Abstract
Cupriavidus metallidurans is a Gram-negative bacterium that has attracted the attention of the scientific community since its discovery back in 1976. It was initially studied as a model organism for bioremediation processes due to its ability to survive in heavy metal-rich environments. However, in recent years, there is evidence that this bacterium can be a potential pathogen for humans. How C. metallidurans can survive in such different environments is unknown and prompted the following work. Its great adaptability could be explained by the structural and conformational studies of the O-antigen portion of the lipopolysaccharide, the main constituent of the outer membrane of Gram-negative bacteria, which is the one in direct contact with the external environment. Therefore, a combination of chemical and spectroscopic analyses was used to define the O-antigen structure, disclosing that it is a polysaccharide constituted of a linear tetrasaccharide repeating unit that does not resemble other structures already reported for bacteria: [4)-α-d-GalNAc-(1→3)-α-d-Qui2NAc4NHBA-(1→3)-α-l-Rha-(1→3)-α-l-Rha-(1→]. Interestingly, the molecular dynamics studies revealed that the three-dimensional structure of the O-antigen is highly flexible: it might adopt three different right-handed helix conformations described by a two, three, or four-fold symmetry. This conformational behavior could represent the reason behind the survival of C. metallidurans in different environments.
1. Introduction
Cupriavidus metallidurans, previously misattributed to the genera Ralstonia, is a Gram-negative bacterium, isolated as far back as 1976 from the wastewater of a zinc factory in Liège, Belgium [1]. Later, this bacterium was found as a dominant microorganism in biofilms on gold grains in the Prophet mine in Queensland, Australia [2]. C. metallidurans is a facultative anaerobe that is unable to use sugars as a carbon source, preferring instead alcohols, carbon dioxide, and hydrogen gas [3]. Interestingly, this bacterium is resistant to toxic heavy metals, and for this reason, it is used as model organism to understand this process and for its possible application in the bioremediation. However, it was also considered as a “modern alchemist” due to its ability to precipitate solid gold from aqueous gold (III) tetrachloride, producing 24 carat gold in a week [2,4].
Recently, the attention on Cupriavidus metallidurans has increased due to growing evidence that this bacterium may be a human pathogen. Indeed, in 2011, there was the first case in which a patient with multiple complications, as result of an invasive surgery, contracted C. metallidurans as a nosocomial infection [5]. Furthermore, two strains of C. metallidurans were also identified in the respiratory secretions of patients with cystic fibrosis [6]. Yet, the role of C. metallidurans in all these cases is unclear [6]. Later, several cases of catheter-related bacteremia by C. metallidurans have been reported [7]. Finally, C. metallidurans is considered as an emerging pathogen responsible for systemic infections not limited to immunocompromised patients [7].
It is clear that C. metallidurans can adapt to quite different environments as those rich in heavy metals or into living organisms during hospital-acquired infection.
In this context, the study of the outer membrane of C. metallidurans, with a special focus on lipopolysaccharides, can shed light on the great adaptability of this bacterium. Indeed, as it is known, lipopolysaccharides (LPSs) are the main constituent of the Gram-negative bacteria’s outer-membrane, and they play crucial roles, such as regulating the inflow and outflow of nutrients, and they act as a protective barrier from the external environment, including antibiotics [8]. In addition, the lipopolysaccharides are also known as endotoxins: they are referred to as Pathogen-Associated Molecular Patterns, and when recognized by specific receptors of the host, collectively named pattern recognition receptors, they activate the innate immune response in multicellular organisms [8]. LPSs are complex macromolecules composed by three domains: lipid A, which presents a disaccharide skeleton made of two units of D-glucosamine and acyl chains; the core oligosaccharide divided into two distinct domains, the inner and outer core; and the O-chain portion, also known as O-antigen, that consists of a polysaccharide built with several copies of repeating units each made of two to eight monosaccharides [9]. However, not all the bacteria have the O-antigen, and this creates a further subdivision into R-type (Rough: the O-antigen is lacking) and S-type (Smooth: the O-antigen is present).
The O-antigen is the most external part of the LPS, and it is in contact with the environment [9]. This portion exhibits a great variability even among bacteria of the same species, and its structure can be modulated in response to environmental factors.
In this study, both the chemical structure and the conformation of the O-antigen from the LPS of C. metallidurans CH34 have been characterized, since they represent a fundamental prerequisite to understand the mechanisms developed by this bacterium to survive in different environments.
2. Materials and Methods
2.1. Growth of C. Metallidurans and Lipopolysaccharide Purification
Cupriavidus metallidurans CH34 was obtained from the DSMZ collection (catalogue number DSM 2839) and it was grown in nutrient broth No. 4 (Cod. 03856-500G, Sigma Aldrich, St. Louis, MO 68178, USA) (16 L) at 28 °C for 48 h under constant shaking. Cells were recovered by centrifugation, 6300 g at 4 °C for 20 min, and freeze-dried (6.7 g).
