Estimation of Growth Parameters of Eustoma grandiflorum Using Smartphone 3D Scanner
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cultivation Conditions
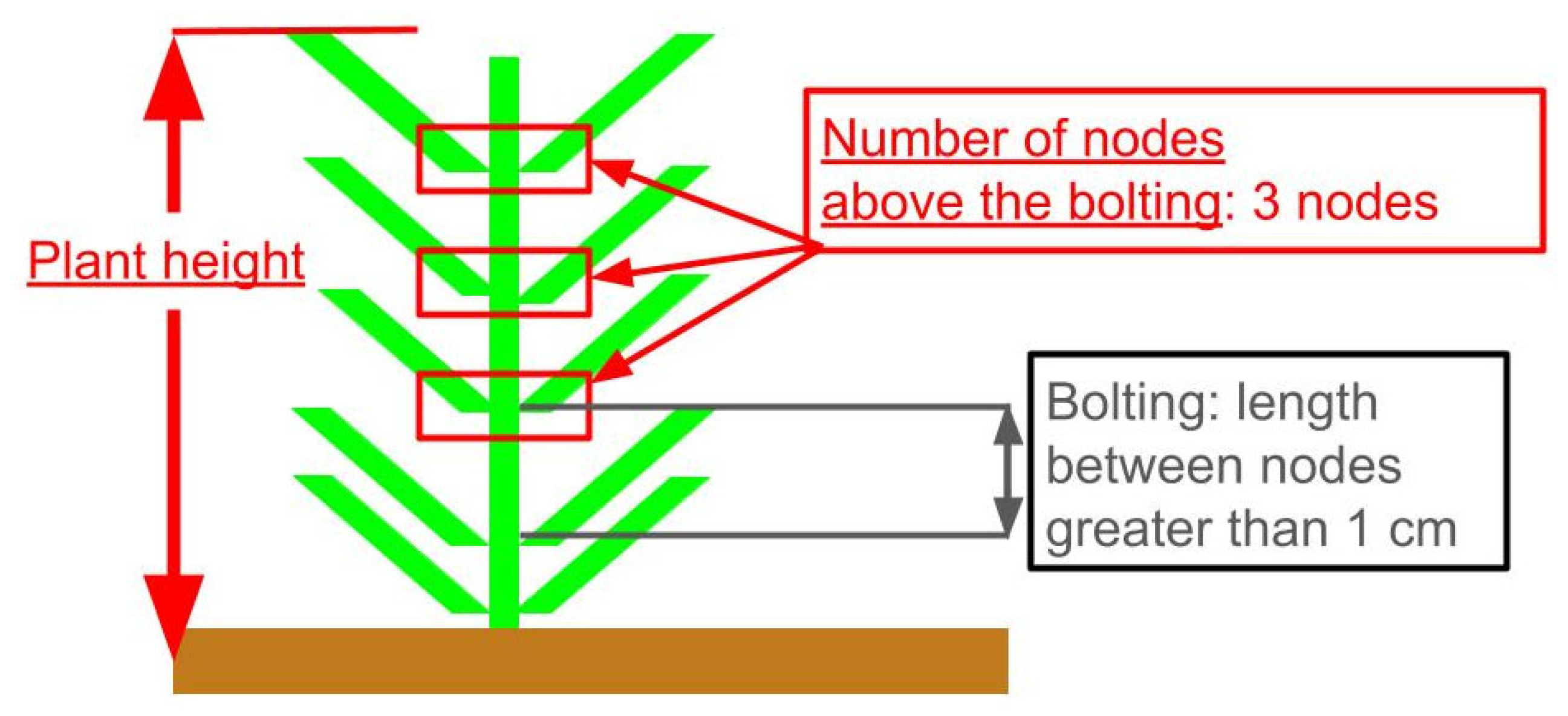
2.2. Growth Survey and Definition of Target Growth Parameters
2.3. Three-Dimensional Data Acquisition Using Smartphone Application
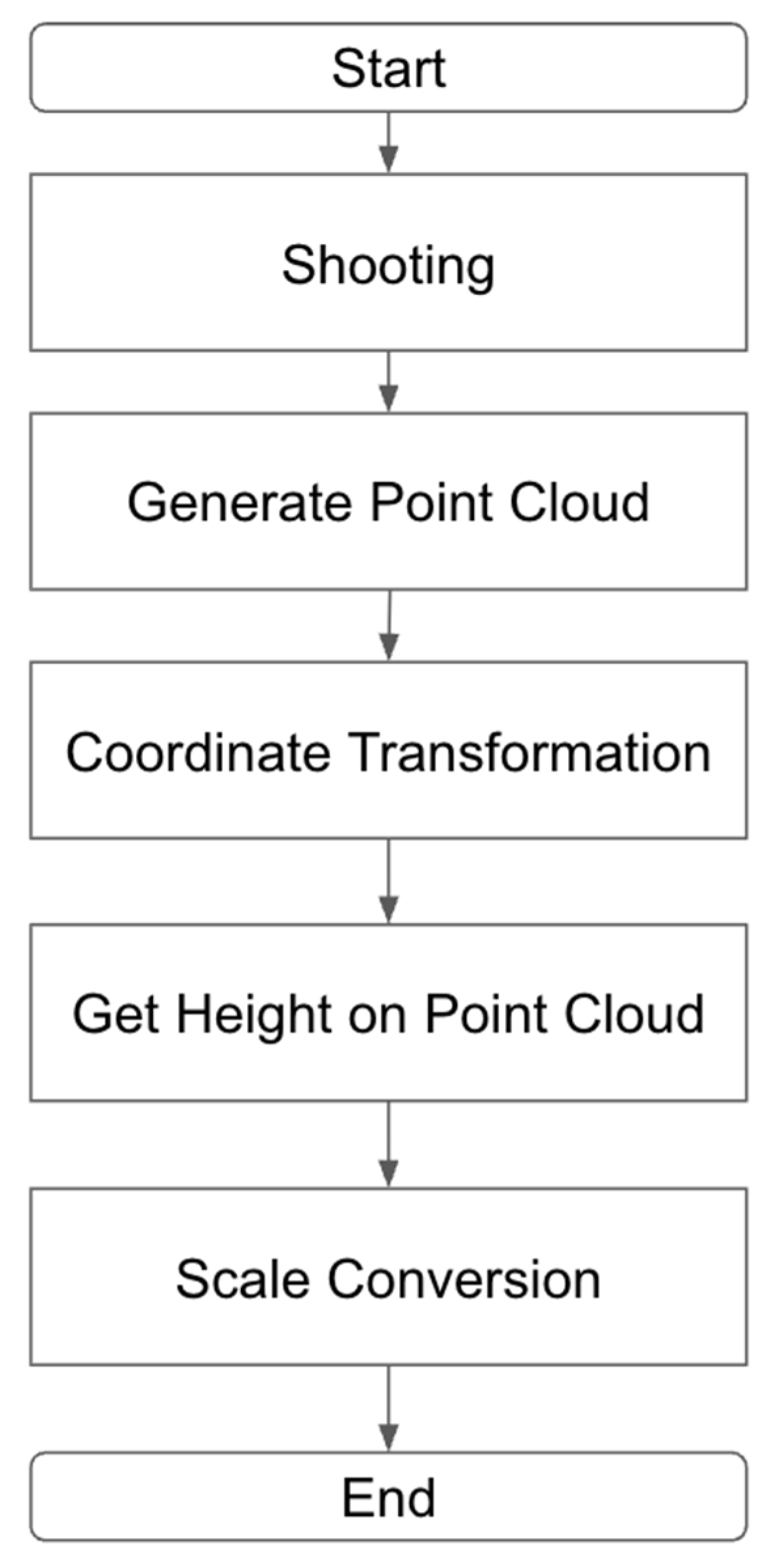
2.4. Estimation of Plant Height from 3D Point Clouds
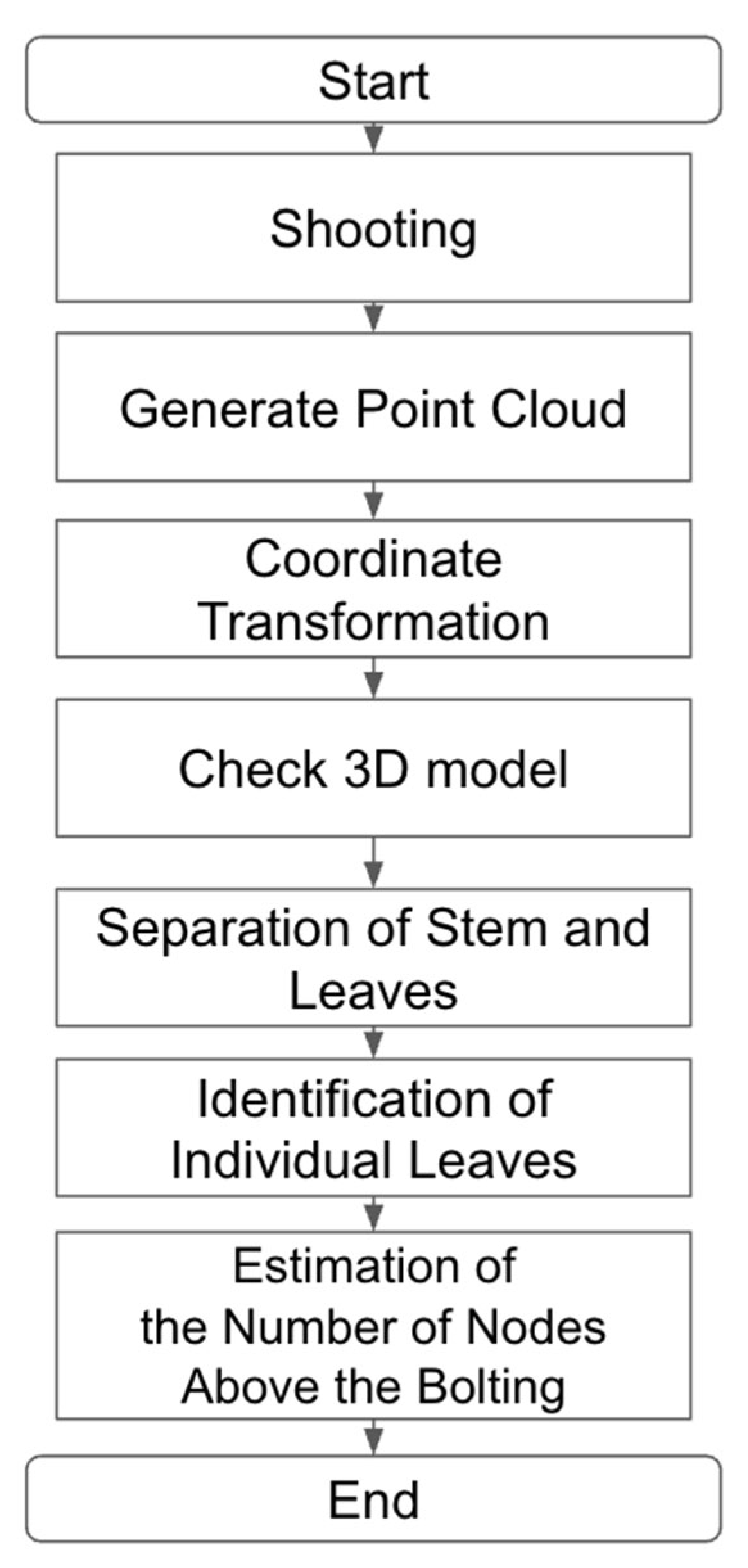
2.5. Estimation of Number of Nodes Above Bolting from 3D Point Clouds
2.5.1. Point Cloud Segmentation and Node Estimation Accuracy
2.5.2. Leaf Clustering and Node Count Estimation
2.6. Evaluation of Estimation Accuracy
3. Results
3.1. Estimation of Plant Height
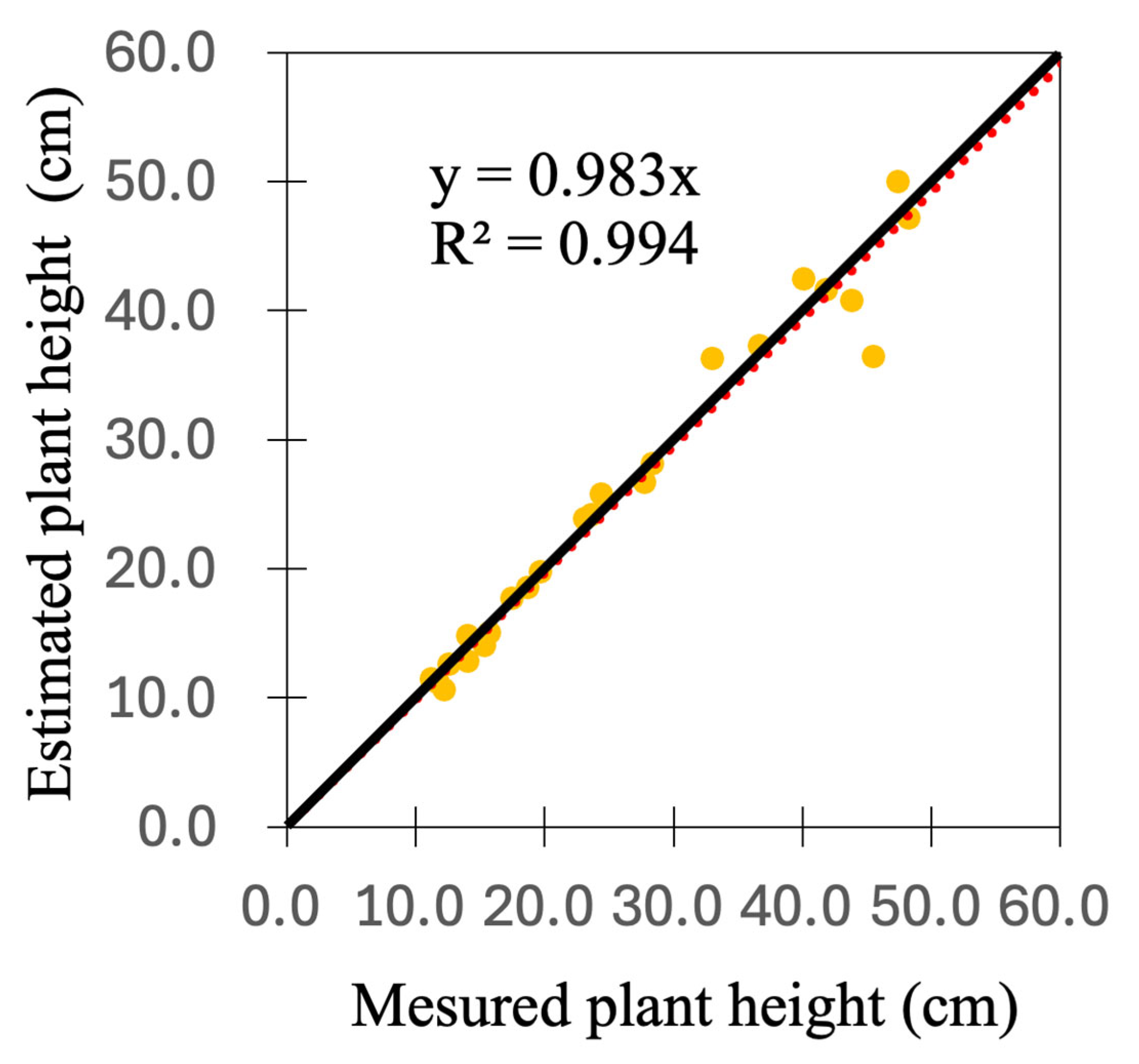
| RMSE (cm) | MAPE (%) | |
|---|---|---|
| Early | 1.0 | 5.0 |
| Middle | 0.7 | 4.5 |
| Late | 3.8 | 6.9 |
| All | 1.2 | 5.3 |
3.2. Separation of Stem and Leaf
