Abstract
Human granulocyte colony-stimulating factor (hG-CSF) is primarily used to treat neutropenia induced by cancer chemotherapy and bone marrow transplantation. The current identification test for hG-CSF relies on Western blot (WB), a labor-intensive and technically demanding method. This study aimed to screen and prepare an anti-hG-CSF nanobody to identify and quantify hG-CSF, with the ultimate goal of developing colloidal gold-labeled nanobody test strips for rapid identification. An alpaca was immunized with hG-CSF, and the VHH gene sequence encoding the anti-hG-CSF nanobody was obtained through sequencing following phage display library construction and multiple rounds of biopanning. The nanobody C68, obtained from screening, was expressed by E. coli, and its physicochemical properties such as molecular weight, isoelectric point, and affinity were characterized after purification. WB analysis demonstrated excellent performance of the nanobody in identification tests in terms of specificity, limit of detection (LOD), applicability with products from various manufacturers, and thermal stability. Additionally, we established an ELISA method for hG-CSF quantification utilizing the nanobody C68 and conducted methodological validation. Finally, colloidal gold-based test strips were constructed using the nanobody C68, with a LOD of 30 μg/mL, achieving rapid identification for hG-CSF. This study represents a novel application of nanobodies in pharmaceutical testing and offers valuable insights for developing identification tests for other recombinant protein drugs.
1. Introduction
Recombinant human granulocyte colony-stimulating factor (rhG-CSF) is a therapeutic biological product primarily used to mitigate chemotherapy-induced neutropenia and restore granulopoiesis after bone marrow transplantation [1]. In recent years, rhG-CSF products have also been explored for their therapeutic potential in spinal cord injuries [2] and reproductive medicine [3].
Identification test is one of the crucial testing items in the quality control of recombinant protein drugs, used to confirm the identity of the product. Key requirements for such tests include high specificity, high sensitivity, good repeatability, simplicity, and efficiency. According to Volume III of the Chinese Pharmacopoeia (2020), identification tests for cytokines typically employ Western blotting or immunospot assays, which rely on antibodies with strong specificity and minimal inter-batch variability. However, commercial monoclonal and polyclonal antibodies often exhibit significant differences between manufacturers and batches, potentially compromising test reproducibility and posing challenges for identification. Furthermore, these conventional antibodies are complex to produce, costly, and require low-temperature storage and transportation. To address these limitations, there is an urgent need for antibodies with strong specificity, high sensitivity, excellent stability, minimal batch variability, and cost-effectiveness as a substitute. In this study, we investigated the application of nanobodies with such advantages in the identification test for hG-CSF, aiming to enhance the efficiency and reliability of this process.
Heavy-chain antibodies (HCAbs) were first discovered in camel serum by C. Hamers-Casterman in 1993 [4] and have since been identified in all camelids and cartilaginous fish, such as sharks. Single-domain antibodies (sdAbs), also known as the variable domain of heavy chain of HCAb (VHH), are derived by cloning the heavy chain variable region of HCAbs. Due to their compact structure, approximately 4 nm in length and 2.5 nm in diameter, these sdAbs are commonly referred to as nanobodies [5]. As the smallest naturally occurring functional antigen-binding fragments, nanobodies possess unique properties. Although the term “nanobody” was initially trademarked by Ablynx in 2003, it has since become a universal label for these sdAbs [6]. Intramolecular disulfide bonds likely contribute to their high thermal stability and resistance to acidic and alkaline conditions [7], making them well-suited for storage and transportation. Furthermore, nanobodies can be expressed in various expression systems, including bacteria [8], fungi [9], plants [10], and animal cells [11].
Over the past three decades since their discovery, nanobodies have been extensively used in various fields, including development of pharmaceuticals and in vitro diagnostic reagents, as well as substance detection related to environmental pollution and food safety [12,13,14,15]. However, their application in testing of recombinant protein drugs has been relatively limited. This study represents an innovative attempt to apply nanobodies in pharmaceutical testing. Using recombinant hG-CSF as a model protein, we successfully screened and developed nanobody C68 demonstrating excellent performance in the identification test for hG-CSF. Building on this, we established an enzyme-linked immunosorbent assay (ELISA) method for quantifying hG-CSF concentrations and finally developed a colloidal gold immunochromatographic test strip for rapid and simple hG-CSF identification. This approach not only reduces detection costs but also enhances detection efficiency, contributing to improved drug testing processes.
2. Results
2.1. Construction of Anti-hG-CSF Nanobody Library and Screening of Nanobodies
The negative control serum and sera from the four immunizations of alpaca were serially diluted to quantify the serum antibody titers using ELISA. The results showed that the antibody titer was significantly increased after the third immunization and was comparable to that after the fourth immunization, the antibody titer was up to 1.28 × 105 after the fourth immunization (Figure 1A). After lymphocyte isolation and total RNA extraction, target fragments (approximately 500 bp) were obtained by reverse transcription PCR and nested PCR (Figure 1B,C). These target fragments were subsequently inserted into the pHEN1 phasmid vector and electroporated into E. coli TG1 competent cells. The insertion efficiency of the nanobody gene was determined to be 100% by colony PCR (Figure 1D), and the final library capacity was 2.3 × 108 CFU/mL.
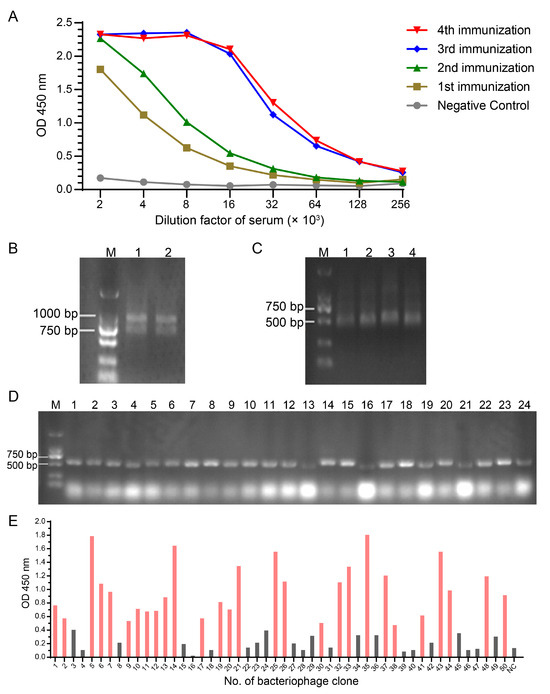
Figure 1.
