Artificial Intelligence, Machine Learning, and Deep Learning in the Diagnosis and Management of Hepatocellular Carcinoma
Abstract
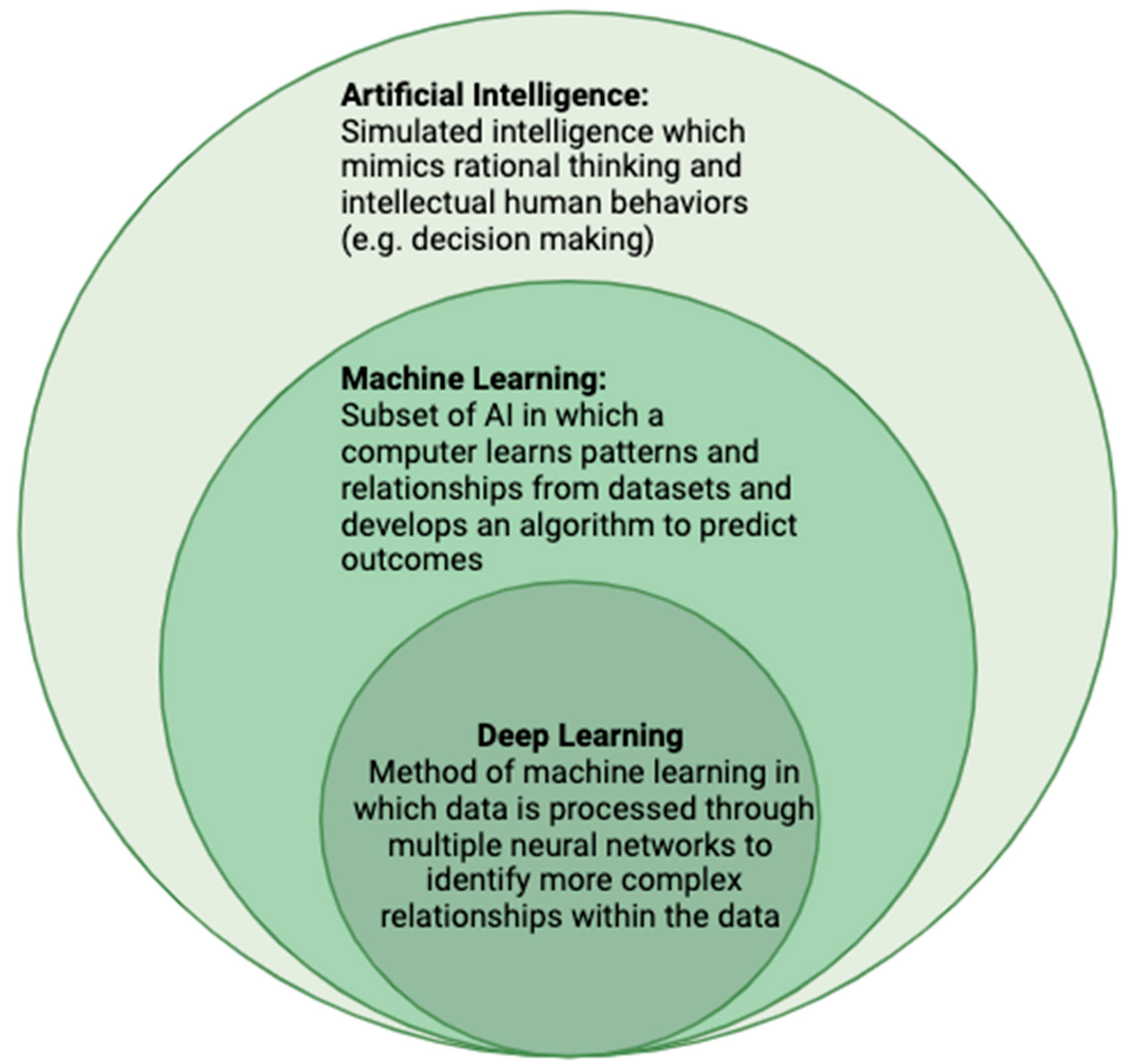
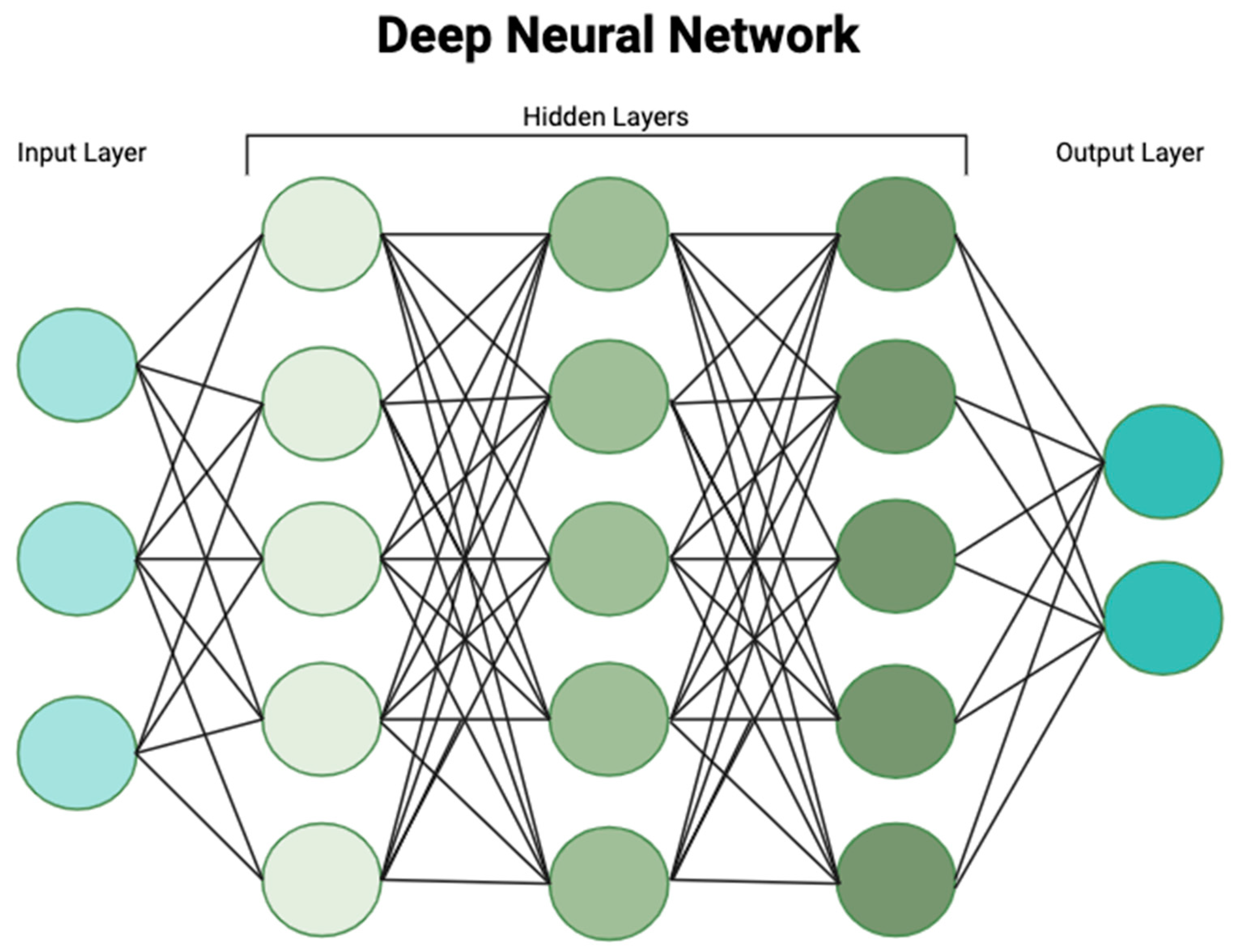
1. Introduction
2. Screening and Detection
| Author, Year | Model Design | Population | AI Methodology | Accuracy |
|---|---|---|---|---|
| Blanes-Vidal et al. (2022) [16] | Prediction of liver fibrosis using clinical data readily available to primary care physicians | Low-prevalence primary care population | Ensemble learning model | AUC: 0.86–0.94 |
| Ioannou et al. (2020) [18] | Identification of patients at high risk of developing HCC by extracting data from electronic medical records | Patients with known Hepatitis C Virus and cirrhosis | Recurrent neural network | AUROC: 0.759 |
| Yasaka et al. (2018) [20] | Differentiation of liver masses on CT, with categorization into HCC, other liver tumors, hemangiomas, or cysts | Patients who had undergone dynamic contrast-enhanced CT for evaluation of liver lesions | Convolutional neural network | AUROC: 0.92 |
| Mokrane et al. (2020) [21] | Diagnosis of liver nodules as HCC vs. non-HCC based on quantitative features extracted from triphasic CT | Patients with cirrhosis and biopsy-proven indeterminate liver nodules | Machine learning-based radiomic signature | AUROC: 0.66 |
| Schmauch et al. (2019) [22] | Detection and characterization of focal liver lesions as benign- vs. malignant-based on ultrasound characteristics | Patients with known liver nodules | Residual neural network | AUROC: 0.935 |
3. Prognosis and Treatment
3.1. HCC Prognosis and Risk of Recurrence
| Author, Year | Model Design | Pertinent Risk Factors | Population | AI Methodology | Accuracy |
|---|---|---|---|---|---|
| Chaudhary et al. (2018) [27] | Predictive model for HCC prognosis based on molecular signature and multi-omic data |
| HCC patients within the Genome Cancer Atlas (TCGA) | Deep learning | C-index: 0.68 |
| Liu et al. (2021) [30] | Prediction of MVI preoperatively based on CT imaging characteristics and patient clinical factors |
| Patients with HCC | Residual Neural Network | AUC: 0.845 |
| Chong et al. (2021) [32] | Creation of radiomic-based nomogram to preoperatively predict risk of MVI and recurrence-free survival, based. on MRI characteristics and clinical data |
| Patients with solitary HCC smaller than 5cm | Random Forrest | AUC: 0.92 |
| Ji et al. (2020) [35] | Creation of radiomic signature with pre- and post-resection features to predict recurrence for early-stage HCC |
| Patients with HCC that met the Milan Criteria and underwent curative intent resection | Machine learning-based radiomic signature | C-index: 0.77 |
3.2. Pathologic Assessment
| Author, Year | Model Design | Population | AI Methodology | Accuracy |
|---|---|---|---|---|
| Qu et al. (2022) [36] | Creation of histological score using whole-slide imaging to predict HCC recurrence | Patients with early-stage HCC who had undergone surgical resection in a single institutional dataset and the TCGA dataset | Convolutional neural network | C-index: 0.804 |
