Targeting Autophagy for Acetaminophen-Induced Liver Injury: An Update
Abstract
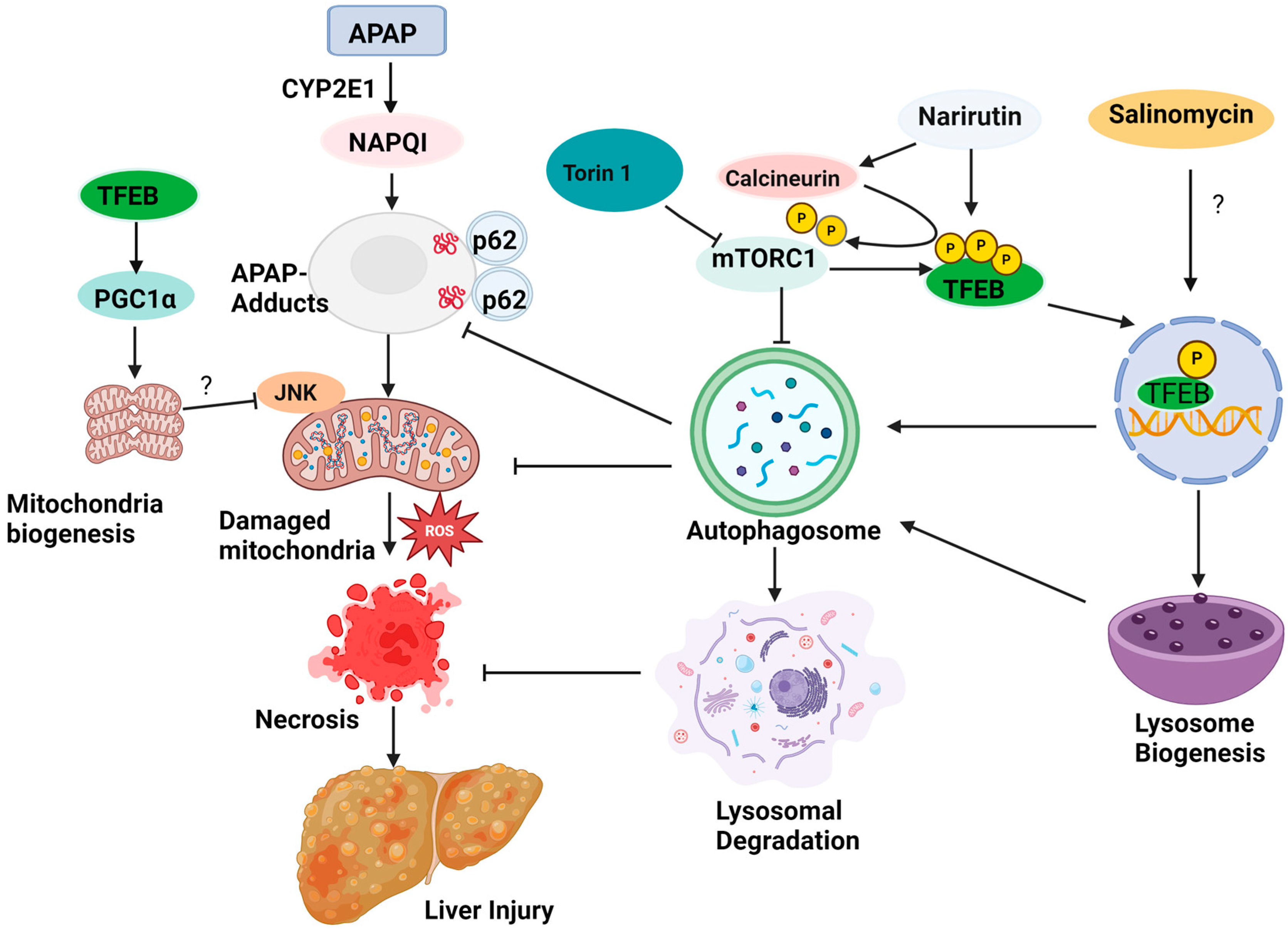
1. Introduction
2. Targeting p62 for AILI
3. Targeting TFEB for AILI
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mowry, J.B.; Spyker, D.A.; Brooks, D.E.; Zimmerman, A.; Schauben, J.L. 2015 Annual Report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 33rd Annual Report. Clin. Toxicol. 2016, 54, 924–1109. [Google Scholar] [CrossRef]
- Lee, W.M. Acetaminophen (APAP) hepatotoxicity—Isn’t it time for APAP to go away? J. Hepatol. 2017, 67, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.M. Acetaminophen hepatotoxicity. Clin. Liver Dis. 2007, 11, 525–548. [Google Scholar] [CrossRef]
- Ostapowicz, G.; Fontana, R.J.; Schiødt, F.V.; Larson, A.; Davern, T.J.; Han, S.H.; McCashland, T.M.; Shakil, A.O.; Hay, J.E.; Hynan, L. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann. Intern. Med. 2002, 137, 947–954. [Google Scholar] [CrossRef]
- Lee, W.M. Acetaminophen-related acute liver failure in the United States. Hepatol. Res. 2008, 38, S3–S8. [Google Scholar] [CrossRef]
- Budnitz, D.S.; Lovegrove, M.C.; Crosby, A.E. Emergency department visits for overdoses of acetaminophen-containing products. Am. J. Prev. Med. 2011, 40, 585–592. [Google Scholar] [CrossRef]
- Dart, R.C.; Mullins, M.E.; Matoushek, T.; Ruha, A.M.; Burns, M.M.; Simone, K.; Beuhler, M.C.; Heard, K.J.; Mazer-Amirshahi, M.; Stork, C.M.; et al. Management of Acetaminophen Poisoning in the US and Canada: A Consensus Statement. JAMA Netw Open 2023, 6, e2327739. [Google Scholar] [CrossRef]
- Heard, K.J. Acetylcysteine for Acetaminophen Poisoning. N. Engl. J. Med. 2008, 359, 285–292. [Google Scholar] [CrossRef]
- Bhushan, B.; Apte, U. Liver Regeneration after Acetaminophen Hepatotoxicity: Mechanisms and Therapeutic Opportunities. Am. J. Pathol. 2019, 189, 719–729. [Google Scholar] [CrossRef]
- McGill, M.R.; Sharpe, M.R.; Williams, C.D.; Taha, M.; Curry, S.C.; Jaeschke, H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J. Clin. Investig. 2012, 122, 1574–1583. [Google Scholar] [CrossRef]
- Hinson, J.A.; Roberts, D.W.; James, L.P. Mechanisms of Acetaminophen-Induced Liver Necrosis. Handb. Exp. Pharmacol. 2010, 196, 369–405. [Google Scholar]
- Yan, M.; Huo, Y.; Yin, S.; Hu, H. Mechanisms of acetaminophen-induced liver injury and its implications for therapeutic interventions. Redox Biol. 2018, 17, 274–283. [Google Scholar] [CrossRef]
- Ramachandran, A.; Jaeschke, H. Acetaminophen Toxicity: Novel Insights Into Mechanisms and Future Perspectives. Gene Expr. 2018, 18, 19–30. [Google Scholar] [CrossRef]
- Ramachandran, A.; Jaeschke, H. Acetaminophen hepatotoxicity: A mitochondrial perspective. Adv. Pharmacol. 2019, 85, 195–219. [Google Scholar]