3.3. Identification of Individual Leaves
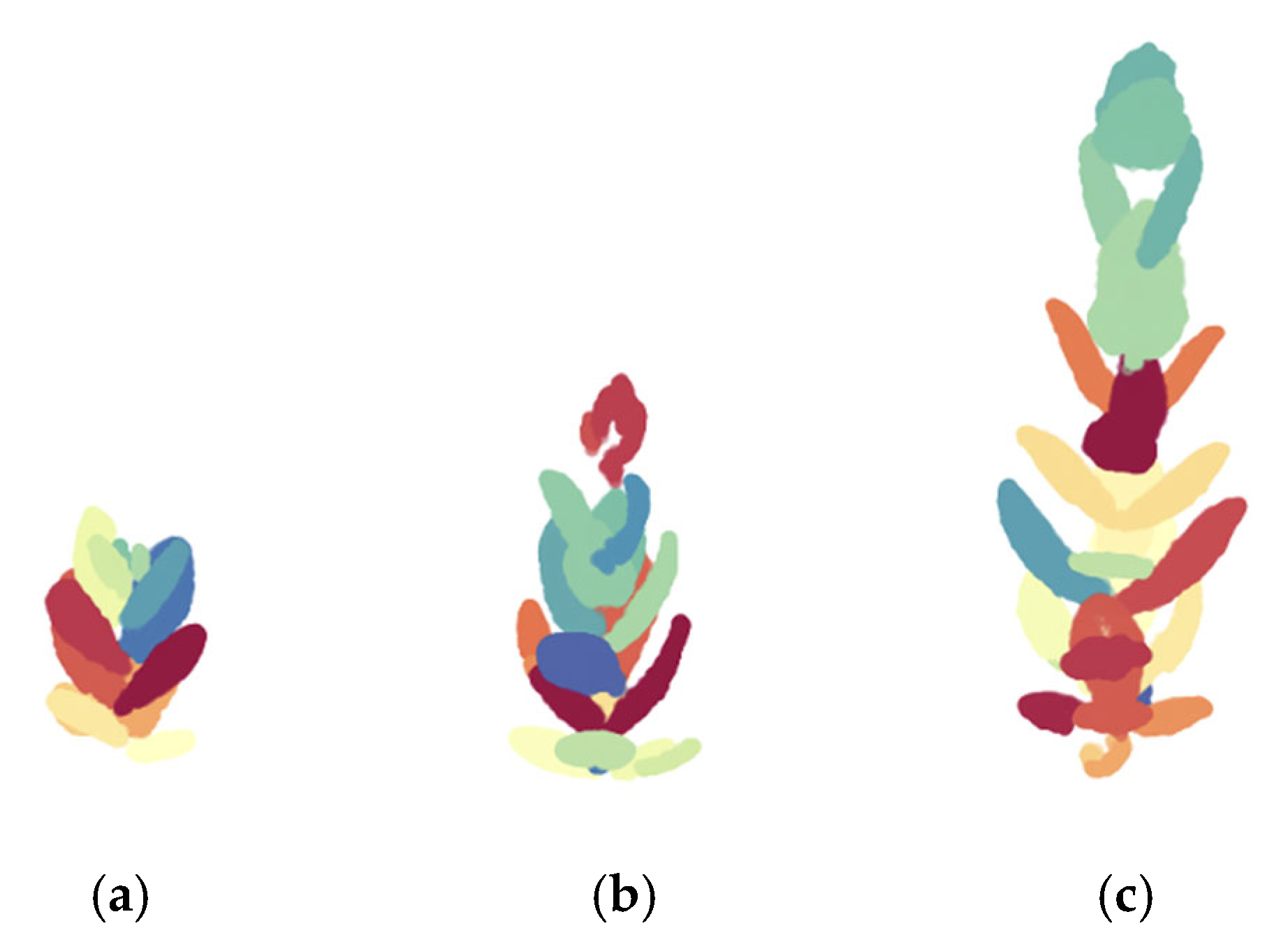
| Success Rate of Individual Leaf (%) | |||
|---|---|---|---|
| Lower | Middle | Upper | |
| Early | 67 | 79 | 13 |
| Middle | 62 | 92 | 43 |
| Late | 28 | 58 | 34 |
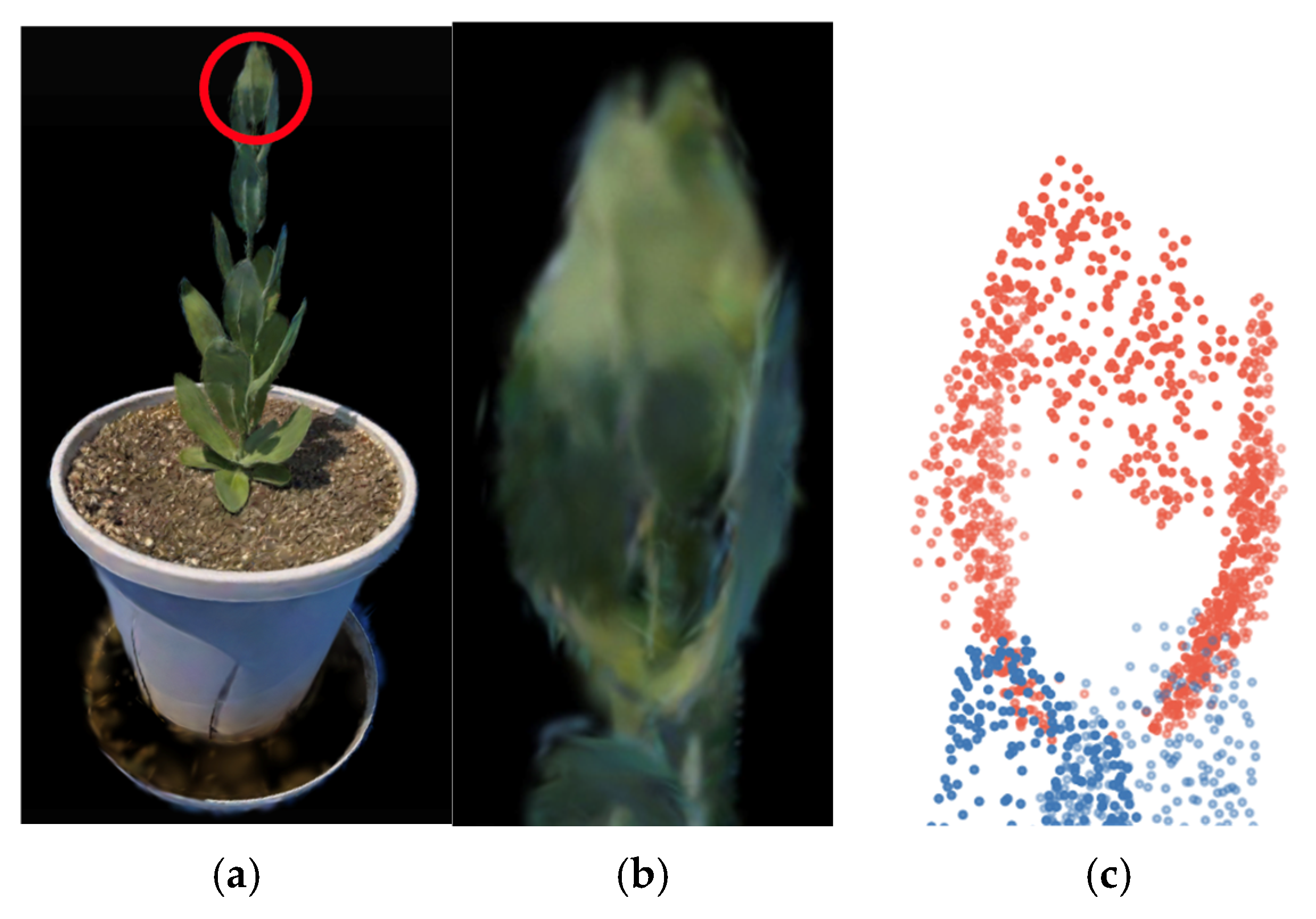
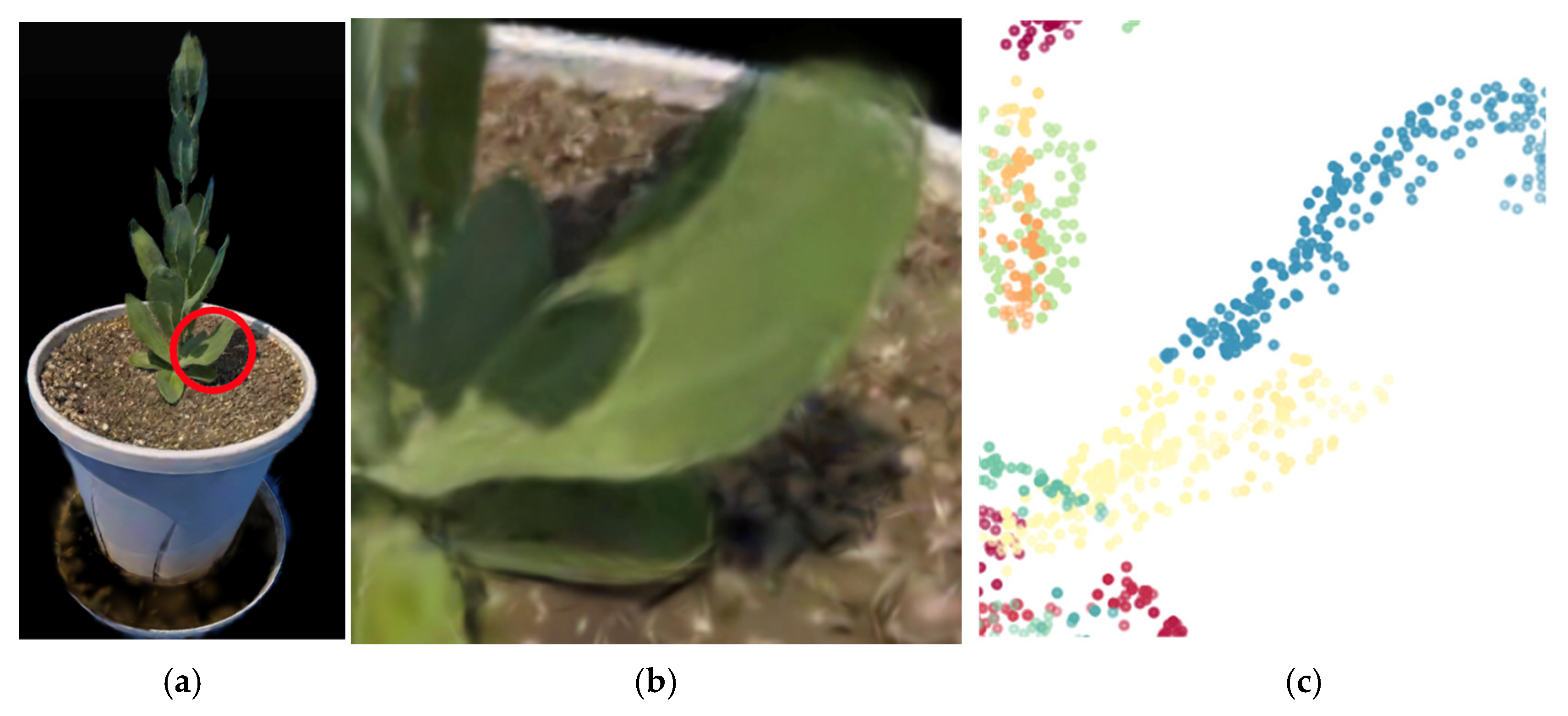
3.4. Estimation of the Number of Nodes Above the Bolting
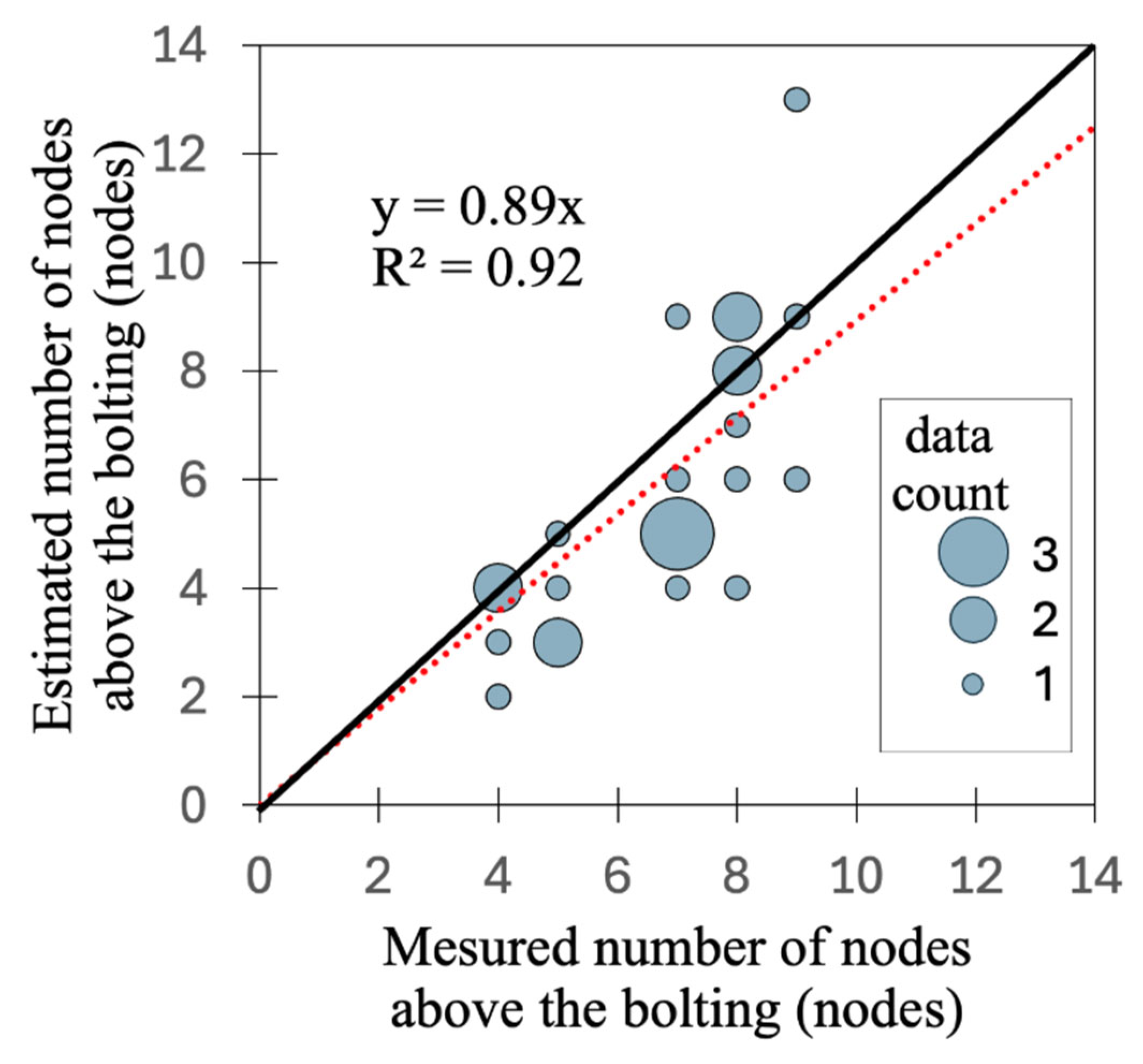
| RMSE (cm) | MAPE (%) | |
|---|---|---|
| Early | 1.3 | 23 |
| Middle | 2.4 | 24 |
| Late | 1.9 | 14 |
| All | 1.2 | 20 |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sato, Y.; Yamashita, Y.; Inaba, O.; Naito, H.; Hoshi, N. Utilization and evaluation of a commuting-based agricultural support system for stock and Eustoma cultivation in areas where farming has resumed. Tohoku Agric. Res. 2023, 76, 91–92. (In Japanese) [Google