Construction and screening of nanobody library against hG-CSF. (A) Determination of serum antibody titers after the alpaca was immunized in four rounds with the hG-CSF antigen. (B) Amplification products of step 1 in nested PCR for VHH fragments. Lanes 1 and 2 were the same PCR product. (C) Amplification products of step 2 in nested PCR for VHH fragments. The size of target fragments was approximately 500 bp. Lanes 1–4 were the same PCR product. (D) Colony PCR identification to determine positive rate of VHH library in E. coli. (E) Identification of positive phage clones with ELISA after screening and enrichment of phage display nanobody library. Red columns represent positive clones, whose OD values were higher than three times of negative control.
After M13KO7 helper phage rescue, the phage display library capacity was 1.13 × 1013 CFU/mL. Positive phages were enriched in the phage display library following four rounds of biopanning with immobilized antigens. Of the 50 clones screened after panning, 28 positive clones were confirmed by ELISA (Figure 1E). The nucleic acid sequences of these positive clones were obtained and translated into amino acid sequences. After multi-sequence analysis, three different VHH sequences were ultimately identified, named C43, C61, and C68.
C68 was selected as the molecule of study in subsequent experiments because of the strongest binding affinity to hG-CSF measured with Octet RED96e as described below. The theoretical amino acid sequence of nanobody C68 is as follows: MQVQLVESGGGLVQAGGSLRLSCTASGRPSNVYDMGWFRRVPGKESRLMAYINWSRGSTYYADSVKGRFTISRDNTNNTVYLQMNSLKPDYCARAARAGPGTPAYNSWGQGTQVTVSSLEHHHHHH. The theoretical molecular weight of C68 is 14,240.83 Da.
2.2. Expression, Purification and Physicochemical Properties of Nanobody C68
After ultrasonication of the bacterial suspension and centrifugation, the supernatant was collected and subjected to electrophoresis to determine the efficiency of C68 expression. C68 accounted for 42.55%, 51.77%, and 39.23% of the total proteins in the three shake flask cultures (500 mL each), respectively (Figure 2A, Supplementary Table S1). The His-tagged nanobody C68 were subsequently purified using Ni-column (Figure 2B) and quantified by Bradford assay (Supplementary Figure S1). The measured quantities of the nanobody were 6.85 mg, 7.07 mg, and 9.19 mg, respectively, with a mean recovery of 74%.
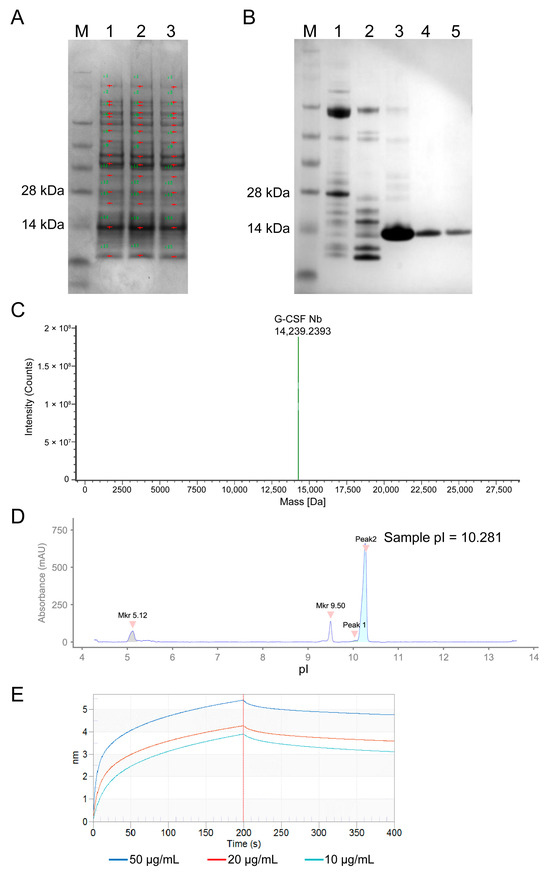
Figure 2.
Expression and physicochemical properties of nanobody C68. (A) Electrophoresis of supernatant from three shake flasks after ultrasonication. (B) Electrophoresis of purified C68 protein eluted with imidazole gradient (Lanes 1–5 are for 50 mM, 80 mM, 100 mM, 150 mM, and 250 mM imidazole, respectively). (C) Molecular weight of the nanobody C68 measured by LC-MS system (Waters). (D) Capillary isoelectric focusing of the nanobody C68 performed with Maurice (ProteinSimple). Two pI markers (5.12 and 9.50) were used. (E) Affinity between C68 and hG-CSF analyzed by following Octet procedures. 10 μg/mL hG-CSF product combined with 50, 20, or 10 μg/mL nanobody C68.
After SDS-PAGE, nanobody C68 was transferred onto a PVDF membrane for N-terminal sequencing. The determined amino acid sequence was 1–16: MQVQLVESGGGLVQAGG, consistent with the theoretical amino acid sequence of C68. The molecular weight of C68 was determined to be 14,239.24 Da (Figure 2C) using the Waters BioAccord LC-MS (liquid chromatography-mass spectrometry) system, consistent with the theoretical value of 14,240.83 Da.
Two repeated measurements using the imaged capillary isoelectric focusing (icIEF) mode on the ProteinSimple Maurice revealed that the pI values of Peak2 were 10.281 (Figure 2D) and 10.279, respectively, indicating that the isoelectric point of C68 is approximately 10.28.
The binding affinity and kinetics of nanobody C68 to hG-CSF were evaluated using the Octet RED96e (Sartorius) (Figure 2E). The equilibrium dissociation constant KD was determined to be 4.4 × 10−8 mol/L, indicating that C68 exhibits a strong affinity for hG-CSF.
2.3. Nanobody C68 Demonstrates Superior Performance in the hG-CSF Identification Test
The specificity of nanobody C68 for hG-CSF was evaluated by WB using cytokines with molecular weights similar to that of hG-CSF (GM-CSF, EPO, TPO, bFGF, and IFN-α2b) as controls. Nanobody C68 was used as the primary antibody, and an HRP-labeled anti-His antibody served as the secondary antibody. The WB results showed that nanobody C68 bound explicitly to hG-CSF but not to other cytokines, demonstrating favorable specificity (Figure 3A).