The lipopolysaccharide was extracted from the dried cells following the PCP procedure, as described elsewhere [10]. All phases were dialyzed against water using a membrane of 12–14 kDa (Cod. 15320762, Fisher Scientific Italia, Rodano, Italy) and then were freeze-dried, yielding two fractions: a precipitate (PCPprec) and a phenol-soluble fraction (PCPsol) in 158 mg and 383 mg yield, respectively. A 15% polyacrylamide gel electrophoresis (SDS-PAGE) analysis, conducted in denaturing conditions [10], and stained with silver nitrate, revealed the presence of the (S)-type LPS in the phenol phase of the PCP extraction only. Therefore, the following analyses were conducted on this fraction.
2.2. LPS Sugar Composition
First, 0.5 mg of LPS were used to analyze the monosaccharides composition as acetylated O-methylglycosides derivatives (AMG), as described [10]. Linkage analysis was carried out on 0.5 mg of LPS derivatized as partially methylated and acetylated alditols (PMAA), as described elsewhere [10].
The absolute configuration of the sugars components, galactosamine (GalNAc) and rhamnose (Rha), was performed as reported [10]. To determine the R/S configuration of hydroxylbutyric acid (HBA), the commercial compound (Cod. H3145, Sigma Aldrich, St. Louis, MO 68178, USA) was derivatized as 2-(-)-octylester and then was trifluoro acetylated with trifluoroacetic anhydride (100 μL, 4 °C, 16 h), as described [11].
All these derivatives were analyzed by GC-MS (GC instrument Agilent 6850 coupled to MS Agilent 5973, Santa Clara, CA 95051, USA) equipped with a SPB-5 capillary column (Supelco, 30 m × 0.25 i.d., flow rate, 0.8 mL min−1), using He as the carrier gas. Electron impact mass spectra were recorded with an ionization energy of 70 eV and an ionizing current of 0.2 mA. The temperature program used for all analyses was set as follows: 150 °C for 5 min, 150→280 °C at 3 °C/min, 300 °C for 5 min; except for the trifluoro acetylated HBA, for which the temperature program was: 90 °C for 3 min 90→150 °C at 5 °C/min, 150 °C for 5 min, 150→300 °C at 25 °C/min, 300 °C for 5 min.
2.3. O-Antigen Purification
The O-antigen was obtained by cleaving the lipid A portion from the LPS under mild acid hydrolysis. Briefly, LPS (18 mg) was heated in 1% acid acetic solution (1.8 mL) for 2 h at 100 °C. Then, the lipid A was recovered as a white solid by centrifugation (4 °C, 514 g, 30 min); instead, the supernatant contained the O-antigen plus the core region fragments (14 mg). The latter was purified by gel permeation chromatography using a Biogel P-10 (fractionation range evaluated for dextrans: 1.5–20 kDa) eluted with 50 mM NH4HCO3 (flow: 17.4 mL/h). The eluate was monitored with an online refractive index detector (Knauer K-2310, Berlin, Germany); the fractions were pooled accordingly and checked via 1H-NMR spectroscopy. The O-antigen (13 mg) was eluted in the column void volume.
2.4. NMR Analysis of the O-Antigen
The 1D and 2D NMR spectra were carried out on a Bruker DRX-600 spectrometer equipped with a cryoprobe. Spectra were recorded in D2O, at 318 K and calibrated with acetone as the internal standard.
The 1D 1H NMR experiment was set at a resolution of 16 k complex data points, and the frequency carrier was placed in correspondence of the D2O signal. For 2D homonuclear NMR experiments (DQ-COSY, TOCSY, NOESY, HSQC, HMBC), 512 FID (Free Induction Decay) of 2 k complex data points were collected, and 24 total scans were acquired. Mixing times of 100 and 200 ms were used for TOCSY and NOESY spectra acquisition, respectively. HSQC and HMBC spectra were acquired with 40 and 64 scans for FIDs, respectively. The elaboration and analysis of spectra was performed with Bruker Topspin 3.0 software.
