A Modular Framework for RGB Image Processing and Real-Time Neural Inference: A Case Study in Microalgae Culture Monitoring
Abstract
1. Introduction
2. Materials and Methods
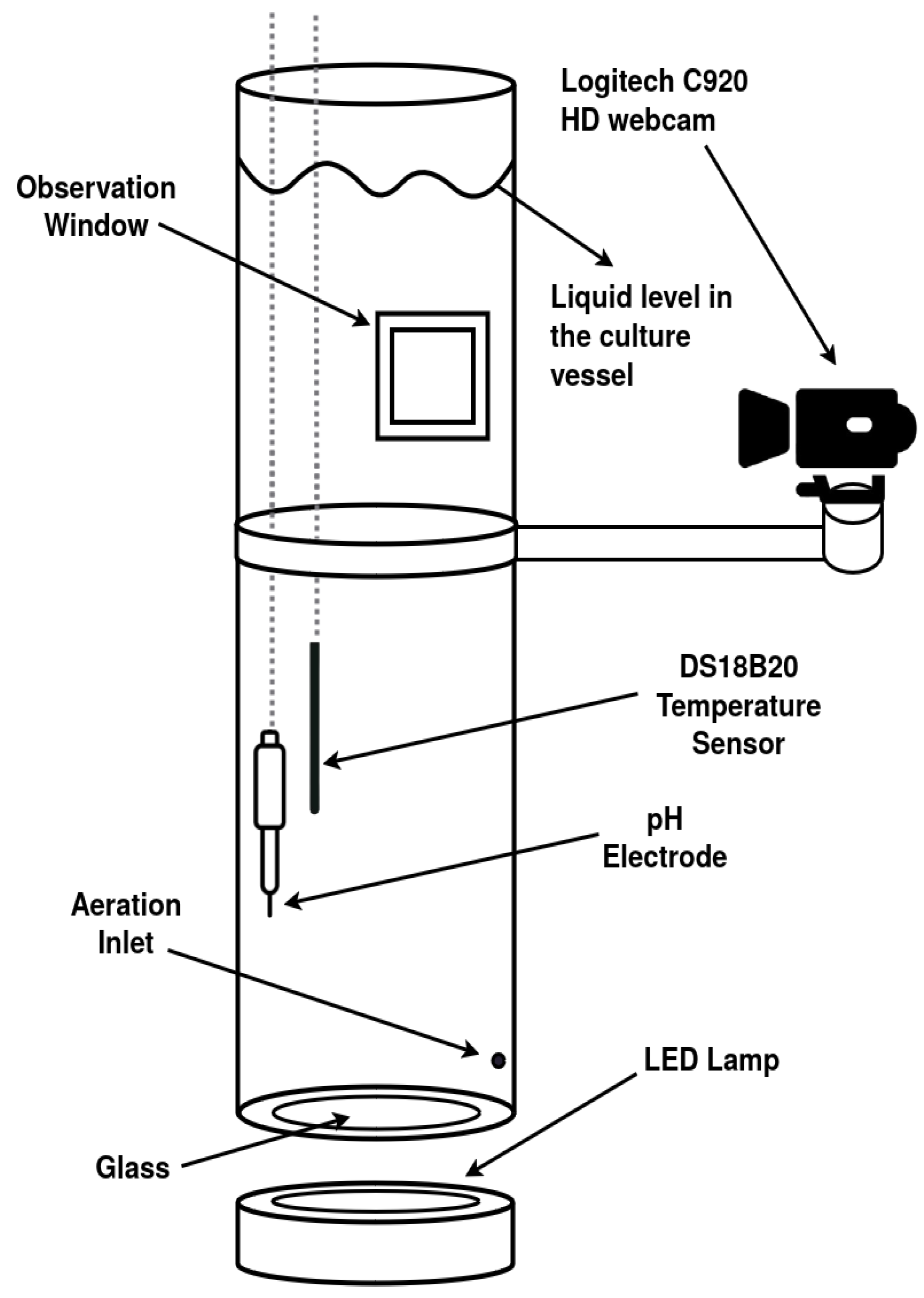
2.1. Experimental System
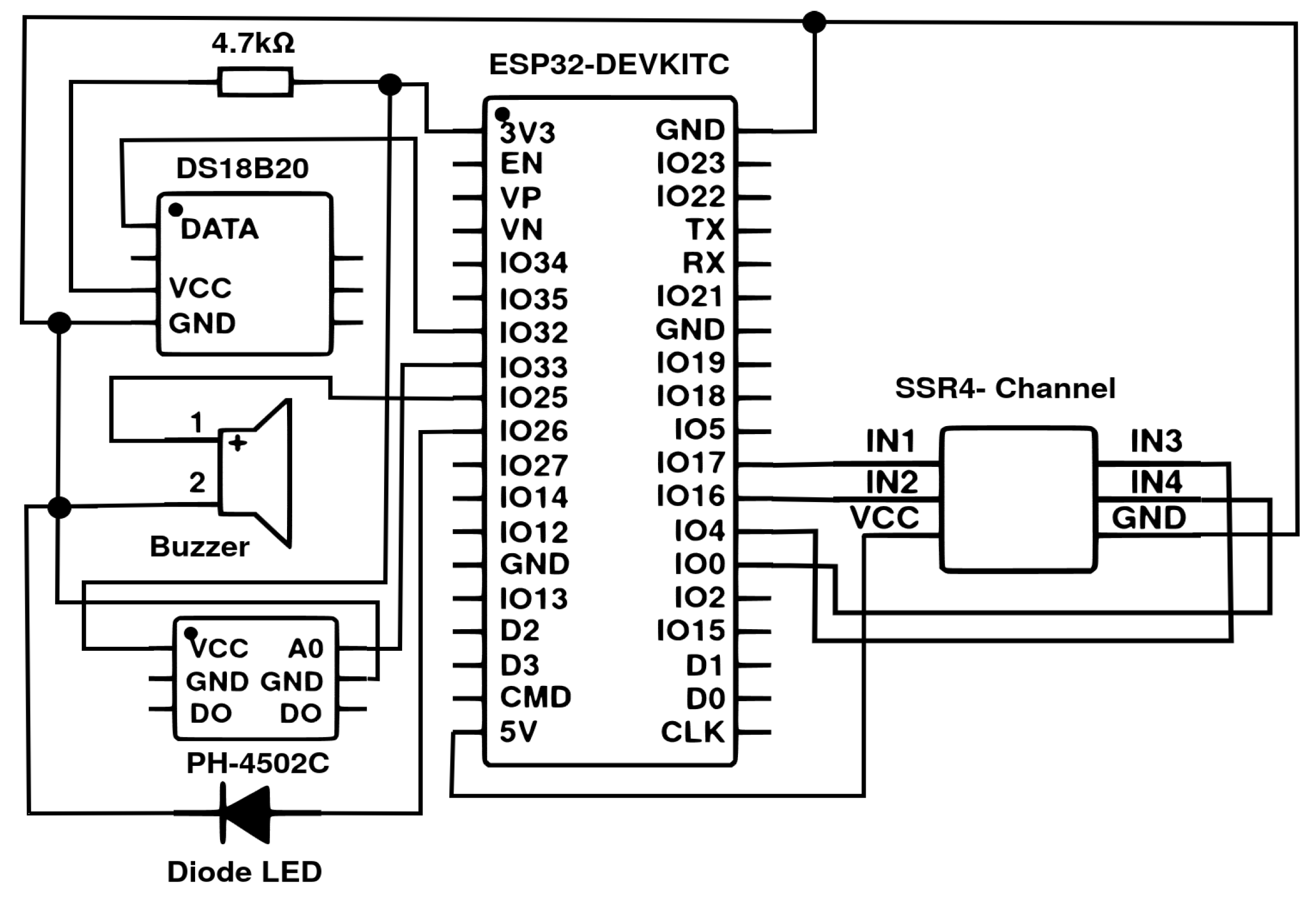
2.2. Framework Architecture
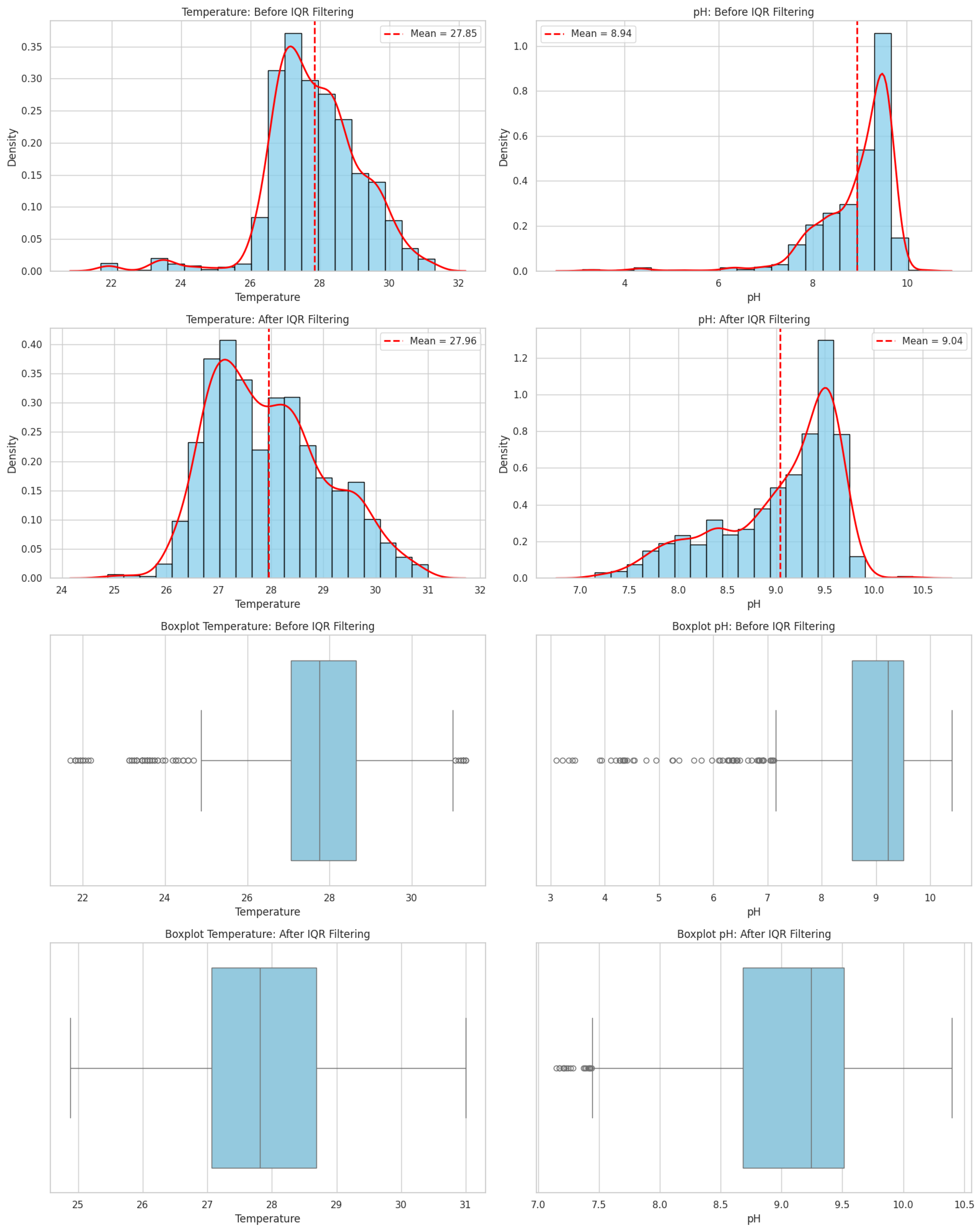
2.3. Data Acquisition and Preprocessing
2.4. Multimodal Feature Construction
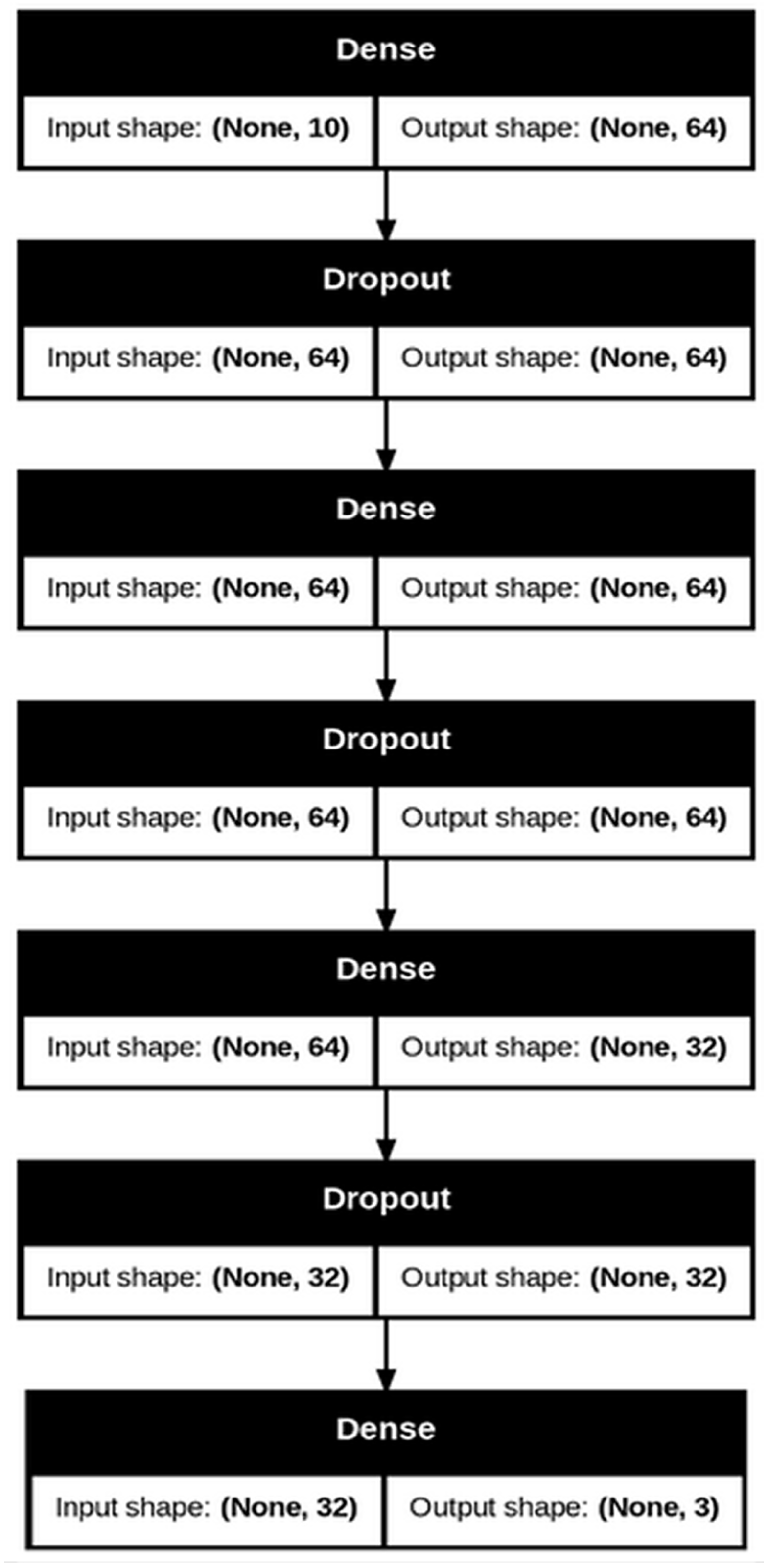
2.5. Neural Network Training
2.6. Implementation and Computing Environment
2.7. Robustness Validation
3. Results
3.1. Overall Classification Performance
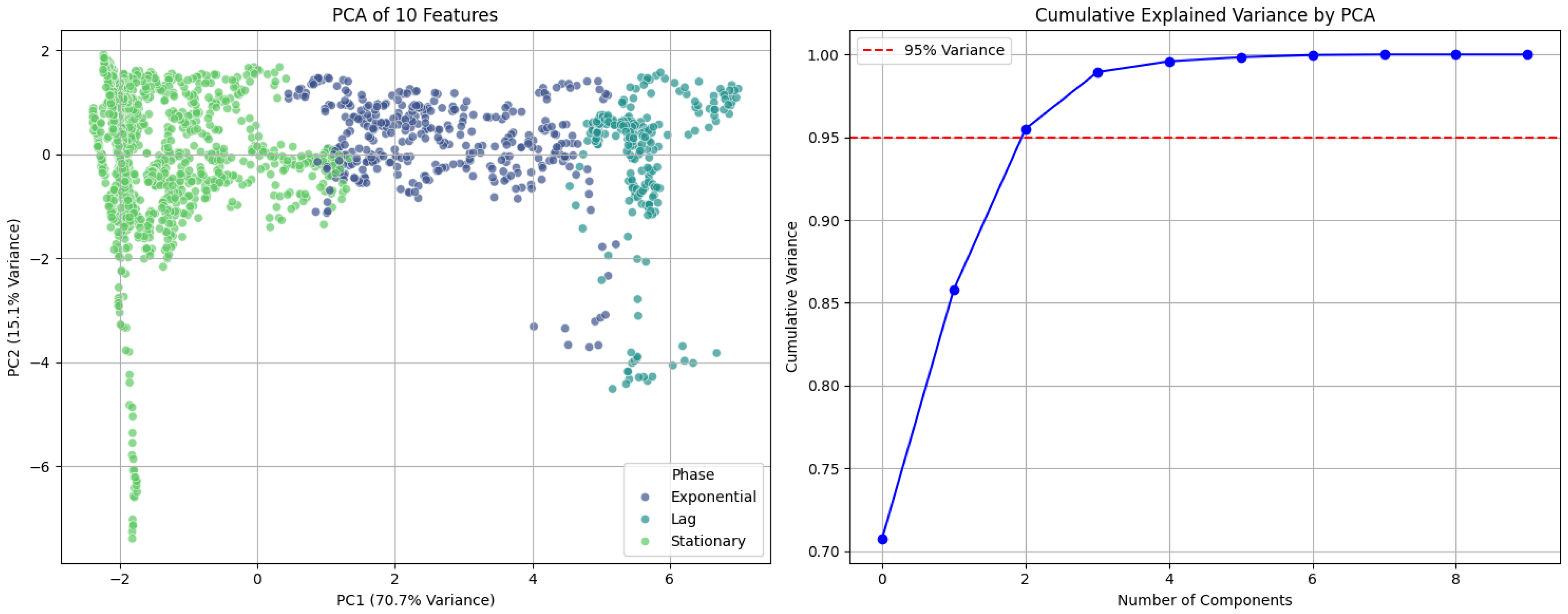
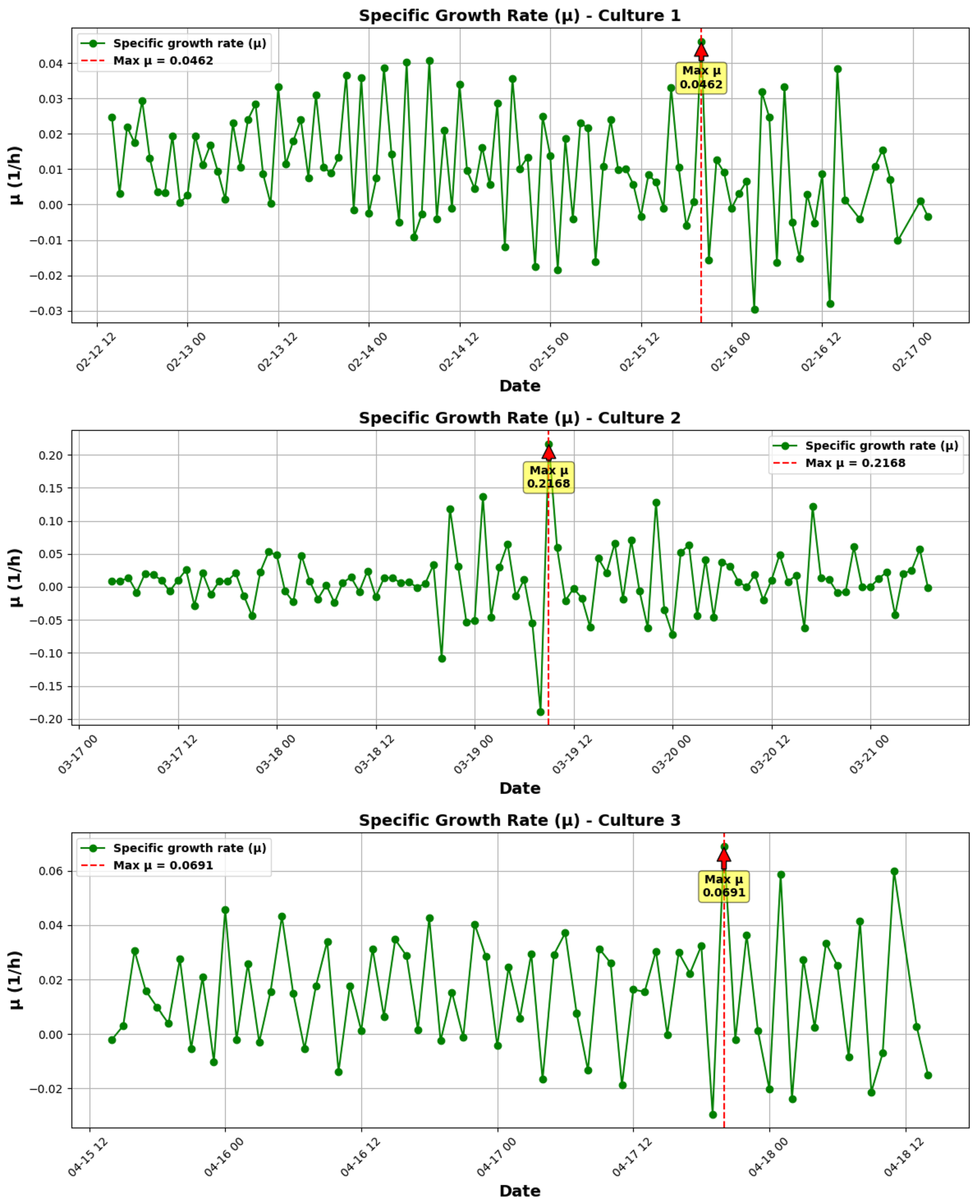
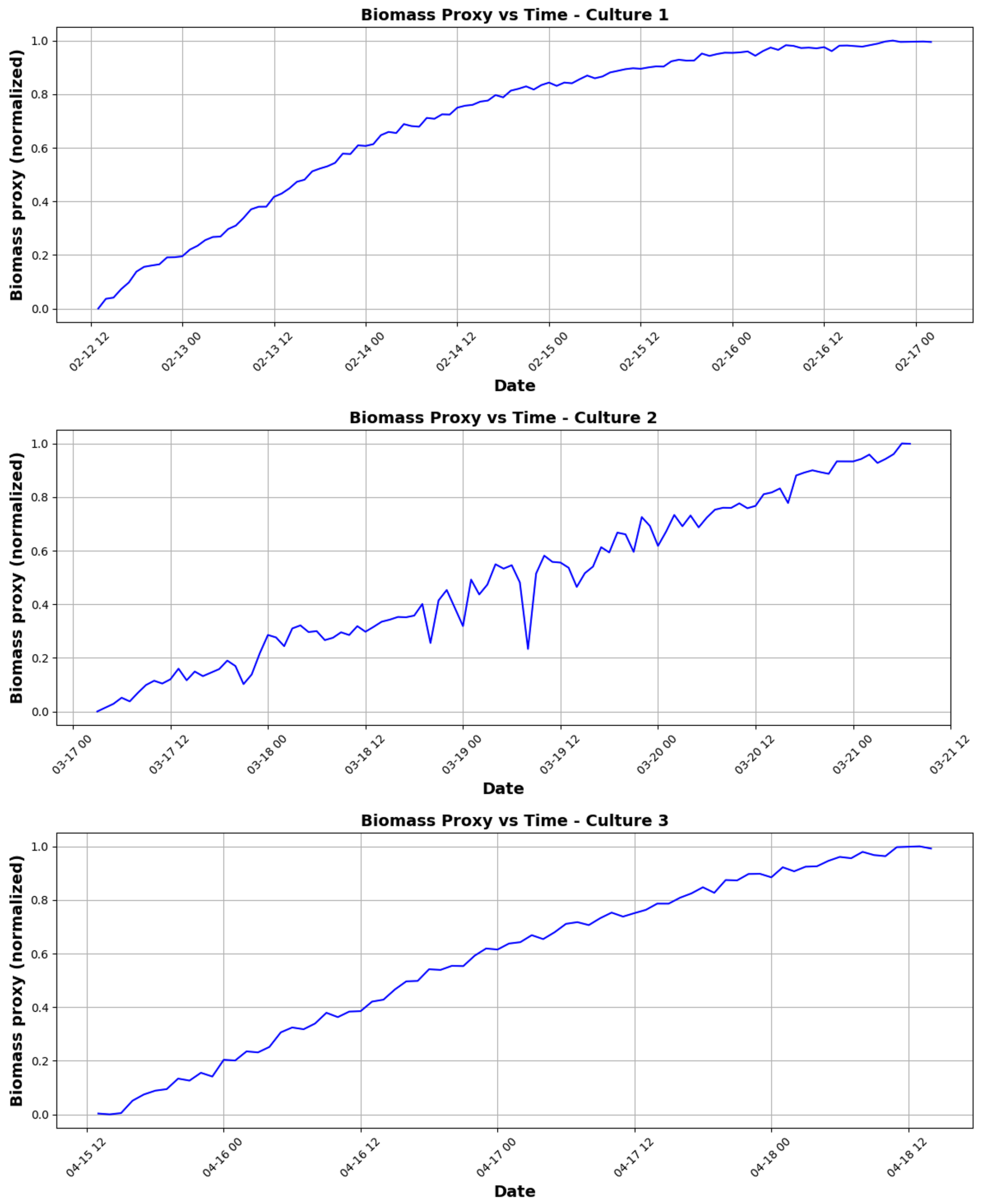