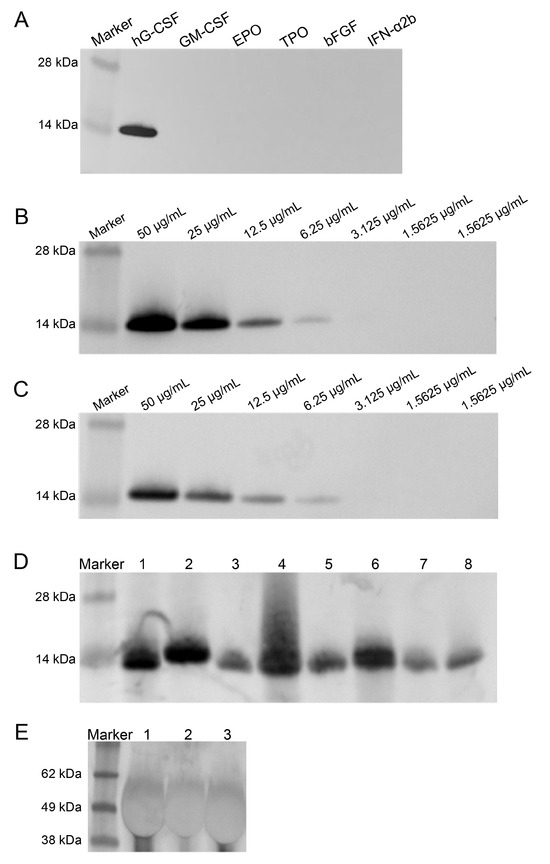
Figure 3.
Specificity, sensitivity, and applicability of nanobody C68 in identifying hG-CSF evaluated by Western blot. (A) Specific binding of C68 to hG-CSF, rather than granulocyte–macrophage colony-stimulating factor (GM-CSF), erythropoietin (EPO), thrombopoietin (TPO), basic fibroblast growth factor (bFGF), or interferon α2b (IFNα2b). (B) Sensitivity of nanobody C68 in detecting different concentration of hG-CSF. (C) Sensitivity of monoclonal antibody against hG-CSF (SANTA CRUZ, sc-53292) in detecting different concentration of hG-CSF. (D) Application of nanobody C68 to detect different rhG-CSF products. Lanes 1–8 were eight different marketed rhG-CSF products from different pharmaceutical companies. (E) Application of nanobody C68 to detect long-acting rhG-CSF products linked with polyethylene glycol (PEG). Lanes 1–3 were three PEGylated hG-CSF products from different companies.
The limits of detection (LOD) for nanobody C68 and anti-hG-CSF monoclonal antibody were compared by WB using HRP-anti-His antibody and HRP-anti-mouse IgG (H+L) as secondary antibodies, with DAB for visualization. The data showed that nanobody C68 and the anti-hG-CSF monoclonal antibody exhibited comparable sensitivity, with an LOD of 81.25 ng (Figure 3B,C).
Next, the compatibility of nanobody C68 with hG-CSF produced by different manufacturers was evaluated by WB. It was found that nanobody C68 effectively recognized hG-CSF expressed by E. coli or CHO cells, as well as PEGylated hG-CSF (Figure 3D,E).
The thermal stability of nanobody C68 was assessed by measuring the changes in LOD after seven days (Figure 4A–C) and 52 days (Figure 4E–G) of storage at 37 °C, room temperature, and 4 °C, respectively, compared to storage at −20 °C (Figure 4D,H). The WB results demonstrated no significant change in LOD at any storage temperature or duration, indicating excellent thermal stability.
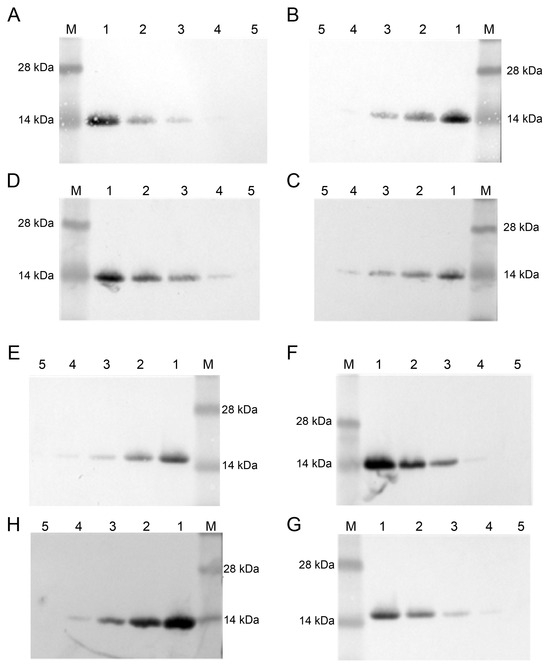
Figure 4.
Thermostability of nanobody C68 evaluated by Western blot. Different concentrations of hG-CSF samples were detected by the nanobody C68, which was stored at 37 °C (A), room temperature (B), 4 °C (C), and −20 °C (D) for 7 days. Similar experiments were performed with C68, which was stored at 37 °C (E), room temperature (F), 4 °C (G), and −20 °C (H) for 52 days. M, marker; 1, 50 μg/mL; 2, 25 μg/mL; 3, 12.5 μg/mL; 4, 6.25 μg/mL; 5, 3.125 μg/mL.
2.4. Nanobody C68 Can Be Used to Quantify hG-CSF Protein Concentration by ELISA
An ELISA method for quantifying hG-CSF using nanobody C68 was established and validated. Six replicate experiments were performed following the established ELISA protocol, and the coefficient of determination (R2) ranged from 0.986 to 1.000 (Supplementary Table S2), demonstrating that the data fit the regression line well (Supplementary Figure S2).
Method specificity was verified through six replicate experiments using GM-CSF as the control group, hG-CSF as the experimental group, and a blank group. The results showed that the OD450 nm values were consistent between the control and blank groups (Table 1), significantly lower than that of the experimental group, indicating good specificity.

Table 1.
Absorbance values of the blank group, control group, and experimental group in ELISA.
Method accuracy was evaluated by measuring the recovery of hG-CSF from samples containing different concentrations of hG-CSF, each spiked with 0.0617 μg/mL of the standard substance. The theoretical values correspond to the concentrations after 1:1 spiking, while the actual measured values are shown in Table 2. The recoveries were 99.87%, 102.11%, and 81.48%, respectively, indicating good accuracy.