2.5. Determination of Qui2NAc4HBA Absolute Configuration and Conformational Analysis of the O-Antigen
To determine the absolute configuration of the Qui2NAc4NHBA residue, the following steps were applied:
∙ Based on the O-antigen structure elucidated by NMR, six different disaccharides were built using the MacroModel (version 9.9, Schrödinger, LLC, New York, NY, USA) package: (1) α-d-GalNAc-(1→3)-α-d-Qui2NAc4NHBA; (2) α-d-GalNAc-(1→3)-α-l-Qui2NAc4NHBA; (3) α-d-Qui2NAc4NHBA-(1→3)-α-l-Rha; (4) α-l-Qui2NAc4NHBA-(1→3)-α-l-Rha; (5) α-l-Rha-(1→3)-α-l-Rha, and (6) α-l-Rha-(1→4)-α-d-GalNAc;
∙ For the glycoside junction of each disaccharide, the optimal value of the dihedral angles Φ/Ψ (Φ = H1-C1-O-C’n; Ψ = C1-O-C’n–H’n) was calculated using the Molecular Mechanics approach with MacroModel 9.9 software, under the Maestro graphical interface (MM3 force field and a dielectric constant ε = 80 as an approximation of the bulk water were set);
∙ Energy maps were calculated employing the Coordinate Scan utility (modulated with the DEBG option 150), by varying incrementally both Φ and Ψ using a grid step of 18 and each (Φ, Ψ) point of the map was optimized using 1000 P.R. conjugate gradients. The plotting and the analysis of the flexible maps was performed with the 2D-plot facility built into the MacroModel package;
∙ The optimal values of the pairs Φ/Ψ (those at minimum energy) of each glycosidic junction were used to construct 2 pentasaccharides: one with the Qui2NAc4NHB with D configuration (D-penta) and one with the same residue with L configuration (L-penta). The behavior of the two oligomers in solution was simulated by Molecular Dynamics simulation using the MM3 force field and approximating the aqueous solvent setting the dielectric constant ε = 80. Each oligomer was placed in a thermal bath at 318 K for 40 ns, the equilibration time was set to 200 ps, and a dynamic time-step of 1.5 fs was applied together with SHAKE protocol to the carbon-hydrogen bonds. An ensemble of 10,000 structures was collected; among them, the structures at lower energy were chosen.
∙ The NOE effects of the two oligosaccharides were simulated with NOEPROM 2000, v. 17.0 program, and compared with the experimental one (obtained from NOESY spectrum), giving information on the configuration of the Qui2NAc4NHBA [12].
Once the configuration of the Qui2NAc4NHBA was established as D, the best conformational parameter of the helix (h: axial rise; n: the number of repeating units for revolution of the helix) were evaluated using POLYS version 1.4 (2004) [13] in order to describe the O-antigen 3D structure.
This program requests as input the residues constituting the repeating unit, their sequence, and the optimal values of the glycosidic junctions (Φ/Ψ), which are expressed in the formalism of the X-ray (Φ = O5-C1-O-C’n; Ψ = C1-O-C’n-C’n + 1). Importantly, before starting, the POLYS database needed to be implemented with the rare Qui2NAc4NHB monosaccharide; therefore, the coordinates of this monosaccharide were converted from the MacroModel format to that of POLYS [13].
The most probable value of Φ/Ψ, expressed in the X-Ray formalism, was deduced by constructing a frequency graph for each angle with OriginPro 8.5 program [14]. The obtained values were given as input to POLYS. As result, POLYS returned non-integer values of n. In the next step, POLYS was instructed to optimize the Φ/Ψ values so that the structure of the pentasaccharide could fit in a helix having an “n” integer value. These new set of values was used to construct oligosaccharides with longer chains and to visualize their folding.
3. Results
3.1. LPS Isolation, Purification and Chemical Analysis
The lipopolysaccharide produced by C. metallidurans CH34 strain was isolated by phenol/chloroform/light petroleum (PCP) extraction [10]. The different fractions were analyzed by SDS-PAGE (Figure S1), revealing the presence of the (S)-type LPS in the phenol phase and traces of (R)-type LPS in the precipitate PCP.
Since the aim of this work was to characterize the O-antigen portion, the attention was focused on the (S)-type LPS only.
The monosaccharide compositional analysis carried out on the LPS revealed the presence of rhamnose (Rha) and galactosamine (GalN) residues as main sugars, while heptose, 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo), and glucose (Glc) occurred as minor constituents. The linkage analysis disclosed that GalN was 4-substituted (4-GalN), while Rha was 3-substituted (3-Rha), together with the presence of an additional sugar residue that eluded the previous analysis: the bacillosamine acylated by a 3-hydroxy-butyrric acid at the 4-amino function (Qui2NAc4NHB) and glycosylated at the position C-3 (Figure S2).
GalN and Rha were derivatized as 2-(-)-octyl glycoside derivatives, showing that their absolute configuration was D and L, respectively, whereas the configuration of the Qui2NAcNHB was inferred by computational analysis. Instead, regarding the 3-hydroxy-butyric acid (HBA), the absolute configuration was S, as proved GC-MS analysis of its as 2-(-)-octylester trifluoracetyl derivative.
In order to fully characterize the structure of the O-antigen, the LPS underwent mild acid hydrolysis to isolate and split the O-antigen from the lipid A portion. The obtained O-antigen was further purified by size exclusion chromatography (Biogel-P10), obtaining a purified O-antigen (13 mg, yield 73%), which was investigated via 1D and 2D NMR [15].
