention. Sci. Transl. Med. 2023, 15, eadh0353. [Google Scholar] [CrossRef]
- Tamborero, D.; Rubio-Perez, C.; Deu-Pons, J.; Schroeder, M.P.; Vivancos, A.; Rovira, A.; Tusquets, I.; Albanell, J.; Rodon, J.; Tabernero, J.; et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. bioRxiv 2017, 140475. [Google Scholar] [CrossRef] [PubMed]
- Turewicz, M.; Frericks-Zipper, A.; Stepath, M.; Schork, K.; Ramesh, S.; Marcus, K.; Eisenacher, M. BIONDA: A free database for a fast information on published biomarkers. Bioinform. Adv. 2021, 1, vbab015. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Bartok, B.; Oler, E.; Liang, K.Y.H.; Budinski, Z.; Berjanskii, M.; Guo, A.; Cao, X.; Wilson, M. MarkerDB: An online database of molecular biomarkers. Nucleic Acids Res. 2021, 49, D1259–D1267. [Google Scholar] [CrossRef]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early detection of cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef]
- Hawkes, N. Cancer survival data emphasise importance of early diagnosis. BMJ 2019, 364, l408. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review (CSR) 1975–2018 National Cancer Institute. 2021. Available online: https://seer.cancer.gov/csr/1975_2018/ (accessed on 9 April 2025).
- Ghimire, M.L.; Cox, B.D.; Winn, C.A.; Rockett, T.W.; Schifano, N.P.; Slagle, H.M.; Gonzalez, F.; Bertino, M.F.; Caputo, G.A.; Reiner, J.E. Selective Detection and Characterization of Small Cysteine-Containing Peptides with Cluster-Modified Nanopore Sensing. ACS Nano 2022, 16, 17229–17241. [Google Scholar] [CrossRef]
- Rockett, T.W.; Almahyawi, M.; Ghimire, M.L.; Jonnalagadda, A.; Tagliaferro, V.; Seashols-Williams, S.J.; Bertino, M.F.; Caputo, G.A.; Reiner, J.E. Cluster-Enhanced Nanopore Sensing of Ovarian Cancer Marker Peptides in Urine. ACS Sens. 2024, 9, 860–869. [Google Scholar] [CrossRef]
- Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959, 82, 70–77. [Google Scholar] [CrossRef]
- Maeda, H.; Matsuno, H.; Ushida, M.; Katayama, K.; Saeki, K.; Itoh, N. 2,4-Dinitrobenzenesulfonyl fluoresceins as fluorescent alternatives to Ellman’s reagent in thiol-quantification enzyme assays. Angew. Chem. Int. Ed. Engl. 2005, 44, 2922–2925. [Google Scholar] [CrossRef]
- Nekrassova, O.; White, P.C.; Threlfell, S.; Hignett, G.; Wain, A.J.; Lawrence, N.S.; Davis, J.; Compton, R.G. An electrochemical adaptation of Ellman’s test. Analyst 2002, 127, 797–802. [Google Scholar] [CrossRef]
- Dhawan, S.; Devnani, H.; Babu, J.; Singh, H.; Haider, M.A.; Khan, T.S.; Ingole, P.P.; Haridas, V. Supersensitive Detection of Anions in Pure Organic and Aqueous Media by Amino Acid Conjugated Ellman’s Reagent. ACS Appl. Bio Mater. 2021, 4, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Guclu, K.; Ozyurek, M.; Gungor, N.; Baki, S.; Apak, R. Selective optical sensing of biothiols with Ellman’s reagent: 5,5′-Dithio-bis(2-nitrobenzoic acid)-modified gold nanoparticles. Anal. Chim. Acta 2013, 794, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Khan, K.A.; Khan, M.K.; Ahmad, A.; Niazi, Z.R.; Mangi, A.; Rehman, F.U.; Shah, K.U.; Gul, R.; Jan, S.U. Thiol-disulfide exchange reactions occurring at modified bovine serum albumin detected using ellman’s reagent (5, 5′-dithiobis (2-itrobenzoic acid). Pak. J. Pharm. Sci. 2020, 33, 2767–2772. [Google Scholar] [PubMed]