| Saillard et al. (2020) [37] | Use of whole-slide imaging to predict risk of HCC recurrence and stratifying it into low- and high-risk subgroups | Patients with HCC who had undergone surgical resection in a single institutional dataset and the TCGA dataset | Convolutional neural network | C-index: 0.72 |
| Yamashita et al. (2021) [40] | Use of whole-slide imaging to formulate a risk score predictive of HCC recurrence | Patients with HCC in the TCGA and Stanford-HCC dataset | Convolutional neural network | C-index: 0.724 |
| Zeng et al. (2022) [42] | Prediction of activation of immune gene signatures based on whole-slide imaging | Patients with HCC who had undergone surgical resection in the TCGA dataset | Clustering-constrained attention multiple instance learning | AUROC: 0.78–0.91 |
3.3. Locoregional Therapies
3.4. Automatic Methods for Liver and Tumor Segmentation
3.5. Surgical Complications
| Author, Year | Model Design | Population | AI Methodology | Accuracy |
|---|---|---|---|---|
| Wu et al. (2017) [42] | Prediction of disease-free survival after radiofrequency ablation based on clinical variables | Patients who underwent CT-guided radiofrequency ablation | Artificial neural network | AUC: 0.75–0.84 |
| Liu et al. (2020) [44] | Prediction of response to first TACE session using contrast- enhanced liver ultrasound | Patients who underwent ultrasound within one week of TACE for HCC | Radiomic-based deep learning | AUC: 0.81–0.93 |
| Meng et al. (2020) [50] | Automatic liver parenchyma and liver tumor segmentation from CT images | Multi-institutional liver tumor segmentation (LiTS) dataset | Dual path multiscale convolutional neural network | Dice: 0.689–0.965 |
| Zheng et al. (2022) [51] | Automatic segmentation of HCC lesions based on dynamic MRI | Patients with HCC who underwent dynamic contrast-enhanced MRI | Convolutional neural network and recurrence neural network | Dice: 0.825 |
| Wang et al. (2022) [52] | Prediction of post-hepatectomy liver failure based on clinical characteristics and surgical variables | Patients with HCC who underwent hepatectomy | Light gradient boosting machine learning | AUC: 0.822–0.944 |
4. Intraoperative Use of Artificial Intelligence
5. Challenges and Future Directions
5.1. Barriers to Clinical Implementation
5.2. The Role of AI in Healthcare Disparities
5.3. Potential Applications
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fitzmaurice, C.; Akinyemiju, T.F.; Al Lami, F.H.; Alam, T.; Alizadeh-Navaei, R.; Allen, C.; Alsharif, U.; Alvis-Guzman, N.; Amini, E.; Anderson, B.O.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2016: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2018, 4, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Chen, M.; Colombo, M.; Roberts, L.R.; Schwartz, M.; Chen, P.J.; Kudo, M.; Johnson, P.; Wagner, S.; Orsini, L.S.; et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: The BRIDGE Study. Liver Int. 2015, 35, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Meyer, T.; Sapisochin, G.; Salem, R.; Saborowski, A. Hepatocellular carcinoma. Lancet 2022, 400, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Choi, R.Y.; Coyner, A.S.; Kalpathy-Cramer, J.; Chiang, M.F.; Campbell, J.P. Introduction to Machine Learning, Neural Networks, and Deep Learning. Transl. Vis. Sci. Technol. 