3.2. Structure Elucidation of the LPS O-antigen
1H NMR spectrum of the purified O-antigen (Figure 1) displayed four signals in the anomeric region (5.3–4.9 ppm) each labeled with a capital letter (A–D), a crowded ring proton region (4.3–3.4 ppm), two methylene protons at 2.32–2.35 ppm assigned to HBA, two sharp peaks at 2.08 and 2.03 ppm related to the N-Acetyl groups, three methyl groups of the 6-deoxy sugars (1.31–1.15), and the methyl group of the HBA at 1.22 ppm.
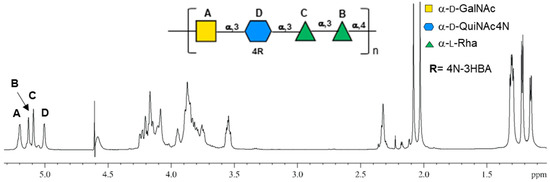
Figure 1.
Proton spectrum of the O-antigen of Cupriavidus metallidurans CH34, measured at 600 MHZ at 318 K in D2O with its chemical structure. Letters used to mark the anomeric signals (5.3–4.9 ppm) are as in Table 1 and are reported on the structure of the linear repeating unit of the O-antigen. The structure is represented according to the symbolic nomenclature for glycans.
All residues were assigned by combining 2D homonuclear and heteronuclear NMR spectra (COSY, TOCSY, NOESY, HSQC and HMBC), and their chemical shift values are reported in Table 1.

Table 1.
(600 MHz, 318 K, D2O) Proton and carbon chemical shift values of the O-antigen of Cupriavidus metallidurans CH34. Both galactosamine (A) and bacillosamine (D) are N-acetylated (1H 2.08 and 2.03 ppm, respectively, 13C 23.7 ppm for both).
Residue A has been attributed to a four-linked α-GalNAc. The chemical shift of the H-1 proton (5.20 ppm) and its small J coupling constant (3JH1,H2 2.9 Hz) suggested its α-configuration. The galacto stereochemistry of this residue was deduced by the typical correlations in the COSY-TOCSY spectra, where the anomeric proton (H-1) of A had only three correlations (Figure 2b) due to the weak H-4/H-5 scalar coupling, which was visible as dipolar coupling only in the NOESY spectrum (Figure 2c).
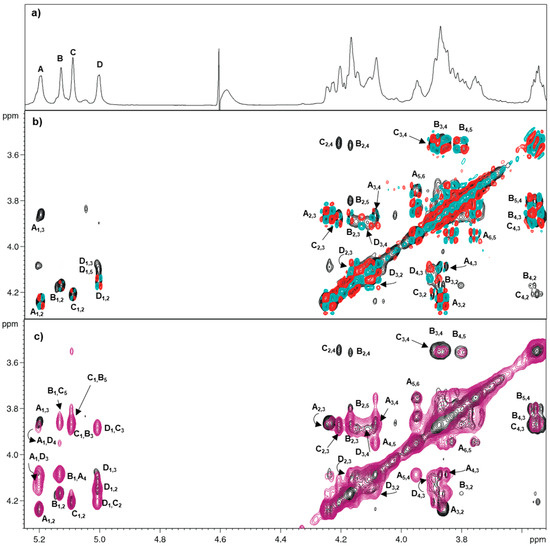
Figure 2.
1H-1H Homonuclear spectra of O-antigen of C. metallidurans CH34. (a) proton spectrum; (b) overlay of TOCSY (black) and COSY (red and cyan) spectra; (c) overlay of TOCSY (black) and NOESY (violet) spectra. Letters used for the annotation of the densities follow the system of Table 1.
HSQC spectrum revealed that C-2 resonated at 50.6 ppm, indicating a nitrogen-bearing carbon, and HMBC correlations showed that the amino group was acetylated (1H 2.083, 13C 23.6). For residue A, the low-field displacement of its C-4 signal (76.5 ppm, Figure S3) compared to the reference value (69.7 ppm) indicated glycosylation at this position [15], which is in agreement with the correlation between H-1 of B with H-4 of A in the NOESY spectrum (Figure 2c).
Residues B and C (H-1 at 5.13 and 5.08 ppm, respectively) were identified as two 3-linked α-Rha units. TOCSY spectrum connectivity (Figure 2b) of H-1 of both B and C correlated with the respective H-2, which in turn displayed the TOCSY cross peaks up to the corresponding H-6, as often occurs for monosaccharides with the manno stereochemistry (Figure 2b). Moreover, the H-6 of B and C resonated at 1.29 and 1.31 ppm, which are values typical of 6-deoxy-sugars, clearly confirming that both residues were rhamnose. Combining TOCSY and COSY information with those from HSQC spectra (Figure 2b and Figure S3) led to the assignment of all proton and carbon chemical shifts (Table 1). Both residues were α-configured at the anomeric center on the basis of their C-5 chemical shift values (70.9 and 70.5 ppm for B and C, respectively) [15]. Furthermore, the low field value of their C-3 (70.1 and 76.7 ppm for B and C, respectively) indicated that both residues were O-substituted at that position [15]. Accordingly, in the NOESY spectrum, H-1 of D correlated with H-3 of C and H-1 of C correlated with H-3 of B.