3.1.1. Growth Kinetics Analysis
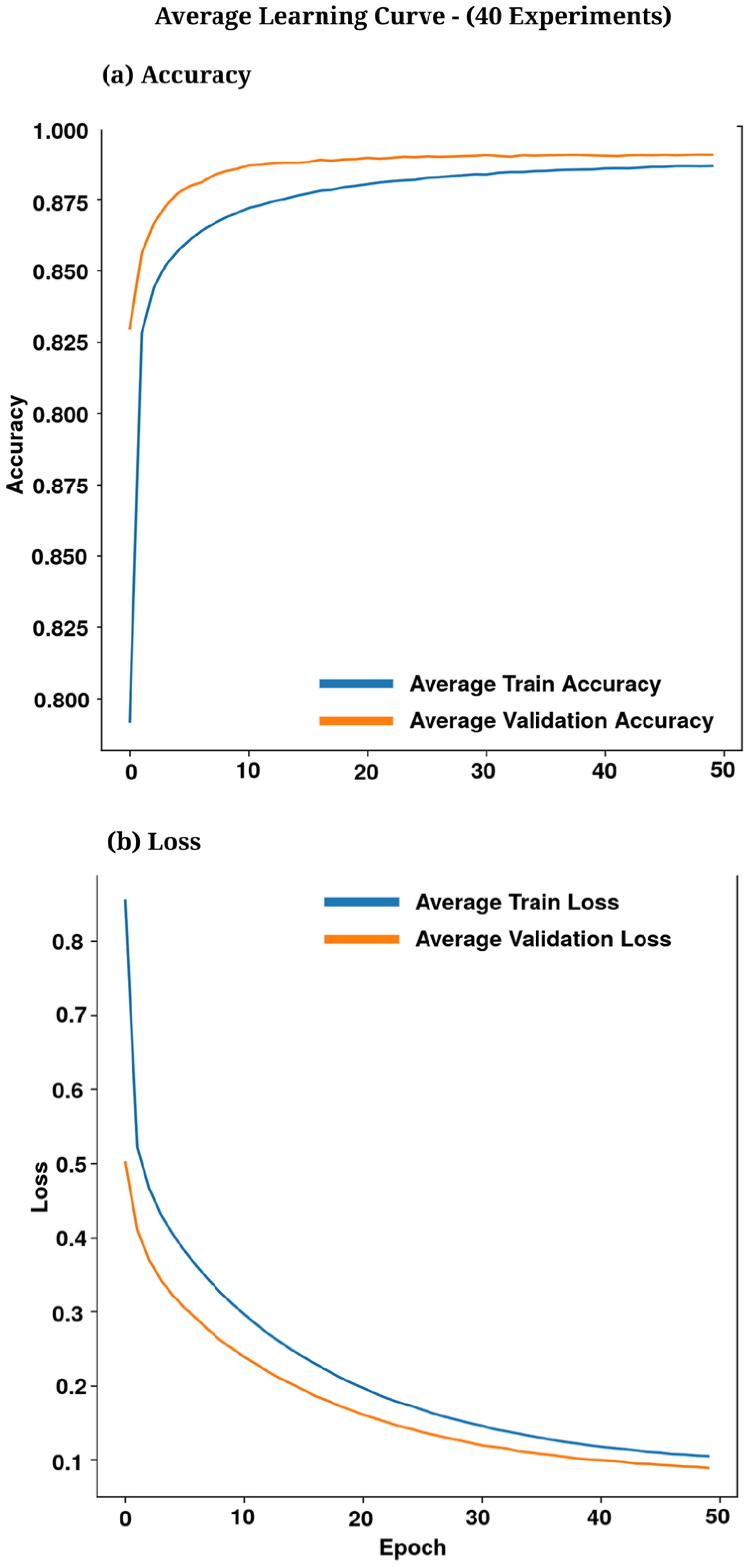
3.2. Training Curves
3.3. Confusion Matrix Analysis
3.4. Robustness Evaluation
3.5. Computational Performance
3.6. Comparison with Previous Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, Y.; Khan, T.M.; Song, Y.; Meijering, E. Edge Deep Learning in Computer Vision and Medical Diagnostics: A Comprehensive Survey. Artif. Intell. Rev. 2025, 58, 93. [Google Scholar] [CrossRef]
- Guo, Y.; Li, B.; Zhang, W.; Dong, W. Multi-Scale Image Edge Detection Based on Spatial-Frequency Domain Interactive Attention. Front. Neurorobot. 2025, 19, 1550939. [Google Scholar] [CrossRef]