Table 2.
Recovery of hG-CSF from spiked samples.
The precision of the method was evaluated by assessing repeatability and inter-day variability. hG-CSF samples were diluted to 0.185 μg/mL, 0.0925 μg/mL, and 0.04625 μg/mL, and the mean concentrations from three repeated measurements were 0.199, 0.0973, and 0.0427, with relative standard deviation (RSD) of 17.6%, 1.19%, and 6.77%, respectively, indicating good repeatability of this method (Table 3).

Table 3.
Repeatability results of hG-CSF quantification by ELISA with nanobody C68.
Inter-day variability was assessed by measuring the concentrations of hG-CSF sample 1 (300 μg/mL) and sample 2 (400 μg/mL) across three consecutive days. As shown in Supplementary Table S3, the RSDs were 3.53% and 4.61%, respectively, demonstrating minimal inter-day variability. Taken together, these data indicate that the method exhibits good precision.
When a four-parameter logistic regression model was used to generate the calibration curve for analyte quantification, measurements outside the two inflection points of the curve showed substantial variation, while those within the inflection points demonstrated higher accuracy. As shown in Table 4, the quantitation range of the method is 0.0069–0.185 μg/mL, within which the measured values closely approximated the theoretical concentrations. In conclusion, this ELISA method with nanobody C68 demonstrates reliable accuracy and precision for quantifying hG-CSF concentrations.

Table 4.
Determination of quantitation range.
2.5. Colloidal Gold-Labeled Nanobody Test Strips for Rapid Identification
Colloidal gold immunochromatographic test strips based on nanobody C68 were developed to evaluate their feasibility for rapid detection (Figure 5A). First, 50 μL of three different diluents (PBS, PBST, and PBS + S9/Tetronic 1307) were added dropwise onto the test strips, which were coated with only the control line, and the results were read after 15 min. A clear band was observed at the C line (Figure 5B), indicating successful colloidal gold labeling of the nanobody. The test strip operates on the principle of competitive binding. In the absence of hG-CSF in the sample, a band will appear at the T line. However, if hG-CSF is present, it binds to the gold-labeled nanobody, preventing the nanobody from binding to the antigen coated on the T line, resulting in no band at the T line (Figure 5C,D).
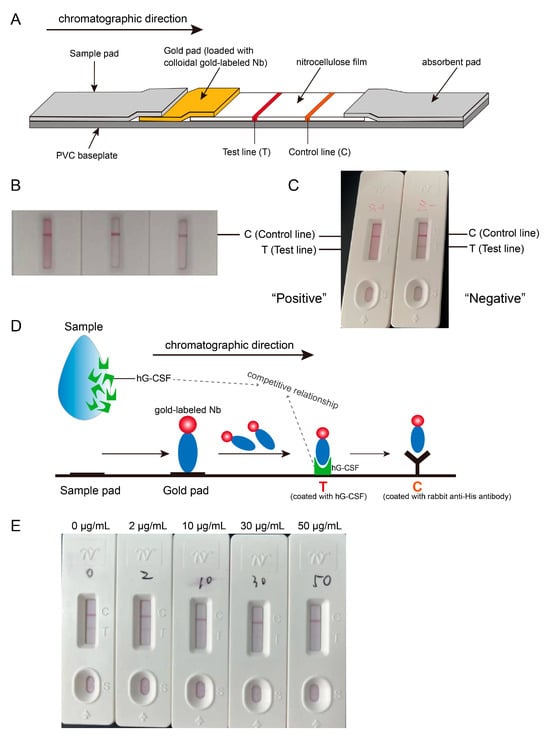
Figure 5.
Rapid identification test for hG-CSF using the developed colloidal gold immunochromatographic test strips. (A) Illustration of test strip assembly. (B) Successful coupling of nanobody with colloidal gold. Purple-red bands always appeared at control line after dropwise adding three different solutions (PBS, PBST, PBS with surfactant S9) on test strips which were only coated control line with rabbit-anti-His antibody. (C) Competition assay was designed in test strip development. If hG-CSF was added to the test strip, it would first bind to the colloidal gold-labeled nanobodies, thereby preventing the nanobodies from binding to the hG-CSF at the test line. Thus, there was no band appearing at the T line, which indicated a positive result. Conversely, if there was no hG-CSF in the sample added on the strip, a band would appear at the T line because of binding of nanobodies to hG-CSF at the T line, reporting a negative result. (D) Schematic representation of the principle of competition assay. (E) Detection limit of the test strip. 0 μg/mL, 2 μg/mL, 10 μg/mL, 30 μg/mL, and 50 μg/mL of hG-CSF samples were separately added on the strips; there was absolutely no band at the T line when hG-CSF concentration ≥30 μg/mL.
To determine the LOD of the test strip, hG-CSF was added to the sample well at concentrations of 0, 2, 10, 30, or 50 μg/mL. It was observed that at least 30 μg/mL of hG-CSF resulted in the absolute absence of a band at the T line (Figure 5E), indicating that the LOD of the colloidal gold-labeled nanobody test strip is 30 μg/mL. Given that the lowest concentration of currently marketed hG-CSF products is 50 μg/mL, this test strip is suitable for the rapid identification for hG-CSF, enhancing ease of operation and reducing test time significantly.
3. Discussion
The anti-hG-CSF nanobody C68 obtained in this study is highly specific, sensitive, and stable at room temperature, making it suitable for the identification test of hG-CSF drugs. Both solid- and liquid-phase panning methods can be used for screening and enriching phage-displayed nanobody libraries during the selection process [16,17,18]. Solid-phase panning is more straightforward, as phage particles carrying specific antibodies bind to the target antigen, while non-specific phage particles are eluted. In this study, nanobodies were enriched through solid-phase panning. Since the coated hG-CSF was adsorbed onto a microplate, nanobodies identified through this approach are more likely to bind to hG-CSF adsorbed on a plate or membrane, which aligns with the immunoblot and immunospot methods commonly used in identification tests. However, antigen exposure is more limited in solid-phase panning compared to liquid-phase screening [19,20], which could affect the nanobody recognition of antigenic epitopes along the immobilized surface, potentially leading to the failure of identifying nanobodies with higher affinity.