Residue D (anomeric proton at 5.00 ppm) was assigned to a three-linked Qui2NAc4NHB α-configured based on the 3JH1,H2 value (3.5 Hz). The TOCSY pattern showed correlations from H-1 of D up to H-6, confirming the relative gluco-stereochemistry of this residue [15]. The 1H-13C HSQC spectrum displayed the correlation of H-2 and H-4 (at 4.15 ppm and 3.88 ppm, respectively) with nitrogen-bearing carbon signals at 53.8 ppm and 58.2 ppm, respectively (data not shown). Notably, the amino group at carbon C-2 was acetylated, as proved by its H-2/C-2 chemical shift values (Table 1) [15], while the one at carbon C-4 had the HBA residue, as detected by linkage analysis (Figure S2). Moreover, the low field value of its C-3 D (73.3 ppm) established that this position was glycosylated [15], while NOESY spectrum indicated that D was linked at that position (Figure 2c).
Combining all the spectroscopic data, the structure of the repeating unit of the O-antigen was determined to be a linear tetrasaccharide repeating unit (Figure 1): [4)-α-d-GalNAc-(1→3)-α-d-Qui2NAc4NHBA-(1→3)-α-l-Rha-(1→3)-α-l-Rha-(1→]n.
Absolute configuration of Qui2NAc4NHBA was evaluated by conformational analysis, as detailed in the next paragraph.
3.3. Determination of the Absolute Configuration of Qui2NAc4NHBA by Molecular Modeling
Chemical analyses did not provide any information about the absolute configuration of the Qui2NAc4NHBA residue.
To gain insight into this issue, the carbon chemical shifts of Qui2NAc4HBA and its NOEs effects were analyzed in view of the magnitudes of the 13C glycosylation effects [16] and of the NOEs’ patterns [17], both depending on the stereochemistry of the residues involved and on how they are linked together. The first type of analysis suggested the D absolute configuration of Qui2NAc4HBA; however, the second approach did not lead to conclusive results, which was probably due to the presence of the two acyl substituents placed on the bacillosamine unit.
Therefore, the molecular mechanics (MM) and the molecular dynamics (MD) approaches were used to complete the characterization of the O-antigen structure.
Since the NOE effects depend on the D or L configuration of the sugars involved, two pentasaccharides were constructed by considering the two possible configurations of Qui2NAc4NHBA. The NOE effects of each oligosaccharide were simulated and compared to those collected experimentally.
Based on the structure of the O-antigen (Figure 1), two complex oligosaccharides were built using MAESTRO program: d-penta, which consisted of the B-A-dD-C-B’ sequence; while l-penta consisted of B-A-lD-C-B’. Capital letters correspond to the labels used during the NMR characterization (Figure 1 and Table 1); while the lowercase letters indicate the absolute configuration of the residue (d means D-configured, l means L). The two pentasaccharides differed for the absolute configuration of the bacillosamine unit, while all other residues had the one found by octyl-glycosidic analysis.
To construct the two pentasaccharides, the preferred dihedral angles (Φ/Ψ) adopted from each glycosidic junction were determined for each of the six following disaccharides: A-dD; A-lD; dD-C; lD-C; C-B; B-A. The Molecular Mechanic approach led to defining the flexible map of each glycosidic junction along with the optimal Φ and Ψ values (Figure S4, Table S1). These dihedral angle values (Table S1) were used to construct d-penta and l-penta, and assigning to HBA the S configuration. Both oligosaccharides were minimized and, subsequently, subjected to MD simulation that returned for each of them an ensemble of 10,000 structures, which were each used to simulate the NOE effects of the molecule with NOEPROM [12] (Table 2). Among all NOE contacts, that between the proton H-1 of GalNAc (residue A) and H-2 of Qui2NAc4NHBA (residue D), labeled A1D2, was discriminant: this NOE effect occurred in l-penta, while it was absent in d-penta (Table 2). However, this correlation did not exist in the experimental NOESY spectrum of the O-antigen; therefore, the D absolute configuration was attributed to D based on the agreement between the experimental and the simulated data. This result was further supported by the A1D4 and A1CH2,HBA correlations that were detected in the experimental NOESY spectrum and in the simulated NOEs of d-penta and not in the simulation of l-penta.

Table 2.