- Wu, L.; Xiao, G.; Huang, D.; Zhang, X.; Ye, D.; Weng, H. Edge Computing-Based Machine Vision for Non-Invasive and Rapid Soft Sensing of Mushroom Liquid Strain Biomass. Agronomy 2025, 15, 242. [Google Scholar] [CrossRef]
- Dudek, P.; Richardson, T.; Bose, L.; Carey, S.; Chen, J.; Greatwood, C.; Mayol-Cuevas, W. Sensor-Level Computer Vision with Pixel Processor Arrays for Agile Robots. Sci. Robot. 2022, 7, eabl7755. [Google Scholar] [CrossRef] [PubMed]
- Mu, C.; Zheng, J.; Chen, C. Beyond Convolutional Neural Networks Computing: New Trends on ISSCC 2023 Machine Learning Chips. J. Semicond. 2023, 44, 050203. [Google Scholar] [CrossRef]
- Olague, G.; Köppen, M.; Cordón, O. Guest Editorial Special Issue on Evolutionary Computer Vision. IEEE Trans. Evol. Comput. 2023, 27, 2–4. [Google Scholar] [CrossRef]
- Kreß, F.; Sidorenko, V.; Schmidt, P.; Hoefer, J.; Hotfilter, T.; Walter, I.; Becker, J. CNNParted: An Open Source Framework for Efficient Convolutional Neural Network Inference Partitioning in Embedded Systems. Comput. Netw. 2023, 229, 109759. [Google Scholar] [CrossRef]
- Alajlan, N.N.; Ibrahim, D.M. TinyML: Enabling of Inference Deep Learning Models on Ultra-Low-Power IoT Edge Devices for AI Applications. Micromachines 2022, 13, 851. [Google Scholar] [CrossRef]
- Mishra, S.; Liu, Y.J.; Chen, C.S.; Yao, D.J. An Easily Accessible Microfluidic Chip for High-Throughput Microalgae Screening for Biofuel Production. Energies 2021, 14, 1817. [Google Scholar] [CrossRef]