Nanobodies have a small molecular weight and can be expressed by various expression systems [8,9,10,11]. They can be produced in prokaryotic cells at higher titers, do not require post-translational modifications, are cost-effective, and exhibit minimal inter-batch variability. Therefore, we used E. coli to express nanobody C68, which was inserted into the pET26b plasmid with six C-terminal His tags to facilitate purification via a nickel column. Both WB and ELISA confirmed that nanobody C68 exhibits excellent specificity for hG-CSF, with sensitivity comparable to that of the anti-hG-CSF monoclonal antibody. The inclusion of the His tag not only facilitates protein purification but also does not compromise the performance of the nanobody. Furthermore, the His tag enables the establishment of a control line in subsequent test strip development.
In this study, we labeled nanobody C68 with colloidal gold and developed an immunochromatographic test strip for rapid hG-CSF detection [21]. The binding between proteins and colloidal gold is primarily driven by electrostatic attraction and hydrophobic forces [22]. To confirm successful colloidal gold labeling of the nanobody, three different solutions (PBS, PBST, or PBS with S9) were applied to test strips coated with the anti-His antibody at the C line but without a T line. Clear bands appeared on the C line, indicating successful labeling. The detection principle of colloidal gold immunochromatographic test strips is typically based on either the competition or sandwich method. The sandwich method is more suited for detecting large molecules because it requires the analyte is simultaneously bound by two antibodies through different epitopes, and the corresponding cost is also higher. In contrast, the competition method is more effective for detecting smaller molecules, offering a more straightforward preparation process and lower cost. In this study, we used the competition method by coating the T line with hG-CSF to develop a complete test strip, enabling effective detection and identification of hG-CSF.
Our findings demonstrated that nanobody C68 specifically recognizes hG-CSF expressed by CHO cells, suggesting that glycosylation does not affect its binding sites on hG-CSF. In future studies, we plan to introduce mutations in the CDR3 region of the nanobody [23] to enhance its specificity and binding affinity, thereby improving its performance in identification test, and providing valuable insights for the screening and mutagenesis of nanobodies targeting other recombinant protein drugs.
4. Materials and Methods
4.1. Recombinant Protein Products and Reagents
The following materials were used in this study: the national reference standard for biological activity assay of hG-CSF (270031-201601, National Institutes for Food and Drug Control, Beijing, China), hG-CSF (marketed product, retained samples in the division of recombinant drugs), granulocyte–macrophage colony-stimulating factor (GM-CSF; marketed product, retained sample in the division of recombinant drugs), erythropoietin (EPO; marketed product, retained sample in the division of recombinant drugs), thrombopoietin (TPO; marketed product, retained sample in the division of recombinant drugs), basic fibroblast growth factor (bFGF; marketed product, retained sample in the division of recombinant drugs), and interferon α2b (IFNα2b; marketed product, retained sample in the division of recombinant drugs).
Freund’s complete adjuvant (FCA) and Freund’s incomplete adjuvant (FIA) (F5881 and F5506, Merck, Billerica, MA, USA), Goat anti-Alpaca IgG(H+L)-HRP (053-404-005, AlpVHHs, Chengdu, China), Human Peripheral Blood Lymphocyte Separation Medium (P8610, Solarbio, Beijing, China), TRIzol Plus RNA Purification Kit (12183555, Invitrogen, Carlsbad, CA, USA), SuperScript III Reverse Transcriptase (18080093, Invitrogen, Carlsbad, CA, USA), DNA Gel Extraction Kit (NA1111, Merck, Billerica, MA, USA), Hot Start Taq DNA Polymerase (M0490S, NEB, Beverly, MA, USA), DNA Ligase (M2622S, NEB, Beverly, MA, USA), restriction enzymes NotI, SfiI, MscI, and XhoI (R0189S, R0123S, R0534S, and R0146S, NEB, Beverly, MA, USA), M13KO7 helper phage (N0315S, NEB, Beverly, MA, USA), PEG-NaCl solution (C7052, Bioss, Beijing, China), carbonate–bicarbonate buffer (C3041-50CAP, Merck, Billerica, MA, USA), ELISA microplate (154041, Thermo Scientific, Waltham, MA, USA), TMB Liquid-1 component (J644, Amresco, Solon, OH, USA), BL21 (DE3) competent cells (EC0114, Thermo Scientific, Waltham, MA, USA), isopropyl β-D-thiogalactopyranoside (IPTG, I5502, Merck, Billerica, MA, USA), High-Performance HisTrap column (GE17-5248-01, Cytiva, Marlborough, MA, USA), imidazole (I5513, Merck, Billerica, MA, USA), NuPAGE precast polyacrylamide gel (NP0321BOX, Invitrogen, Carlsbad, CA, USA), DAB Substrate Kit (SK-4105, Vector Laboratories, Burlingame, CA, USA), Coomassie Brilliant Blue (P0017B, Beyotime, Shanghai, China), G-CSF monoclonal antibody (sc-53292, SANTA CRUZ Biotechnology, Santa Cruz, CA, USA).
4.2. Alpaca Immunization
A healthy adult male alpaca was immunized four times with hG-CSF, with a 2-week interval between each immunization. For the first immunization, 800 μg of hG-CSF was diluted to 1 mL in PBS, mixed with 1 mL of FCA, and injected along the base of the neck across 6 to 10 sites. For the second through fourth immunizations, 400 μg of hG-CSF was diluted to 1 mL in PBS, mixed with 1 mL of FIA, and injected along the base of the neck across 6 to 10 sites. Peripheral blood (5 mL) was collected prior to the first immunization as a negative control, and at least 5 mL of peripheral blood was collected seven days after each immunization to determine serum antibody titers. Additionally, 20 mL of peripheral blood was collected seven days after the last immunization for the determination of serum antibody titers and lymphocyte isolation.
4.3. Determination of Serum Antibody Titers
hG-CSF was diluted to 2 μg/mL in coating buffer (one capsule of carbonate–bicarbonate buffer (C3041-50CAP, Merck, Billerica, MA, USA) dissolved in 100 mL deionized water), added to a microplate at 100 μL/well, and incubated overnight at 4 °C. The plate was washed three times with PBST, blocked with 4% skim milk at room temperature for 1 h on a shaker, and washed five times with PBST. Alpaca antisera and negative control sera were serially diluted 2-fold into eight concentrations, each added to the plate at 100 μL/well. The plate was incubated at room temperature for 1 h, washed five times with PBST, incubated with goat anti-alpaca IgG(H+L)-HRP (1:10,000) at room temperature for 1 h, washed five times with PBST, and incubated with 100 μL/well of TMB solution for 15 min in the dark, followed by the addition of 100 μL/well of 1 mol/L H2SO4 stop solution. Absorbance at 450 nm was measured using a microplate reader (SpectraMax iD3, Molecular Devices, Sunnyvale, CA, USA). If the OD value of the test well is >0.1 and more than three times that of the negative control well at the corresponding dilution, the result is considered positive. The highest dilution showing a positive result is considered the serum antibody titer.