- Huebener, P.; Pradere, J.P.; Hernandez, C.; Gwak, G.Y.; Caviglia, J.M.; Mu, X.; Loike, J.D.; Schwabe, R.F. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J. Clin. Investig. 2015, 125, 539–550. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Feng, D.; Li, M.; Gao, Y.; Ramirez, T.; Cao, H.; Kim, S.J.; Yang, Y.; Cai, Y.; Ju, C.; et al. Hepatic mitochondrial DNA/Toll-like receptor 9/MicroRNA-223 forms a negative feedback loop to limit neutrophil overactivation and acetaminophen hepatotoxicity in mice. Hepatology 2017, 66, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Ganey, P.E.; Luyendyk, J.P.; Newport, S.W.; Eagle, T.M.; Maddox, J.F.; Mackman, N.; Roth, R.A. Role of the coagulation system in acetaminophen-induced hepatotoxicity in mice. Hepatology 2007, 46, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Holt, M.P.; Cheng, L.; Ju, C. Identification and characterization of infiltrating macrophages in acetaminophen-induced liver injury. J. Leukoc. Biol. 2008, 84, 1410–1421. [Google Scholar] [CrossRef]
- Groeneveld, D.; Cline-Fedewa, H.; Baker, K.S.; Williams, K.J.; Roth, R.A.; Mittermeier, K.; Lisman, T.; Palumbo, J.S.; Luyendyk, J.P. Von Willebrand factor delays liver repair after acetaminophen-induced acute liver injury in mice. J. Hepatol. 2020, 72, 146–155. [Google Scholar] [CrossRef]
- Ni, H.M.; Bockus, A.; Boggess, N.; Jaeschke, H.; Ding, W.X. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology 2012, 55, 222–232. [Google Scholar] [CrossRef]
- Williams, J.A.; Ni, H.M.; Haynes, A.; Manley, S.; Li, Y.; Jaeschke, H.; Ding, W.X. Chronic Deletion and Acute Knockdown of Parkin Have Differential Responses to Acetaminophen-induced Mitophagy and Liver Injury in Mice. J. Biol. Chem. 2015, 290, 10934–10946. [Google Scholar] [CrossRef]
- Wang, H.; Ni, H.M.; Chao, X.; Ma, X.; Rodriguez, Y.A.; Chavan, H.; Wang, S.; Krishnamurthy, P.; Dobrowsky, R.; Xu, D.X.; et al. Double deletion of PINK1 and Parkin impairs hepatic mitophagy and exacerbates acetaminophen-induced liver injury in mice. Redox Biol. 2019, 22, 101148. [Google Scholar] [CrossRef]
- Ni, H.M.; McGill, M.R.; Chao, X.; Du, K.; Williams, J.A.; Xie, Y.; Jaeschke, H.; Ding, W.X. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J. Hepatol. 2016, 65, 354–362. [Google Scholar] [CrossRef]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Ma, X.; McKeen, T.; Zhang, J.; Ding, W.X. Role and Mechanisms of Mitophagy in Liver Diseases. Cells 2020, 9, 837. [Google Scholar] [CrossRef] [PubMed]
- Lamark, T.; Johansen, T. Mechanisms of Selective Autophagy. Annu. Rev. Cell Dev. Biol. 2021, 37, 143–169. [Google Scholar] [CrossRef]
- Settembre, C.; Fraldi, A.; Medina, D.L.; Ballabio, A. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013, 14, 283–296. [Google Scholar] [CrossRef]
- Chao, X.; Wang, S.; Zhao, K.; Li, Y.; Williams, J.A.; Li, T.; Chavan, H.; Krishnamurthy, P.; He, X.C.; Li, L.; et al. Impaired TFEB-mediated Lysosome Biogenesis and Autophagy Promote Chronic Ethanol-induced Liver Injury and Steatosis in Mice. Gastroenterology 2018, 155, 865–879.e12. [Google Scholar] [CrossRef]
- Settembre, C.; De Cegli, R.; Mansueto, G.; Saha, P.K.; Vetrini, F.; Visvikis, O.; Huynh, T.; Carissimo, A.; Palmer, D.; Klisch, T.J.; et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013, 15, 647–658. [Google Scholar] [CrossRef]