Comparison of simulated NOE values of both d-penta (B-A-dD-C-B’) and l-penta (B-A-lD-C-B’) with the experimental ones, which were obtained from the NOESY spectrum recorded for the O-antigen isolated from Cupriavidus metallidurans CH34. The intensities of the NOE are classified as strong (+++), medium (++), and weak (+) based on the following interprotonic distances (in Å): (+++) 2.5–2.0; (++) 3.0–2.5; (+) 4.0–3.0. The discriminating NOEs for the evaluation of the absolute configuration of bacillosamine are in bold and underlined. Capital letters correspond to the labels used during the NMR characterization (Figure 1 and Table 1).
3.4. Conformational Analysis of the O-Antigen
The three-dimensional structure of the O-antigen polysaccharide was evaluated by analyzing the MD data of d-penta with POLYS [13], which is a program that is able to define which the helix conformation is more probable for the polysaccharide examined.
First, the most probable values of the dihedral angles of each glycosidic junction of d-penta were determined by plotting the corresponding frequency graphs (Figure S5). This approach showed that each glycosidic junction had only one distribution of values, as inferred by the fact that both Φ/Ψ had only one maximum. On the contrary, the C–B junction existed in two possible states due to the presence of two different sets of Φ/Ψ values, −70.1/−96.7 and −70.1/−164.7 (Figure S5, Table S2). Then, two oligosaccharides defined from the different Φ/Ψ values of the C–B glycosidic junction were built: d-penta-A (Φ/Ψ −70.1/−96.7) and d-penta-B (Φ/Ψ −70.1/−164.7). The values of all the other glycosidic junctions were set to those with maximum probability in the frequency count graphs (Figure S5, Table S2).
Feeding POLYS with the values of each glycosidic junction distinctive of d-penta-A and d-penta-B gave as a result that each oligosaccharide adopted a right-handed helix conformation and that these helices differed for the number of repeating units necessary to define the helix revolution parameter (n) or the periodicity of the helix.
For d-penta-A, POLYS gave as a first unoptimized (or raw) value n = 3.71; that indicated that these dihedral values conferred to the glycan a helix fold with a periodicity close to four repeating units per turn. Then, the Φ and Ψ of all junctions were optimized by POLYS to fit the structure into a regular helix. Accordingly, n was set to the closest integer value, 4, which is indicative of a regular helix with four repeating units per turn (Table S2).
By applying the same approach to d-penta-B, the program returned a value of n = 2.41, which is well in between 2 and 3; thus, the Φ/Ψ values were optimized by taking into account both n = 2 or 3 (Table S2).
First, the analysis of the Φ/Ψ values associated to the three optimized helices (n = 2, 3 or 4, Table S2) revealed that they were within the boundaries defined from the frequency counts graphics (Table S2, Figure S5); therefore, these three types of helices could occur during the MD simulation. Clearly, the O-antigen could adopt such conformations along with others where the regularity of the helix was not perfect.
Then, these new dihedral values were used to build oligomers made of six repeating units, 24 monosaccharide residues in total, to appreciate their tridimensional form (Figure 3). By looking the three of them from the top (Figure 3a), it was possible to appreciate the regularity of the helix; indeed, the one with n = 4 appeared as a square with an appreciable inner cavity, while that with n = 3 resembled a triangle with little space in the inner side. As for n = 2, no inner cavity was observed, which was expected for this type of multiplicity. When the three molecules were observed from the side, it appeared immediately that the one with n = 4 had a very compressed shape compared to the other three, as evidenced from different values measured for the axial rise per repeating unit, which were 9.1, 12.6, and 14.0 Å for n = 4, 3 and 2, respectively.
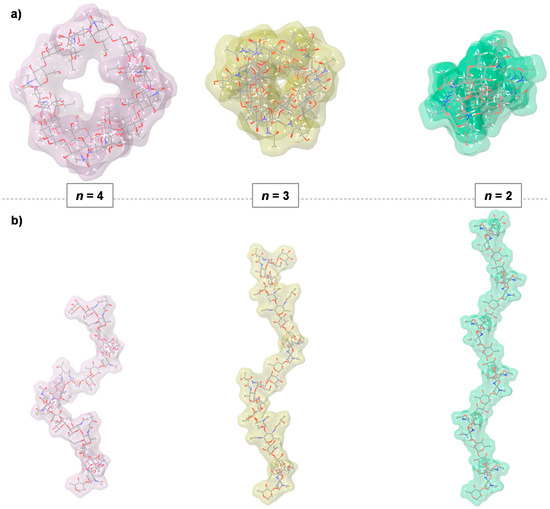
Figure 3.
Conformational analysis of the O-antigen of C. metallidurans CH34. (a) Top view evidencing the symmetry of the three helices; (b) Lateral view. The O-antigen assumed a right-handed helix conformation with a certain number of repeating units for the turn of helix: n = 2 (light green), n = 3 (light yellow), or n = 4 (light pink). The axial rise per repeating unit is of 9.1, 12.6 and 14.0 Å, for n = 4, 3 and 2, respectively.