4.4. Total RNA Extraction from Lymphocytes
Peripheral blood lymphocytes were isolated using the Lymphocyte Separation Medium according to the manufacturer’s instructions. Total RNA was extracted from the isolated lymphocytes by adding 1 mL of TRIzol per 5.0 × 106 cells and incubating the cells at room temperature for 5 min to allow complete cell lysis. The suspension was centrifuged at 12,000 rpm for 5 min, and the supernatant was collected and incubated with 1/5 volume of chloroform at room temperature for 15 min. After centrifugation at 12,000 rpm and 4 °C, the upper aqueous phase was transferred to a clean centrifuge tube, incubated with 1/2 volume of isopropanol at room temperature for 10 min to precipitate RNA, and centrifuged at 12,000 rpm and 4 °C for 15 min. The pellet was resuspended in 75% ethanol, centrifuged at 10,000 rpm and 4 °C for 5 min, and air-dried at room temperature. The RNA was dissolved in deionized water, and total RNA concentration was measured using a Nanodrop.
4.5. Construction of Nanobody Library
The RNA was reverse transcribed into cDNA using the SuperScript III Reverse Transcriptase according to the manufacturer’s instructions. The VHH fragments were amplified using two-step nested PCR: (1) 20 cycles of 98 °C for 10 s, 50 °C for 30 s, and 72 °C for 1 min, followed by 98 °C for 10 s, 68 °C for 1 min, and 72 °C for 10 min using the Vh-LD (5′-CTTGGTGGTCCTGGCTGC-3′) and CH2-R (5′-GGTACGTGCTGTTGAACTGTTCC-3′) primers; (2) five cycles of 98 °C for 10 s, 50 °C for 20 s, and 72 °C for 40 s, followed by 30 cycles of 98 °C for 10 s and 68 °C for 40 s, then 72 °C for 10 min using the VHHF-SfiI (5′-TCGCggcccagccggccatggccCAGKTGCAGCTCGTGGAGTCNGGNGG-3′) and VHHR-NotI (5′-CGAGTgcggccgcGGGTCTTCGCTGTGGTGCG-3′) primers. The target band (approximately 500 bp) was collected by gel extraction.
The pHEN1 plasmid and target VHH fragments were double-digested with NotI and SfiI, incubated with T4 ligase overnight at 16 °C, and heat-inactivated at 65 °C for 10 min. The recombinant vector was electroporated into competent TG1 cells and cultured for 2.5 h at 37 °C and 160 rpm. A 100 μL aliquot of the recovered bacterial suspension was serially diluted to generate 105-, 106-, and 107-fold dilutions and 100 μL of each diluted suspension was plated on LB agar plates supplemented with ampicillin (Amp). The plates were incubated overnight at 37 °C, and library capacity was assessed. Twenty-four colonies were randomly selected from plates containing fewer than 300 clones for PCR identification using the M13-R (5′-AGCGGATAACAATTTCACACAGGA-3′) and pHEN1-R (5′-GCCCCATTCAGATCCTCTTC-3′) primers to determine the positive rate.
4.6. Preparation of Phage Display Library
The recovered bacterial suspension was inoculated into 5 mL of 2× YT medium (containing 2% glucose and 100 μg/mL Amp) and cultured until the OD600 nm reached approximately 0.5. M13KO7 helper phage was added at a multiplicity of infection (MOI) ratio of 20:1 for rescue. The suspension was incubated at 37 °C for 20 min, followed by incubation at 37 °C and 210 rpm for 1.5 h. The bacterial suspension was then centrifuged at 5000 rpm and 4 °C for 25 min, resuspended in 50 mL of 2× YT medium (containing 100 μg/mL Amp and 50 μg/mL Kan), and cultured at 30 °C and 210 rpm for 6–8 h. The suspension was centrifuged at 6000 rpm and 4 °C for 15 min, incubated with 1/5 volume of the original suspension volume of PEG-NaCl solution overnight at 4 °C, centrifuged again at 10,000 rpm and 4 °C for 20 min, and washed once with 5 mL of PBS. The pellet was resuspended in 2 mL of PBS to obtain the nanobody phage display library. A 20 μL aliquot was used for titer determination, and the remaining solution was mixed with an equal volume of sterile glycerol (final concentration: 50%) and stored at −80 °C for subsequent screening.
4.7. Determination of Phage Display Library Titer
The test phage was serially diluted in PBS. 10 μL of each diluted phage (or 10 μL of PBS as a negative control) was added to 190 μL of E. coli TG1 suspension and incubating at 37 °C for 15 min. The suspension was spread-plated on LB agar plates containing Amp and incubated at 37 °C for 1 h, followed by incubation in an inverted position for 12 h. The number of colonies was counted to determine the library titer (CFU/mL).
4.8. Screening and Enrichment of Nanobodies by Repetitive Biopanning with Immobilized Antigen
Four rounds of nanobody biopanning were performed. hG-CSF was diluted in coating buffer to 100 μg/mL, 50 μg/mL, 25 μg/mL, and 10 μg/mL and added to microplate wells at 100 μL/well. The plate was coated overnight at 4 °C, washed five times with PBS, blocked with 150 μL/well of 3% BSA (in PBS) at 37 °C for 2 h, washed five times with PBS, incubated with 100 μL/well of phage display library at 37 °C for 1 h, washed five times with PBS, and incubated with 100 μL/well of Gly-HCl (pH = 2.2) at 37 °C for 6–8 min. The solution in each well was gently pipetted several times, aspirated, and mixed with 15 μL of neutralization buffer (Tris-HCl, pH = 8.0). A 10 μL aliquot was used for titer determination, and the remaining solution was amplified for the next round of biopanning.