- Sinko, G.; Calic, M.; Bosak, A.; Kovarik, Z. Limitation of the Ellman method: Cholinesterase activity measurement in the presence of oximes. Anal. Biochem. 2007, 370, 223–227. [Google Scholar] [CrossRef]
- Zhu, J.; Dhimitruka, I.; Pei, D. 5-(2-Aminoethyl)dithio-2-nitrobenzoate as a more base-stable alternative to Ellman’s reagent. Org. Lett. 2004, 6, 3809–3812. [Google Scholar] [CrossRef]
- Riddles, P.W.; Blakeley, R.L.; Zerner, B. Reassessment of Ellman’s reagent. Methods Enzymol. 1983, 91, 49–60. [Google Scholar] [CrossRef]
- Kraus, D. Consolidated data analysis and presentation using an open-source add-in for the Microsoft Excel® spreadsheet software. Med. Writ. 2014, 23, 25–28. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [PubMed]
- Bhensdadia, N.M.; Hunt, K.J.; Lopes-Virella, M.F.; Michael Tucker, J.; Mataria, M.R.; Alge, J.L.; Neely, B.A.; Janech, M.G.; Arthur, J.M. Urine haptoglobin levels predict early renal functional decline in patients with type 2 diabetes. Kidney Int. 2013, 83, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- El Atab, O.; Gupta, B.; Han, Z.; Stribny, J.; Asojo, O.A.; Schneiter, R. Alpha-1-B glycoprotein (A1BG) inhibits sterol-binding and export by CRISP2. J. Biol. Chem. 2024, 300, 107910. [Google Scholar] [CrossRef] [PubMed]
- Kentsis, A.; Ahmed, S.; Kurek, K.; Brennan, E.; Bradwin, G.; Steen, H.; Bachur, R. Detection and diagnostic value of urine leucine-rich alpha-2-glycoprotein in children with suspected acute appendicitis. Ann. Emerg. Med. 2012, 60, 78–83.e1. [Google Scholar] [CrossRef]





| Name | Amino Acid Sequence | # Amino Acids | Molecular Weight | Net Charge b | GRAVY a |
|---|---|---|---|---|---|
| 5C1 | CASEW | 5 | 594.64 | −1 | −0.180 |
| 7C1NH2 | CLSASEW-NH2 | 7 | 793.9 | 0 | 0.300 |
| 9C5 | AESLCASEW | 9 | 995.08 | −2 | 0.044 |
| CP-2 | AVYYCQQY | 8 | 1037.15 | 0 | −0.300 |
| IRP2P | RssHCQREEAGGRD * | 14 | 1747.65 | 1 | n/c |
| Cp20L | WCFGPDGTGPNILTDITKGV | 20 | 2091.37 | −1 | 0.005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ginet, S.R.; Gonzalez, F.; Marano, M.L.; Salecha, M.D.; Reiner, J.E.; Caputo, G.A. Evaluation of the Ellman’s Reagent Protocol for Free Sulfhydryls Under Protein Denaturing Conditions. Analytica 2025, 6, 18. https://doi.org/10.3390/analytica6020018
Ginet SR, Gonzalez F, Marano ML, Salecha MD, Reiner JE, Caputo GA. Evaluation of the Ellman’s Reagent Protocol for Free Sulfhydryls Under Protein Denaturing Conditions. Analytica. 2025; 6(2):18. https://doi.org/10.3390/analytica6020018
Chicago/Turabian StyleGinet, Sophia R., Frank Gonzalez, Maxine L. Marano, Megha D. Salecha, Joseph E. Reiner, and Gregory A. Caputo. 2025. "Evaluation of the Ellman’s Reagent Protocol for Free Sulfhydryls Under Protein Denaturing Conditions" Analytica 6, no. 2: 18. https://doi.org/10.3390/analytica6020018
APA StyleGinet, S. R., Gonzalez, F., Marano, M. L., Salecha, M. D., Reiner, J. E., & Caputo, G. A. (2025). Evaluation of the Ellman’s Reagent Protocol for Free Sulfhydryls Under Protein Denaturing Conditions. Analytica, 6(2), 18. https://doi.org/10.3390/analytica6020018