Scholar]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, Camera, Action: High-Throughput Plant Phenotyping Is Ready for a Close-Up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Teng, P.; Aono, M.; Shimizu, Y.; Hosoi, F.; Omasa, K. 3D Monitoring for Plant Growth Parameters in Field with a Single Camera by Multi-View Approach. J. Agric. Meteorol. 2018, 74, 129–139. [Google Scholar] [CrossRef]
- Boogaard, F.P.; van Henten, E.J.; Kootstra, G. The Added Value of 3D Point Clouds for Digital Plant Phenotyping—A Case Study on Internode Length Measurements in Cucumber. Biosyst. Eng. 2023, 234, 1–12. [Google Scholar] [CrossRef]
- Bao, Y.; Tang, L.; Srinivasan, S.; Schnable, P.S. Field-Based Architectural Traits Characterisation of Maize Plant Using Time-of-Flight 3D Imaging. Biosyst. Eng. 2019, 178, 86–101. [Google Scholar] [CrossRef]
- Hosoi, F.; Nakabayashi, K.; Omasa, K. 3-D Modeling of Tomato Canopies Using a High-Resolution Portable Scanning Lidar for Extracting Structural Information. Sensors 2011, 11, 2166–2174. [Google Scholar] [CrossRef]
- Fang, K.; Xu, K.; Wu, Z.; Huang, T.; Yang, Y. Three-Dimensional Point Cloud Segmentation Algorithm Based on Depth Camera for Large Size Model Point Cloud Unsupervised Class Segmentation. Sensors 2023, 24, 112. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Xiang, L.; Wu, Q.; Jiang, H. Automatic Non-Destructive Growth Measurement of Leafy Vegetables Based on Kinect. Sensors 2018, 18, 806. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Zhou, Y.; Wang, H.; Li, M. Banana Pseudostem Width Detection Based on Kinect V2 Depth Sensor. Comput. Intell. Neurosci. 2022, 2022, 3083647. [Google Scholar] [CrossRef]
- Andújar, D.; Ribeiro, A.; Fernández-Quintanilla, C.; Dorado, J. Using Depth Cameras to Extract Structural Parameters to Assess the Growth State and Yield of Cauliflower Crops. Comput. Electron. Agric. 