4.9. Identification of Positive Phage Clones
After the final round of biopanning, the phage solution was plated and cultured overnight, and 50 clones were selected from the plate and seeded into 96-well plates. After the expansion culture with rescue using the helper phage, the supernatant was collected for ELISA to identify positive clones. hG-CSF was diluted to 5 μg/mL in coating buffer and added to a microplate. The plate was incubated overnight at 4 °C, washed five times with PBST (300 μL/well), blocked with 250 μL/well 1× Blocking Buffer at 37 °C for 2 h, washed five times with PBST (300 μL/well), incubated with 100 μL/well of phage at 37 °C for 1 h, and washed five times with PBST (300 μL/well). The plate was then incubated with 100 μL/well of HRP-anti-M13 antibody (1:5000 in 1× Blocking Buffer) at 37 °C for 1 h, washed five times with PBST (300 μL/well), and incubated with 100 μL/well of TMB solution at 37 °C for 10 min. The reaction was terminated by adding 100 μL of 1 mol/L H2SO4 to each well, and OD450 nm was measured. A display phage well with an OD ratio ≥ 3 compared to the negative control well was considered a positive clone. All identified positive clones were sequenced to obtain their nucleic acid sequences, which were then translated into amino acid sequences to identify the various VHH sequences.
4.10. Construction of Expression Plasmid and Expression and Purification of Nanobodies
The VHH gene was inserted into the pET26b prokaryotic expression plasmid following MscI and XhoI double digestion, and the recombinant plasmid was expressed in E. coli BL21 (DE3) cells. Once the OD600 nm of the LB broth containing kanamycin reached 0.6, IPTG was added to a final concentration of 0.1 mM to induce expression at 19 °C for 15 h. Subsequently, the bacterial cells from three flasks (500 mL of culture in each flask) were parallelly collected by centrifugation at 8000 rpm and 4 °C for 30 min, resuspended in equilibration buffer, and sonicated on ice. The supernatant was collected, purified using a nickel column (HisTrap column (GE17-5248-01, Cytiva, Marlborough, MA, USA)) through the principle of binding of nickel column with His tag in the nanobody, and eluted using an imidazole gradient (50 mM, 80 mM, 100 mM, 150 mM, 250 mM) to obtain the nanobodies.
4.11. Western Blot
The corresponding samples and controls were selected based on the experimental objective and diluted to the appropriate concentrations. A 26 μL aliquot of each sample was mixed with 10 μL of 4× LDS sample buffer and 4 μL of 10× reduction agent, bringing the total volume to 40 μL. The mixture was heated in a boiling water bath for 8 min and centrifuged at 10,000 rpm for 5 min. A total of 20 μL of each sample was loaded onto a 4–12% NuPAGE Bis-Tris precast gel and electrophoresed at 200 V and 120 mA for 30 min. The proteins were then transferred onto a PVDF membrane using the Invitrogen iBlot2 transfer device. The membrane was blocked in about 25 mL of PBS containing 5% skim milk powder at room temperature for 70 min on a shaker. After three washes with PBS (5 min per wash), the membrane was incubated with the HRP-conjugated nanobodies (in PBS containing 5% skim milk) at room temperature for 1 h on a shaker, washed again three times with PBS (5 min per wash), and immersed in DAB staining solution. Once clear bands were observed, the PVDF membrane was rinsed with water to stop color development.
4.12. hG-CSF Quantification by ELISA with Nanobody
The nanobody (C43) was diluted to 5 μg/mL in coating buffer and added to a microplate at 100 μL/well. The plate was coated overnight at 4 °C, washed five times with PBST, blocked with 5% BSA on a shaker at room temperature for 1 h, and washed five times with PBST. The hG-CSF product and reference standard were serially diluted 3-fold into eight concentrations, starting from 5 μg/mL, and added to the plate at 100 μL/well. After incubation at room temperature for 1 h on a shaker, the plate was washed five times with PBST, incubated with HRP-conjugated nanobody (C68) (1:8000) at room temperature for 1 h on a shaker, and rewashed five times with PBST. The plate was then incubated with 100 μL/well TMB substrate solution for 15 min, and the reaction was stopped by adding 1 mol/L of H2SO4 to each well. Absorbance was measured at 450 nm, and the data were fitted to a four-parameter curve. Then, specificity, accuracy, precision, and quantification range of this quantification method were validated one by one.
4.13. Preparation of Colloidal Gold-Labeled Nanobody Test Strips
A 1 mL aliquot of colloidal gold (particle size: 40 nm, concentration: 1 mg/mL) was transferred to a 1.5 mL centrifuge tube, mixed with 2 μL of 0.2 mol/L potassium carbonate, and incubated with 10 μg of His-tagged nanobody for 15 min. The mixture was then incubated with 50 μL of 8% BSA for 15 min and centrifuged at 12,000 rpm for 20 min. The supernatant was discarded, and the pellet was reconstituted in 500 μL of gold reconstitution solution. The solution was added onto a 7 mm × 150 mm gold pad and dried in an oven at 37 °C. Rabbit anti-His antibody was diluted to 0.7 mg/mL in diluent and applied onto a nitrocellulose membrane at 1 μL/cm as the control line. hG-CSF was diluted to 1.5 mg/mL in diluent and applied onto the nitrocellulose membrane as the test line, and dried at 37 °C. The test pad was assembled in the chromatographic direction with the following sequence: sample chromatography pad, gold pad, nitrocellulose film, absorbent pad. The assembled pad was then cut into test strips of appropriate width using a die cutter and combined with the base plate for detection.
5. Conclusions
In this study, anti-hG-CSF nanobody sequences were obtained through steps such as alpaca immunization, nanobody gene library construction, phage display library preparation, and solid-phase panning. The nanobody was expressed in E. coli and its basic physicochemical properties were characterized. The resulting nanobody demonstrated an LOD comparable to that of the monoclonal antibody while offering advantages such as simpler expression and purification processes, reduced cost, lower inter-batch variability, and excellent thermal stability. This nanobody is highly suitable for the identification of hG-CSF produced by different manufacturers, including hG-CSF derived from E. coli and CHO cells, as well as PEGylated hG-CSF. The established and validated ELISA method exhibited strong specificity, accuracy, and precision for quantifying hG-CSF concentrations. Finally, the rapid detection test strip, prepared by labeling the His-tagged nanobody with colloidal gold, enables fast and efficient identification of hG-CSF within 15 min.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/biophysica5040047/s1.