- Moscat, J.; Diaz-Meco, M.T. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 2009, 137, 1001–1004. [Google Scholar] [CrossRef]
- Komatsu, M.; Kageyama, S.; Ichimura, Y. p62/SQSTM1/A170: Physiology and pathology. Pharmacol. Res. 2012, 66, 457–462. [Google Scholar] [CrossRef]
- Manley, S.; Williams, J.A.; Ding, W.X. Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp. Biol. Med. 2013, 238, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Ding, W.X. SQSTM1/p62 and Hepatic Mallory-Denk Body Formation in Alcohol-Associated Liver Disease. Am. J. Pathol. 2023, 193, 1415–1426. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 2011, 192, 17–27. [Google Scholar] [CrossRef]
- Ren, J.; Wang, J.; Wang, Z.; Wu, J. Structural and biochemical insights into the homotypic PB1-PB1 complex between PKCζ and p62. Sci. China Life Sci. 2014, 57, 69–80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sanchez-Martin, P.; Saito, T.; Komatsu, M. p62/SQSTM1: ‘Jack of all trades’ in health and cancer. FEBS J. 2019, 286, 8–23. [Google Scholar] [CrossRef]
- Nakamura, K.; Kimple, A.J.; Siderovski, D.P.; Johnson, G.L. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-κB activation. J. Biol. Chem. 2010, 285, 2077–2089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mun, S.R.; Linares, J.F.; Ahn, J.; Towers, C.G.; Ji, C.H.; Fitzwalter, B.E.; Holden, M.R.; Mi, W.; Shi, X. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat. Commun. 2018, 9, 4373. [Google Scholar] [CrossRef]
- Sanz, L.; Sanchez, P.; Lallena, M.J.; Diaz-Meco, M.T.; Moscat, J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999, 18, 3044–3053. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Cha-Molstad, H.; Sung, K.S.; Hwang, J.; Kim, K.A.; Yu, J.E.; Yoo, Y.D.; Jang, J.M.; Han, D.H.; Molstad, M.; Kim, J.G.; et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat. Cell Biol. 2015, 17, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Cha-Molstad, H.; Lee, S.H.; Kim, J.G.; Sung, K.W.; Hwang, J.; Shim, S.M.; Ganipisetti, S.; McGuire, T.; Mook-Jung, I.; Ciechanover, A.; et al. Regulation of autophagic proteolysis by the N-recognin SQSTM1/p62 of the N-end rule pathway. Autophagy 2018, 14, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Yoon, H.S.; Heo, A.J.; Jung, E.J.; Ji, C.H.; Mun, S.R.; Lee, M.J.; Kwon, Y.T.; Park, J.W. Mitophagy and endoplasmic reticulum-phagy accelerated by a p62 ZZ ligand alleviates paracetamol-induced hepatotoxicity. Br. J. Pharmacol. 2023, 180, 1247–1266. [Google Scholar] [CrossRef] [PubMed]
- Schimmack, G.; Schorpp, K.; Kutzner, K.; Gehring, T.; Brenke, J.K.; Hadian, K.; Krappmann, D. YOD1/TRAF6 association balances p62-dependent IL-1 signaling to NF-kappaB. Elife 2017, 6, e22416. [Google Scholar] [CrossRef] [PubMed]