- Sunoj, S.; Hammed, A.; Igathinathane, C.; Eshkabilov, S.; Simsek, H. Identification, Quantification, and Growth Profiling of Eight Different Microalgae Species Using Image Analysis. Algal Res. 2021, 60, 102487. [Google Scholar] [CrossRef]
- Xu, J.; Li, Y.; Zhang, X.; Wang, Y.; Liu, J. Automated Classification of Microalgae Using Deep Learning and Spectral Imaging. Algal Res. 2021, 58, 102366. [Google Scholar] [CrossRef]
- Zhou, S.; Jiang, J.; Hong, X.; Fu, P.; Yan, H. Vision Meets Algae: A Novel Way for Microalgae Recognization and Health Monitor. Front. Mar. Sci. 2023, 10, 1105545. [Google Scholar] [CrossRef]
- Abdullah; Ali, S.; Khan, Z.; Hussain, A.; Athar, A.; Kim, H.C. Computer Vision Based Deep Learning Approach for the Detection and Classification of Algae Species Using Microscopic Images. Water 2022, 14, 2219. [Google Scholar] [CrossRef]
- Ilniyaz, O.; Du, Q.; Shen, H.; He, W.; Feng, L.; Azadi, H.; Kurban, A.; Chen, X. Leaf Area Index Estimation of Pergola-Trained Vineyards in Arid Regions Using Classical and Deep Learning Methods Based on UAV-Based RGB Images. Comput. Electron. Agric. 2023, 207, 107723. [Google Scholar] [CrossRef]
- Cao, M.; Wang, J.; Chen, Y.; Wang, Y. Detection of Microalgae Objects Based on the Improved YOLOv3 Model. Environ. Sci. Process. Impacts 2021, 23, 1516–1530. [Google Scholar] [CrossRef]
- Xu, K.; Shu, L.; Xie, Q.; Song, M.; Zhu, Y.; Cao, W.; Ni, J. Precision Weed Detection in Wheat Fields for Agriculture 4.0: A Survey of Enabling Technologies, Methods, and Research Challenges. Comput. Electron. Agric. 