Author Contributions
Conceptualization, C.L., X.Q. and J.W.; methodology, Q.M., L.Z., X.L., D.P., L.Y. and X.S.; validation, Q.M. and L.Z.; investigation, Q.M., L.Z. and X.Q.; resources, X.Q.; data curation, Q.M. and L.Z.; writing—original draft preparation, Q.M. and L.Z.; writing—review and editing, Q.M., C.L., X.Q. and J.W.; visualization, Q.M. and L.Z.; supervision, Y.Z. and Z.F.; project administration, Z.F., C.L. and J.W.; funding acquisition, X.S., Y.Z., C.L. and J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by National Key Research and Development Program of China, grant number 2021YFF0600804.
Institutional Review Board Statement
The animal study protocol was approved by the laboratory animal care and ethics committee of Guangdong Laidi Biomedical Research Institute Co., LTD (protocol code 2022030, 10 August 2022).
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data of this study are presented in the article. Additional data will be available upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Morstyn, G.; Foote, M.; Perkins, D.; Vincent, M. The clinical utility of granulocyte colony-stimulating factor: Early achievements and future promise. Stem Cells 1994, 12 (Suppl. S1), 213–227; discussion 218–227. [Google Scholar] [CrossRef]
- Khorasanizadeh, M.; Eskian, M.; Vaccaro, A.R.; Rahimi-Movaghar, V. Granulocyte Colony-Stimulating Factor (G-CSF) for the Treatment of Spinal Cord Injury. CNS Drugs 2017, 31, 911–937. [Google Scholar] [CrossRef] [PubMed]
- Wurfel, W. Treatment with granulocyte colony-stimulating factor in patients with repetitive implantation failures and/or recurrent spontaneous abortions. J. Reprod. Immunol. 2015, 108, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Muyldermans, S.; Baral, T.N.; Retamozzo, V.C.; De Baetselier, P.; De Genst, E.; Kinne, J.; Leonhardt, H.; Magez, S.; Nguyen, V.K.; Revets, H.; et al. Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 2009, 128, 178–183. [Google Scholar] [CrossRef]
- Muyldermans, S. A guide to: Generation and design of nanobodies. FEBS J. 2021, 288, 2084–2102. [Google Scholar] [CrossRef]
- Goldman, E.R.; Liu, J.L.; Zabetakis, D.; Anderson, G.P. Enhancing Stability of Camelid and Shark Single Domain Antibodies: An Overview. Front. Immunol. 2017, 8, 865. [Google Scholar] [CrossRef]
- Iwaki, T.; Hara, K.; Umemura, K. Nanobody production can be simplified by direct secretion from Escherichia coli. Protein Expr. Purif. 2020, 170, 105607. [Google Scholar] [CrossRef]
- Chen, Q.; Zhou, Y.; Yu, J.; Liu, W.; Li, F.; Xian, M.; Nian, R.; Song, H.; Feng, D. An efficient constitutive expression system for Anti-CEACAM5 nanobody production in the yeast Pichia pastoris. Protein Expr. Purif. 2019, 155, 43–47. [Google Scholar] [CrossRef]
- Modarresi, M.; Javaran, M.J.; Shams-Bakhsh, M.; Zeinali, S.; Behdani, M.; Mirzaee, M. Transient expression of anti-VEFGR2 nanobody in Nicotiana tabacum and N. benthamiana. 3 Biotech 2018, 8, 484. [Google Scholar] [CrossRef]
- Shokrollahi, N.; Habibi-Anbouhi, M.; Jahanian-Najafabadi, A.; Alirahimi, E.; Behdani, M. Expressing of Recombinant VEGFR2-specific Nanobody in Baculovirus Expression System. Iran. J. Biotechnol. 2021, 19, e2783. [Google Scholar] [CrossRef]
- Muyldermans, S. Applications of Nanobodies. Annu. Rev. Anim. Biosci. 2021, 9, 401–421. [Google Scholar] [CrossRef] [PubMed]
- Jovcevska, I.; Muyldermans, S. The Therapeutic Potential of Nanobodies. BioDrugs 2020, 34, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zeng, W.; Yu, F.; Xu, P.; Chen, C.Y.; Chen, W.; Dong, Y.; Wang, F.; Ma, L. Visual and High-Efficiency Secretion of SARS-CoV-2 Nanobodies with Escherichia coli. Biomolecules 2025, 15, 111. [Google Scholar] [CrossRef]
- Hao, Z.; Dong, X.; Zhang, Z.; Qin, Z. A Nanobody of PEDV S1 Protein: Screening and Expression in Escherichia coli. Biomolecules 2024, 14, 1116. [Google Scholar] [CrossRef]
- Aujame, L.; Geoffroy, F.; Sodoyer, R. High affinity human antibodies by phage display. Hum. Antibodies 1997, 8, 155–168. [Google Scholar] [CrossRef]
- Zhu, M.; Gong, X.; Hu, Y.; Ou, W.; Wan, Y. Streptavidin-biotin-based directional double Nanobody sandwich ELISA for clinical rapid and sensitive detection of influenza H5N1. J. Transl. Med. 2014, 12, 352. [Google Scholar] [CrossRef]
- Zhong, N.; Lei, W.; Liu, Z.; Xie, X.; Zhang, L.; Jin, T.; Cao, M.; Chen, Y. Screening and characterization of camelid-derived nanobodies against hemoglobin. Sheng Wu Gong Cheng Xue Bao 2025, 41, 1515–1534. [Google Scholar] [CrossRef]
- Kobayashi, N.; Karibe, T.; Goto, J. Dissociation-independent selection of high-affinity anti-hapten phage antibodies using cleavable biotin-conjugated haptens. Anal. Biochem. 2005, 347, 287–296. [Google Scholar] [CrossRef]
- Pini, A.; Ricci, C.; Bracci, L. Phage display and colony filter screening for high-throughput selection of antibody libraries. Comb. Chem. High Throughput Screen. 2002, 5, 503–510. [Google Scholar] [CrossRef]
- Qin, X.; Duan, M.; Pei, D.; Lin, J.; Wang, L.; Zhou, P.; Yao, W.; Guo, Y.; Li, X.; Tao, L.; et al. Development of novel-nanobody-based lateral-flow immunochromatographic strip test for rapid detection of recombinant human interferon alpha2b. J. Pharm. Anal. 2022, 12, 308–316. [Google Scholar] [CrossRef]
- Hughes, D. Preparation of colloidal gold probes. Methods Mol. Biol. 2005, 295, 155–172. [Google Scholar] [CrossRef]
- Kunert, R.; Reinhart, D. Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 2016, 100, 3451–3461. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).