2020, 9, 14. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Rudin, C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Taroni, J.N.; Allaway, R.J.; Prasad, D.V.; Guinney, J.; Greene, C. Machine learning in rare disease. Nat. Methods 2023, 20, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.Y.; Liu, X.C.; Nejatian, N.P.; Nasir-Moin, M.; Wang, D.; Abidin, A.; Eaton, K.; Riina, H.A.; Laufer, I.; Punjabi, P.; et al. Health system-scale language models are all-purpose prediction engines. Nature 2023, 619, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.; Zheng, Z.; Fang, X.; Jiang, H.; Zhu, M.; Yu, J.; Zhao, H.; Zhang, L.; Yao, J.; Lu, L.; et al. Artificial Intelligence to Predict Lymph Node Metastasis at CT in Pancreatic Ductal Adenocarcinoma. Radiology 2023, 306, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M. Management of Hepatocellular Carcinoma in Japan as a World-Leading Model. Liver Cancer 2018, 7, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Blanes-Vidal, V.; Lindvig, K.P.; Thiele, M.; Nadimi, E.S.; Krag, A. Artificial intelligence outperforms standard blood-based scores in identifying liver fibrosis patients in primary care. Sci. Rep. 2022, 12, 2914. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Mukherjee, A.; Elmunzer, B.J.; Higgins, P.D.; Lok, A.S.; Zhu, J.; Marrero, J.A.; Waljee, A.K. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am. J. Gastroenterol. 2013, 108, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, G.N.; Tang, W.; Beste, L.A.; Tincopa, M.A.; Su, G.L.; Van, T.; Tapper, E.B.; Singal, A.G.; Zhu, J.; Waljee, A.K. Assessment of a Deep Learning Model to Predict Hepatocellular Carcinoma in Patients with Hepatitis C Cirrhosis. JAMA Netw. Open 2020, 3, e2015626. [Google Scholar] [CrossRef]
- Mitchell, D.G.; Bruix, J.; Sherman, M.; Sirlin, C.B. LI-RADS (Liver Imaging Reporting and Data System): Summary, discussion, and consensus of the LI-RADS Management Working Group and future directions. Hepatology 2015, 61, 1056–1065. [Google Scholar] [CrossRef]
- Yasaka, K.; Akai, H.; Abe, O.; Kiryu, S. Deep Learning with Convolutional Neural Network for Differentiation of Liver Masses at Dynamic Contrast-enhanced CT: A Preliminary Study. Radiology 2018, 286, 887–896. [Google Scholar] [CrossRef]
- Mokrane, F.Z.; Lu, L.; Vavasseur, A.; Otal, P.; Peron, J.M.; Luk, L.; Yang, H.; Ammari, S.; Saenger, Y.; Rousseau, H.; et al. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur. Radiol. 2020, 30, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Schmauch, B.; Herent, P.; Jehanno, P.; Dehaene, O.; Saillard, C.; Aubé, C.; Luciani, A.; Lassau, N.; Jégou, S. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn. Interv. Imaging 2019, 100, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Wei, J.; Hao, X.; Kong, D.; Yu, X.; Jiang, T.; Xi, J.; Cai, W.; Luo, Y.; Jing, X.; et al. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: A multicentre study. EBioMedicine 2020, 56, 102777. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.J.A.; Kuijf, H.J.; Veldhuis, W.B.; Wessels, F.J.; Viergever, M.A.; Pluim, J.P.W. Automatic classification of focal liver lesions based on MRI and risk factors. PLoS ONE 2019, 14, e0217053. [Google Scholar] [CrossRef] [PubMed]