2016, 122, 67–73. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Slaughter, D.C.; Max, N.; Maloof, J.N.; Sinha, N. Structured Light-Based 3D Reconstruction System for Plants. Sensors 2015, 15, 18587–18612. [Google Scholar] [CrossRef]
- Shi, W.; van de Zedde, R.; Jiang, H.; Kootstra, G. Plant-Part Segmentation Using Deep Learning and Multi-View Vision. Biosyst. Eng. 2019, 187, 81–95. [Google Scholar] [CrossRef]
- Shangpeng, S.; Li, C.; Chee, P.W.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Adhikari, J.; Shehzad, T. Three-Dimensional Photogrammetric Mapping of Cotton Bolls In Situ Based on Point Cloud Segmentation and Clustering. ISPRS J. Photogramm. Remote Sens. 2020, 160, 195–207. [Google Scholar] [CrossRef]
- Yang, Z.; Han, Y. A Low-Cost 3D Phenotype Measurement Method of Leafy Vegetables Using Video Recordings from Smartphones. Sensors 2020, 20, 6068. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, X.; Xie, P.; Guo, Z.; Fang, H.; Fu, H.; Hu, X.; Sun, Z.; Cen, H. PanicleNeRF: Low-Cost, High-Precision in-Field Phenotypingof Rice Panicles with Smartphone. Plant Phenomics 2024, 6, 0279. [Google Scholar] [CrossRef]
- Bar-Sella, G.; Gavish, M.; Moshelion, M. From Selfies to Science—Precise 3D Leaf Measurement with iPhone 13 and Its Implications for Plant Development and Transpiration. bioRxiv 2024. bioRxiv:2023.12.30.573617. [Google Scholar]
- Dutagaci, H.; Rasti, P.; Galopin, G.; Rousseau, D. ROSE-X: An Annotated Data Set for Evaluation of 3D Plant Organ Segmentation Methods. Plant Methods 2020, 16, 28. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, H.; Chen, Y. An Automated Phenotyping Method for Chinese Cymbidium Seedlings Based on 3D Point Cloud. Plant Methods 2024, 20, 151. [Google Scholar] [CrossRef]
- Kawabata, S.; Nii, K.; Yokoo, M. Three-Dimensional Formation of Corolla Shapes in Relation to the Developmental Distortion of Petals in Eustoma grandiflorum. Sci. Hortic. 2011, 132, 66–70. [Google Scholar] [CrossRef]
- Ijiri, T.; Yokoo, M.; Kawabata, S.; Igarashi, T. Surface-Based Growth Simulation for Opening Flowers. In Proceedings of the Graphics Interface 2008, Windsor, ON, Canda, 28–30 May 2008; Canadian Information Processing Society: Mississauga, ON, Canada, 2008; pp. 227–234. [Google Scholar]
- Kerbl, B.; Kopanas, G.; Leimkühler, T.; Drettakis, G. 3D Gaussian Splatting for Real-Time Radiance Field Rendering. ACM Trans. Graph. 2023, 42, 139:1–139:14. [Google Scholar] [CrossRef]
- Meier, U. (Ed.) Growth Stages of Mono- and Dicotyledonous Plants: BBCH Monograph, 2nd ed.; Federal Biological Research Centre for Agriculture and Forestry: Bonn, Germany, 2001; Available online: https://www.reterurale.it/downloads/BBCH_engl_2001.pdf (accessed on 26 August 2025).