It should be noted that these evaluations suggested which type of helix was compatible with dihedral angle values of the glycosidic junction, which are not the same as what is expected to exist in solution. Indeed, the glycan does not exist in a pure helical conformation, but it is very likely that some portions of the whole chain adopt the dihedral angles of the helix with n = 4, while others adopt those of n = 3, and others adopt those of n = 2.
In essence, the O-antigen should be considered as a flexible entity, with portions that temporarily adopt a certain type of helix conformation and that convert into each other as a consequence of a dynamic process. Each of these motifs is a right-handed helix with a different number of repeating units, namely 2, 3 or 4 that enable the corresponding part of the molecule to assume an overall shape that varies from being rather tight (n = 4) to very extended (n = 2, Figure 3).
3.5. Discussion
It is known that C. metallidurans CH34 lives in different environments, such as those rich in heavy metals [1,2,3] and the human body after nosocomial infection [5,6,7]. However, how this bacterium can adapt itself in the extreme variable conditions is still unknown. Since the LPS is the major constituent of the outer membrane of the Gram-negative bacteria, with the O-antigen portion directly in contact with the external environment, the study of the O-antigen structure and its conformation is a prerequisite to get closer at the molecular level to the adaptability process developed by this bacterium.
To the best of our knowledge, C. metallidurans CH34 represents the first example of a Gram-negative bacterium of the genus Cupriavidus for which the structure and the conformation of the O-antigen were determined.
A combination of the chemical, spectroscopic, and molecular mechanics and dynamics approaches allowed the identification of the O-antigen polysaccharide structure, which is defined as a linear tetrasaccharide repeating unit made by [4)-α-d-GalNAc-(1→3)-α-d-Qui2NAc4NHBA-(1→3)-α-l-Rha-(1→3)-α-l-Rha-(1→]n. A search in the Carbohydrate Structure Database revealed that the identified structure is new, as no match with other bacterial structures was found [18].
Importantly, in the O-antigen of C. metallidurans, there is a rare aminosugar, the α-d-Qui2NAc4NHBA that is only found in few bacterial LPS, including the O-antigen of Pseudomonas fluorescens [19], where this residue is linked to D-GalNAc, as in this case.
In the literature, it is reported that polysaccharides can adopt different conformations that can considerably influence their properties, such as the stability and recognition process [20].
This study showed that the O-antigen polysaccharide of C. metallidurans CH34 is highly flexible and can adopt three different right-handed helix conformations described by two, three, four-fold symmetry (Figure 3). Interestingly, the four-folded helix offers an inner cavity, which may play an important role in the complex formation with small molecules as nutrient or metals. On the contrary, the three/two-folded helices are much more compact compared to the four-folded helix, especially the two-folded helix in which there is no inner cavity.
Concluding, the overall structural features of the O-antigen polysaccharide could be the necessary expedient at the basis of the adaptability process of C. metallidurans CH34 to different environments, perhaps shielding the bacterium from the uptake of harmful molecules there present.
Supplementary Materials
The following information are available online at https://www.mdpi.com/article/10.3390/polysaccharides3010009/s1, Figure S1: SDS-PAGE (15%) of the fraction of the PCP extractions, Figure S2: EI-MS spectrum of the Qui2NAc4NHBA (retention time 36 min), a sugar component of the O-antigen of Cupriavidus metallidurans CH34, Figure S3: 1H and 13C heteronuclear spectra of O-antigen of C. metallidurans CH34 (600 MHZ, 318 K, in D2O), Figure S4: Relaxed energy maps for the glycosidic junctions of the six disaccharides constructed on the basis of the O-antigen structure from C. metallidurans CH34, Figure S5: Frequency count graphs of Φ and Ψ values of the glycosidic junction of D-penta, Table S1: Values found for the glycosidic linkages of six disaccharides by using a molecular mechanic approach, Table S2: Comparison of simulated NOE values of both D-penta and L-penta with the experimental ones, obtained from the NOESY spectrum recorded for the O-antigen isolated from C. metallidurans CH34.