- McMahon, B.; Cohen, C.; Brown, R.S., Jr.; El-Serag, H.; Ioannou, G.N.; Lok, A.S.; Roberts, L.R.; Singal, A.G.; Block, T. Opportunities to address gaps in early detection and improve outcomes of liver cancer. JNCI Cancer Spectr. 2023, 7, pkad034. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiao, C.; Yue, K.; Chen, M.; Zhou, H.; Yan, X. Identification of multi-omics biomarkers and construction of the novel prognostic model for hepatocellular carcinoma. Sci. Rep. 2022, 12, 12084. [Google Scholar] [CrossRef]
- Chaudhary, K.; Poirion, O.B.; Lu, L.; Garmire, L.X. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin. Cancer Res. 2018, 24, 1248–1259. [Google Scholar] [CrossRef]
- Lee, S.; Kang, T.W.; Song, K.D.; Lee, M.W.; Rhim, H.; Lim, H.K.; Kim, S.Y.; Sinn, D.H.; Kim, J.M.; Kim, K.; et al. Effect of Microvascular Invasion Risk on Early Recurrence of Hepatocellular Carcinoma after Surgery and Radiofrequency Ablation. Ann. Surg. 2021, 273, 564–571. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, S.; Xu, Y.; Wu, J. Diagnostic Accuracy of Artificial Intelligence Based on Imaging Data for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 763842. [Google Scholar] [CrossRef]
- Liu, S.C.; Lai, J.; Huang, J.Y.; Cho, C.F.; Lee, P.H.; Lu, M.H.; Yeh, C.C.; Yu, J.; Lin, W.C. Predicting microvascular invasion in hepatocellular carcinoma: A deep learning model validated across hospitals. Cancer Imaging 2021, 21, 56. [Google Scholar] [CrossRef]
- Sun, S.W.; Xu, X.; Liu, Q.P.; Chen, J.N.; Zhu, F.P.; Liu, X.S.; Zhang, Y.D.; Wang, J. LiSNet: An artificial intelligence -based tool for liver imaging staging of hepatocellular carcinoma aggressiveness. Med. Phys. 2022, 49, 6903–6913. [Google Scholar] [CrossRef]
- Chong, H.H.; Yang, L.; Sheng, R.F.; Yu, Y.L.; Wu, D.J.; Rao, S.X.; Yang, C.; Zeng, M.S. Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤ 5 cm. Eur. Radiol. 2021, 31, 4824–4838. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; He, N.; Yang, J.J.; Xiao, J.J.; Zhang, Y.; Du, J.; Zuo, S.; Li, H.Y.; Gu, H. Prediction of 3-year recurrence rate of hepatocellular carcinoma after resection based on contrast-enhanced CT: A single-centre study. Br. J. Radiol. 2023, 96, 20220702. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Zhang, X.; Zhang, B.; Geng, Z.; Xie, C.; Yang, W.; Zhang, S.; Qi, Z.; Lin, T.; Ke, Q.; et al. Deep learning nomogram based on Gd-EOB-DTPA MRI for predicting early recurrence in hepatocellular carcinoma after hepatectomy. Eur. Radiol. 2023, 33, 4949–4961. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.W.; Zhu, F.P.; Xu, Q.; Wang, K.; Wu, M.Y.; Tang, W.W.; Li, X.C.; Wang, X.H. Radiomic Features at Contrast-enhanced CT Predict Recurrence in Early Stage Hepatocellular Carcinoma: A Multi-Institutional Study. Radiology 2020, 294, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.F.; Tian, M.X.; Qiu, J.T.; Guo, Y.C.; Tao, C.Y.; Liu, W.R.; Tang, Z.; Qian, K.; Wang, Z.X.; Li, X.Y.; et al. Exploring pathological signatures for predicting the recurrence of early-stage hepatocellular carcinoma based on deep learning. Front. Oncol. 2022, 12, 968202. [Google Scholar] [CrossRef] [PubMed]