- Duran, A.; Amanchy, R.; Linares, J.F.; Joshi, J.; Abu-Baker, S.; Porollo, A.; Hansen, M.; Moscat, J.; Diaz-Meco, M.T. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell 2011, 44, 134–146. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kumanomidou, T.; Sou, Y.S.; Mizushima, T.; Ezaki, J.; Ueno, T.; Kominami, E.; Yamane, T.; Tanaka, K.; Komatsu, M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008, 283, 22847–22857. [Google Scholar] [CrossRef] [PubMed]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.-A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Curr. Opin. Toxicol. 2016, 1, 54–61. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Ichimura, Y.; Waguri, S.; Sou, Y.S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Lamark, T.; Sjottem, E.; Larsen, K.B.; Awuh, J.A.; Overvatn, A.; McMahon, M.; Hayes, J.D.; Johansen, T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010, 285, 22576–22591. [Google Scholar] [CrossRef] [PubMed]
- Inami, Y.; Waguri, S.; Sakamoto, A.; Kouno, T.; Nakada, K.; Hino, O.; Watanabe, S.; Ando, J.; Iwadate, M.; Yamamoto, M. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011, 193, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.M.; Boggess, N.; McGill, M.R.; Lebofsky, M.; Borude, P.; Apte, U.; Jaeschke, H.; Ding, W.X. Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol. Sci. 2012, 127, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.M.; Woolbright, B.L.; Williams, J.; Copple, B.; Cui, W.; Luyendyk, J.P.; Jaeschke, H.; Ding, W.X. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J. Hepatol. 2014, 61, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Isogai, S.; Morimoto, D.; Arita, K.; Unzai, S.; Tenno, T.; Hasegawa, J.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; Shirakawa, M.; et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 2011, 286, 31864–31874. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ni, H.M.; Jaeschke, H.; Ding, W.X. Chlorpromazine protects against acetaminophen-induced liver injury in mice by modulating autophagy and c-Jun N-terminal kinase activation. Liver Res. 2019, 3, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Bai, Q.; Yang, X.; Akakpo, J.Y.; Ji, L.; Yang, L.; Rulicke, T.; Zatloukal, K.; Jaeschke, H.; Ni, H.M.; et al. Dual roles of p62/SQSTM1 in the injury and recovery phases of acetaminophen-induced liver injury in mice. Acta Pharm. Sin. B 2021, 11, 3791–3805. [Google Scholar] [CrossRef]
- Cha-Molstad, H.; Yu, J.E.; Feng, Z.W.; Lee, S.H.; Kim, J.G.; Yang, P.; Han, B.; Sung, K.W.; Yoo, Y.D.; Hwang, J.; et al. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 2017, 8, 102. [Google Scholar] [CrossRef]
- Ji, C.H.; Kim, H.Y.; Heo, A.J.; Lee, S.H.; Lee, M.J.; Kim, S.B.; Srinivasrao, G.; Mun, S.R.; Cha-Molstad, H.; Ciechanover, A.; et al. The N-Degron Pathway Mediates ER-phagy. Mol. Cell 2019, 75, 1058–1072.e9. [Google Scholar] [CrossRef]
- Miller, S.; Muqit, M.M.K. Therapeutic approaches to enhance PINK1/Parkin mediated mitophagy for the treatment of Parkinson’s disease. Neurosci. Lett. 2019, 705, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Wang, X.J.; Zhao, F.; Villeneuve, N.F.; Wu, T.; Jiang, T.; Sun, Z.; White, E.; Zhang, D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Mol. Cell. Biol. 2010, 30, 3275–3285. [Google Scholar] [CrossRef] [PubMed]