- Mariga, L. pyRANSAC-3D; 2022. Available online: https://leomariga.github.io/pyRANSAC-3D/ (accessed on 26 August 2025).
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Ohashi, Y.; Ishigami, Y.; Goto, E. Monitoring the Growth and Yield of Fruit Vegetables in a Greenhouse Using a Three-Dimensional Scanner. Sensors 2020, 20, 5270. [Google Scholar] [CrossRef]
- Zhang, D.; Gajardo, J.; Medic, T.; Katircioglu, I.; Boss, M.; Kirchgessner, N.; Walter, A.; Roth, L. Wheat3DGS: In-Field 3D Reconstruction, Instance Segmentation and Phenotyping of Wheat Heads with Gaussian Splatting. In Proceedings of the Computer Vision and Pattern Recognition Conference, Nashville, TN, USA, 11–15 June 2025. [Google Scholar]
- Zhou, H.; Zhou, Y.; Long, W.; Wang, B.; Zhou, Z.; Chen, Y. A Fast Phenotype Approach of 3D Point Clouds of Pinus Massoniana Seedlings. Front. Plant Sci. 2023, 14, 1146490. [Google Scholar] [CrossRef]
- Turgut, K.; Dutagaci, H.; Galopin, G.; Rousseau, D. Segmentation of Structural Parts of Rosebush Plants with 3D Point-Based Deep Learning Methods. Plant Methods 2022, 18, 20. [Google Scholar] [CrossRef]
- Li, H.; Wu, G.; Tao, S.; Yin, H.; Qi, K.; Zhang, S.; Guo, W.; Ninomiya, S.; Mu, Y. Automatic Branch–Leaf Segmentation and Leaf Phenotypic Parameter Estimation of Pear Trees Based on Three-Dimensional Point Clouds. Sensors 2023, 23, 4572. [Google Scholar] [CrossRef]
- Bae, S.-J.; Kim, J.-Y. Indoor Clutter Object Removal Method for an As-Built Building Information Model Using a Two-Dimensional Projection Approach. Appl. Sci. 2023, 13, 9636. [Google Scholar] [CrossRef]
- Hu, J.S.K.; Kuai, T.; Waslander, S.L. Point Density-Aware Voxels for LiDAR 3D Object Detection. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Guo, R.; Xie, J.; Zhu, J.; Cheng, R.; Zhang, Y.; Zhang, X.; Gong, X.; Zhang, R.; Wang, H.; Meng, F. Improved 3D Point Cloud Segmentation for Accurate Phenotypic Analysis of Cabbage Plants Using Deep Learning and Clustering Algorithms. Comput. Electron. Agric. 2023, 211, 108014. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Yang, J.; Ren, G.; Wang, W.; Zhang, W.; Li, F. A Method for Tomato Plant Stem and Leaf Segmentation and Phenotypic Extraction Based on Skeleton Extraction and Supervoxel Clustering. Agronomy 2024, 14, 198. [Google Scholar] [CrossRef]

| IoU | mIoU | Acc | ||
|---|---|---|---|---|
| Leaf | Stem | |||
| Early | 0.933 | 0.435 | 0.684 | 0.937 |
| Middle | 0.924 | 0.616 | 0.770 | 0.932 |
| Late | 0.954 | 0.694 | 0.824 | 0.958 |
| Average | 0.937 | 0.581 | 0.759 | 0.942 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanagita, R.; Naito, H.; Yamashita, Y.; Hosoi, F. Estimation of Growth Parameters of Eustoma grandiflorum Using Smartphone 3D Scanner. Eng 2025, 6, 232. https://doi.org/10.3390/eng6090232
Yanagita R, Naito H, Yamashita Y, Hosoi F. Estimation of Growth Parameters of Eustoma grandiflorum Using Smartphone 3D Scanner. Eng. 2025; 6(9):232. https://doi.org/10.3390/eng6090232
Chicago/Turabian StyleYanagita, Ryusei, Hiroki Naito, Yoshimichi Yamashita, and Fumiki Hosoi. 2025. "Estimation of Growth Parameters of Eustoma grandiflorum Using Smartphone 3D Scanner" Eng 6, no. 9: 232. https://doi.org/10.3390/eng6090232
APA StyleYanagita, R., Naito, H., Yamashita, Y., & Hosoi, F. (2025). Estimation of Growth Parameters of Eustoma grandiflorum Using Smartphone 3D Scanner. Eng, 6(9), 232. https://doi.org/10.3390/eng6090232