- Saillard, C.; Schmauch, B.; Laifa, O.; Moarii, M.; Toldo, S.; Zaslavskiy, M.; Pronier, E.; Laurent, A.; Amaddeo, G.; Regnault, H.; et al. Predicting Survival after Hepatocellular Carcinoma Resection Using Deep Learning on Histological Slides. Hepatology 2020, 72, 2000–2013. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Heij, L.R.; Grabsch, H.I.; Loeffler, C.; Echle, A.; Muti, H.S.; Krause, J.; Niehues, J.M.; Sommer, K.A.J.; Bankhead, P.; et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat. Cancer 2020, 1, 789–799. [Google Scholar] [CrossRef]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef]
- Yamashita, R.; Long, J.; Saleem, A.; Rubin, D.L.; Shen, J. Deep learning predicts postsurgical recurrence of hepatocellular carcinoma from digital histopathologic images. Sci. Rep. 2021, 11, 2047. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, B.; Topatana, W.; Cao, J.; Zhu, H.; Juengpanich, S.; Mao, Q.; Yu, H.; Cai, X. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis. Oncol. 2020, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Klein, C.; Caruso, S.; Maille, P.; Laleh, N.G.; Sommacale, D.; Laurent, A.; Amaddeo, G.; Gentien, D.; Rapinat, A.; et al. Artificial intelligence predicts immune and inflammatory gene signatures directly from hepatocellular carcinoma histology. J. Hepatol. 2022, 77, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.F.; Wu, Y.J.; Liang, P.C.; Wu, C.H.; Peng, S.F.; Chiu, H.W. Disease-free survival assessment by artificial neural networks for hepatocellular carcinoma patients after radiofrequency ablation. J. Formos. Med. Assoc. 2017, 116, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Liu, F.; Xie, X.; Su, L.; Liu, M.; Xie, X.; Kuang, M.; Huang, G.; Wang, Y.; Zhou, H.; et al. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur. Radiol. 2020, 30, 2365–2376. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Kloeckner, R.; Mähringer-Kunz, A.; Stoehr, F.; Düber, C.; Arnhold, G.; Gairing, S.J.; Foerster, F.; Weinmann, A.; Galle, P.R.; et al. Fully automated AI-based splenic segmentation for predicting survival and estimating the risk of hepatic decompensation in TACE patients with HCC. Eur. Radiol. 2022, 32, 6302–6313. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, X.; Lv, P.; Wang, H.; Cheng, Y. Automatic Liver Segmentation Using EfficientNet and Attention-Based Residual U-Net in CT. J. Digit. Imaging 2022, 35, 1479–1493. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Wu, F.; Kong, D.; Mao, X. Automatic liver segmentation using 3D convolutional neural networks with a hybrid loss function. Med. Phys. 2021, 48, 1707–1719. [Google Scholar] [CrossRef] [PubMed]
- Özcan, F.; Uçan, O.N.; Karaçam, S.; Tunçman, D. Fully Automatic Liver and Tumor Segmentation from CT Image Using an AIM-Unet. Bioengineering 2023, 10, 215. [Google Scholar] [CrossRef]
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Şahin, Y.; Özkan, S.; Baydar, B.; Yüksel, U.; Kılıkçıer, Ç.; Olut, Ş.; Bozdağı Akar, G.; et al. Comparison of semi-automatic and deep learning-based automatic methods for liver segmentation in living liver transplant donors. Diagn. Interv. Radiol. 2020, 26, 11–21. [Google Scholar] [CrossRef]