Author Contributions
Conceptualization, C.D.C. and A.M.; methodology, A.N., A.V. and I.S.; formal analysis, C.D.C.; investigation, C.D.C.; writing—original draft preparation, A.N., A.V. and I.S.; writing—review and editing, C.D.C. and A.M.; visualization, C.D.C.; supervision, C.D.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Goris, J.; De Vos, P.; Coenye, T.; Hoste, B.; Janssens, D.; Brim, H.; Diels, L.; Mergeay, M.; Kersters, K.; Vandamme, P. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int. J. Syst. Evol. Microbiol. 2001, 51, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Reith, F. Biomineralization of Gold: Biofilms on Bacterioform Gold. Science 2006, 313, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Van Houdt, R. Cupriavidus metallidurans CH34, a historical perspective on its discovery, characterization and metal resistance. FEMS Microbiol. Ecol. 2021, 97, fiaa247. [Google Scholar] [CrossRef] [PubMed]
- Lal, D.; Nayyar, N.; Kohli, P.; Lal, R. Cupriavidus metallidurans: A Modern Alchemist. Indian J. Microbiol. 2013, 53, 114–115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Langevin, S.; Vincelette, J.; Bekal, S.; Gaudreau, C. First Case of Invasive Human Infection Caused by Cupriavidus metallidurans. J. Clin. Microbiol. 2011, 49, 744–745. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Spilker, T.; Reik, R.; Vandamme, P.; LiPuma, J.J. Use of PCR Analyses to Define the Distribution of Ralstonia Species Recovered from Patients with Cystic Fibrosis. J. Clin. Microbiol. 2005, 43, 3463–3466. [Google Scholar] [CrossRef] [PubMed]
- D’Inzeo, T.; Santangelo, R.; Fiori, B.; De Angelis, G.; Conte, V.; Giaquinto, A.; Palucci, I.; Scoppettuolo, G.; Di Florio, V.; Giani, T.; et al. Catheter-related bacteremia by Cupriavidus metallidurans. Diagn. Microbiol. Infect. Dis. 2015, 81, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; De Castro, C.; Silipo, A.; Molinaro, A. Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 2019, 43, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; Duda, K.A.; Lanzetta, R.; Silipo, A.; De Castro, C.; Molinaro, A. A Journey from Structure to Function of Bacterial Lipopolysaccharides. Chem. Rev. 2021. [Google Scholar] [CrossRef] [PubMed]
- De Castro, C.; Parrilli, M.; Holst, O.; Molinaro, A. Microbe-Associated Molecular Patterns in Innate Immunity. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2010; Volume 480, pp. 89–115. [Google Scholar]
- Kenne, L.; Lindberg, B.; Rahman, M.M.; Mosihuzzaman, M. Structural studies of the Vibrio mimicus W-26768 O-antigen polysaccharide. Carbohydr. Res. 1993, 243, 131–138. [Google Scholar] [CrossRef]
- Asensio, J.L.; Jimenez-Barbero, J. The use of the AMBER force field in conformational analysis of carbohydrate molecules: Determination of the solution conformation of methyl α-lactoside by NMR spectroscopy, assisted by molecular mechanics and dynamics calculations. Biopolymers 1995, 35, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Engelsen, S.B.; Cros, S.; Mackie, W.; Pérez, S. A molecular builder for carbohydrates: Application to polysaccharides and complex carbohydrates. Biopolymers 1996, 39, 417–433. [Google Scholar] [CrossRef]
- Edwards, P.M. Origin 7.0: Scientific Graphing and Data Analysis Software. J. Chem. Inf. Comput. Sci. 2002, 42, 1270–1271. [Google Scholar] [CrossRef]
- Speciale, I.; Notaro, A.; Garcia-Vello, P.; Di Lorenzo, F.; Armiento, S.; Molinaro, A.; Marchetti, R.; Silipo, A.; De Castro, C. Liquid-state NMR spectroscopy for complex carbohydrate structural analysis: A hitchhiker’s guide. Carbohydr. Polym. 2022, 277, 118885. [Google Scholar] [CrossRef] [PubMed]
- Shashkov, A.S.; Lipkind, G.M.; Knirel, Y.A.; Kochetkov, N.K. Stereochemical factors determining the effects of glycosylation on the 13C chemical shifts in carbohydrates. Magn. Reason. Chem. 1988, 26, 735–747. [Google Scholar] [CrossRef]
- Lipkind, G.M.; Shashkov, A.S.; Mamyan, S.S.; Kochetkov, N.K. The nuclear overhauser effect and structural factors determining the conformations of disaccharide glycosides. Carbohydr. Res. 1988, 181, 1–12. [Google Scholar] [CrossRef]
- Toukach, P.V.; Egorova, K.S. Carbohydrate structure database merged from bacterial, archaeal, plant and fungal parts. Nucleic Acids Res. 2016, 44, D1229–D1236. [Google Scholar] [CrossRef] [PubMed]
- Shashkov, A.S.; Paramonov, N.A.; Veremeychenko, S.P.; Grosskurth, H.; Zdorovenko, G.M.; Knirel, Y.A.; Kochetkov, N.K. Somatic antigens of pseudomonads: Structure of the O-specific polysaccharide of Pseudomonas fluorescens biovar B, strain IMV 247. Carbohydr. Res. 1998, 306, 297–303. [Google Scholar] [CrossRef]
- Fittolani, G.; Seeberger, P.H.; Delbianco, M. Helical polysaccharides. Pept. Sci. 2020, 112, e24124. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).