- Reisman, S.A.; Csanaky, I.L.; Aleksunes, L.M.; Klaassen, C.D. Altered disposition of acetaminophen in Nrf2-null and Keap1-knockdown mice. Toxicol. Sci. Off. J. Soc. Toxicol. 2009, 109, 31–40. [Google Scholar] [CrossRef]
- Gum, S.I.; Cho, M.K. Recent updates on acetaminophen hepatotoxicity: The role of nrf2 in hepatoprotection. Toxicol. Res. 2013, 29, 165–172. [Google Scholar] [CrossRef]
- Moscat, J.; Karin, M.; Diaz-Meco, M.T. p62 in Cancer: Signaling Adaptor Beyond Autophagy. Cell 2016, 167, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, G.; Ballabio, A. TFEB at a glance. J. Cell Sci. 2016, 129, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Diab, H.I.; Brady, O.A.; Puertollano, R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016, 35, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Scotto-Rosato, A.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef]
- Wang, S.; Ni, H.M.; Chao, X.; Ma, X.; Kolodecik, T.; De Lisle, R.; Ballabio, A.; Pacher, P.; Ding, W.X. Critical Role of TFEB-Mediated Lysosomal Biogenesis in Alcohol-Induced Pancreatitis in Mice and Humans. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 59–81. [Google Scholar] [CrossRef]
- Chao, X.; Niu, M.; Wang, S.; Ma, X.; Yang, X.; Sun, H.; Hu, X.; Wang, H.; Zhang, L.; Huang, R.; et al. High-throughput screening of novel TFEB agonists in protecting against acetaminophen-induced liver injury in mice. Acta Pharm. Sin. B 2024, 14, 190–206. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Ni, H.-M.; McCracken, J.; Akakpo, J.Y.; Fulte, S.; Mckeen, T.; Jaeschke, H.; Wang, H.; Ding, W.X. Liver-specific deletion of mechanistic target of rapamycin does not protect against acetaminophen-induced liver injury in mice. Liver Res. 2021, 5, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Xu, Y.; Liu, G.; Shao, Q.; Niu, X.; Tai, W.; Shen, T.; Fan, M.; Chen, M.; Lei, L.; et al. Narirutin activates TFEB (transcription factor EB) to protect against Acetaminophen-induced liver injury by targeting PPP3/calcineurin. Autophagy 2023, 19, 2240–2256. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinz, K.; Niu, M.; Ni, H.-M.; Ding, W.-X. Targeting Autophagy for Acetaminophen-Induced Liver Injury: An Update. Livers 2024, 4, 377-387. https://doi.org/10.3390/livers4030027
Hinz K, Niu M, Ni H-M, Ding W-X. Targeting Autophagy for Acetaminophen-Induced Liver Injury: An Update. Livers. 2024; 4(3):377-387. https://doi.org/10.3390/livers4030027
Chicago/Turabian StyleHinz, Kaitlyn, Mengwei Niu, Hong-Min Ni, and Wen-Xing Ding. 2024. "Targeting Autophagy for Acetaminophen-Induced Liver Injury: An Update" Livers 4, no. 3: 377-387. https://doi.org/10.3390/livers4030027
APA StyleHinz, K., Niu, M., Ni, H.-M., & Ding, W.-X. (2024). Targeting Autophagy for Acetaminophen-Induced Liver Injury: An Update. Livers, 4(3), 377-387. https://doi.org/10.3390/livers4030027