- Meng, L.; Tian, Y.; Bu, S. Liver tumor segmentation based on 3D convolutional neural network with dual scale. J. Appl. Clin. Med. Phys. 2020, 21, 144–157. [Google Scholar] [CrossRef]
- Zheng, R.; Wang, Q.; Lv, S.; Li, C.; Wang, C.; Chen, W.; Wang, H. Automatic Liver Tumor Segmentation on Dynamic Contrast Enhanced MRI Using 4D Information: Deep Learning Model Based on 3D Convolution and Convolutional LSTM. IEEE Trans. Med. Imaging 2022, 41, 2965–2976. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, T.; Liao, Y.; Geng, S.; Li, J.; Zhang, Z.; Shang, D.; Liu, C.; Yu, P.; Huang, Y.; et al. Machine learning prediction model for post- hepatectomy liver failure in hepatocellular carcinoma: A multicenter study. Front. Oncol. 2022, 12, 986867. [Google Scholar] [CrossRef] [PubMed]
- Laino, M.E.; Fiz, F.; Morandini, P.; Costa, G.; Maffia, F.; Giuffrida, M.; Pecorella, I.; Gionso, M.; Wheeler, D.R.; Cambiaghi, M.; et al. A virtual biopsy of liver parenchyma to predict the outcome of liver resection. Updates Surg. 2023, 75, 1519–1531. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Q.; Du, B.; Wang, X.; Xue, Q.; Gao, W. A meta-analysis of the three-dimensional reconstruction visualization technology for hepatectomy. Asian J. Surg. 2023, 46, 669–676. [Google Scholar] [CrossRef] [PubMed]
- Takamoto, T.; Ban, D.; Nara, S.; Mizui, T.; Nagashima, D.; Esaki, M.; Shimada, K. Automated Three-Dimensional Liver Reconstruction with Artificial Intelligence for Virtual Hepatectomy. J. Gastrointest. Surg. 2022, 26, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Barash, Y.; Klang, E.; Lux, A.; Konen, E.; Horesh, N.; Pery, R.; Zilka, N.; Eshkenazy, R.; Nachmany, I.; Pencovich, N. Artificial intelligence for identification of focal lesions in intraoperative liver ultrasonography. Langenbecks Arch. Surg. 2022, 407, 3553–3560. [Google Scholar] [CrossRef] [PubMed]
- Takamoto, T.; Nara, S.; Ban, D.; Mizui, T.; Murase, Y.; Esaki, M.; Shimada, K. Enhanced Recognition Confidence of Millimeter-Sized Intrahepatic Targets by Real-Time Virtual Sonography. J. Ultrasound Med. 2023, 42, 1789–1797. [Google Scholar] [CrossRef] [PubMed]
- Golse, N.; Petit, A.; Lewin, M.; Vibert, E.; Cotin, S. Augmented Reality during Open Liver Surgery Using a Markerless Non-rigid Registration System. J. Gastrointest. Surg. 2021, 25, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, J.; Zhou, W.; An, P.; Shen, L.; Liu, J.; Jiang, X.; Huang, X.; Mu, G.; Wan, X.; et al. Randomised controlled trial of WISENSE, a real-time quality improving system for monitoring blind spots during esophagogastroduodenoscopy. Gut 2019, 68, 2161–2169. [Google Scholar] [CrossRef]
- Wang, P.; Berzin, T.M.; Glissen Brown, J.R.; Bharadwaj, S.; Becq, A.; Xiao, X.; Liu, P.; Li, L.; Song, Y.; Zhang, D.; et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: A prospective randomised controlled study. Gut 2019, 68, 1813–1819. [Google Scholar] [CrossRef]
- Titano, J.J.; Badgeley, M.; Schefflein, J.; Pain, M.; Su, A.; Cai, M.; Swinburne, N.; Zech, J.; Kim, J.; Bederson, J.; et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat. Med. 2018, 24, 1337–1341. [Google Scholar] [CrossRef] [PubMed]
- INFANT Collaborative Group. Computerised interpretation of fetal heart rate during labour (INFANT): A randomised controlled trial. Lancet 2017, 389, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Rocher, L.; Hendrickx, J.M.; de Montjoye, Y.A. Estimating the success of re-identifications in incomplete datasets using generative models. Nat Commun. 2019, 10, 3069. [Google Scholar] [CrossRef] [PubMed]
- Tom, E.; Keane, P.A.; Blazes, M.; Pasquale, L.R.; Chiang, M.F.; Lee, A.Y.; Lee, C.S.; AAO Artificial Intelligence Task Force. Protecting Data Privacy in the Age of AI-Enabled Ophthalmology. Transl. Vis. Sci. Technol. 2020, 9, 36. [Google Scholar] [CrossRef]
- Wu, G.; Segovis, C.S.; Nicola, L.P.; Chen, M.M. Current Reimbursement Landscape of Artificial Intelligence. J. Am. Coll. Radiol. 2023, 20, 957–961. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef]
- Yang, X.; Amgad, M.; Cooper, L.A.D.; Du, Y.; Fu, H.; Ivanov, A.A. High expression of MKK3 is associated with worse clinical outcomes in African American breast cancer patients. J. Transl. Med. 2020, 18, 334. [Google Scholar] [CrossRef]
- Pierson, E.; Cutler, D.M.; Leskovec, J.; Mullainathan, S.; Obermeyer, Z. An algorithmic approach to reducing unexplained pain disparities in underserved populations. Nat. Med. 2021, 27, 136–140. [Google Scholar] [CrossRef]
- Brar Prayaga, R.; Agrawal, R.; Nguyen, B.; Jeong, E.W.; Noble, H.K.; Paster, A.; Prayaga, R.S. Impact of Social Determinants of Health and Demographics on Refill Requests by Medicare Patients Using a Conversational Artificial Intelligence Text Messaging Solution: Cross-Sectional Study. JMIR Mhealth Uhealth 2019, 7, e15771. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Z.; Wu, J.; Liu, K. An Intelligent Decision-Making Support System for the Detection and Staging of Prostate Cancer in Developing Countries. Comput. Math. Methods Med. 2020, 2020, 5363549. [Google Scholar] [CrossRef]
- Uthoff, R.D.; Song, B.; Sunny, S.; Patrick, S.; Suresh, A.; Kolur, T.; Keerthi, G.; Spires, O.; Anbarani, A.; Wilder-Smith, P.; et al. Point-of-care, smartphone-based, dual-modality, dual-view, oral cancer screening device with neural network classification for low-resource communities. PLoS ONE 2018, 13, e0207493. [Google Scholar] [CrossRef] [PubMed]
- Sidey-Gibbons, C.; Pfob, A.; Asaad, M.; Boukovalas, S.; Lin, Y.L.; Selber, J.C.; Butler, C.E.; Offodile, A.C., 2nd. Development of Machine Learning Algorithms for the Prediction of Financial Toxicity in Localized Breast Cancer Following Surgical Treatment. JCO Clin. Cancer Inform. 2021, 5, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Choo, J.M.; Ryu, H.S.; Kim, J.S.; Cheong, J.Y.; Baek, S.J.; Kwak, J.M.; Kim, J. Conversational artificial intelligence (chatGPT™) in the management of complex colorectal cancer patients: Early experience. ANZ J. Surg. 2023, 13, 1502. [Google Scholar] [CrossRef]
- Griewing, S.; Gremke, N.; Wagner, U.; Lingenfelder, M.; Kuhn, S.; Boekhoff, J. Challenging ChatGPT 3.5 in Senology-An Assessment of Concordance with Breast Cancer Tumor Board Decision Making. J. Pers. Med. 2023, 13, 1502. [Google Scholar] [CrossRef]
- Park, Y.E.; Chae, H. The Fidelity of Artificial Intelligence to Multidisciplinary Tumor Board Recommenda-tions for Patients with Gastric Cancer: A Retrospective Study. J. Gastrointest. Cancer 2023. [Google Scholar] [CrossRef]
- Vela Ulloa, J.; King Valenzuela, S.; Riquoir Altamirano, C.; Urrejola Schmied, G. Artificial intelligence-based decision-making: Can ChatGPT replace a multidisciplinary tumour board? Br. J. Surg. 2023, 110, 1543–1544. [Google Scholar] [CrossRef]
- Ng, S.S.T.; Oehring, R.; Ramasetti, N.; Roller, R.; Thomas, P.; Chen, Y.; Moosburner, S.; Winter, A.; Maurer, M.M.; Auer, T.A.; et al. Concordance of a decision algorithm and multidisciplinary team meetings for patients with liver cancer-a study protocol for a randomized controlled trial. Trials 2023, 24, 577. [Google Scholar] [CrossRef]
- Shen, M.; Zou, Z.; Bao, H.; Fairley, C.K.; Canfell, K.; Ong, J.J.; Hocking, J.; Chow, E.P.F.; Zhuang, G.; Wang, L.; et al. Cost-effectiveness of artificial intelligence-assisted liquid-based cytology testing for cervical cancer screening in China. Lancet Reg. Health West Pac. 2023, 34, 100726. [Google Scholar] [CrossRef]
- Mital, S.; Nguyen, H.V. Cost-effectiveness of using artificial intelligence versus polygenic risk score to guide breast cancer screening. BMC Cancer 2022, 22, 501. [Google Scholar] [CrossRef]
- Ziegelmayer, S.; Graf, M.; Makowski, M.; Gawlitza, J.; Gassert, F. Cost-Effectiveness of Artificial Intelligence Support in Computed Tomography-Based Lung Cancer Screening. Cancers 2022, 14, 1729. [Google Scholar] [CrossRef]
- Valvert, F.; Silva, O.; Solórzano-Ortiz, E.; Puligandla, M.; Siliézar Tala, M.M.; Guyon, T.; Dixon, S.L.; López, N.; López, F.; Carías Alvarado, C.C.; et al. Low-cost transcriptional diagnostic to accurately categorize lymphomas in low- and middle-income countries. Blood Adv. 2021, 5, 2447–2455. [Google Scholar] [CrossRef] [PubMed]
- Wolff, J.; Pauling, J.; Keck, A.; Baumbach, J. The Economic Impact of Artificial Intelligence in Health Care: Systematic Review. J. Med. Internet Res. 2020, 22, e16866. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larrain, C.; Torres-Hernandez, A.; Hewitt, D.B. Artificial Intelligence, Machine Learning, and Deep Learning in the Diagnosis and Management of Hepatocellular Carcinoma. Livers 2024, 4, 36-50. https://doi.org/10.3390/livers4010004
Larrain C, Torres-Hernandez A, Hewitt DB. Artificial Intelligence, Machine Learning, and Deep Learning in the Diagnosis and Management of Hepatocellular Carcinoma. Livers. 2024; 4(1):36-50. https://doi.org/10.3390/livers4010004
Chicago/Turabian StyleLarrain, Carolina, Alejandro Torres-Hernandez, and Daniel Brock Hewitt. 2024. "Artificial Intelligence, Machine Learning, and Deep Learning in the Diagnosis and Management of Hepatocellular Carcinoma" Livers 4, no. 1: 36-50. https://doi.org/10.3390/livers4010004
APA StyleLarrain, C., Torres-Hernandez, A., & Hewitt, D. B. (2024). Artificial Intelligence, Machine Learning, and Deep Learning in the Diagnosis and Management of Hepatocellular Carcinoma. Livers, 4(1), 36-50. https://doi.org/10.3390/livers4010004