2023, 212, 108106. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, H.; Forsberg, E.; Hou, S.; Li, S.; Xu, Z.; Chen, X.; Sun, X.; He, S. Confocal Hyperspectral Microscopic Imager for the Detection and Classification of Individual Microalgae. Opt. Express 2021, 29, 37281–37301. [Google Scholar] [CrossRef]
- Solovchenko, A. Seeing Good and Bad: Optical Sensing of Microalgal Culture Condition. Algal Res. 2023, 71, 103071. [Google Scholar] [CrossRef]
- Liu, L.; Wang, L.; Ma, Z. Improved Lightweight YOLOv5 Based on ShuffleNet and Its Application on Traffic Signs Detection. PLoS ONE 2024, 19, e0310269. [Google Scholar] [CrossRef]
- Xie, Y.; Zhao, Y. Lightweight Improved YOLOv5 Algorithm for PCB Defect Detection. J. Supercomput. 2025, 81, 261. [Google Scholar] [CrossRef]
- Dantas, P.V.; Silva, W.S., Jr.; Cordeiro, L.C.; Carvalho, C.B. A Comprehensive Review of Model Compression Techniques in Machine Learning. Appl. Intell. 2024, 54, 11804–11844. [Google Scholar] [CrossRef]
- Lopes, A.; Santos, F.P.; Oliveira, D.; Schiezaro, M.; Pedrini, H. Computer Vision Model Compression Techniques for Embedded Systems: A Survey. Comput. Graph. 2024, 123, 104015. [Google Scholar] [CrossRef]
- Thottempudi, P.; Jambek, A.B.B.; Kumar, V.; Acharya, B.; Moreira, F. Resilient Object Detection for Autonomous Vehicles: Integrating Deep Learning and Sensor Fusion in Adverse Conditions. Eng. Appl. Artif. Intell. 2025, 151, 110563. [Google Scholar] [CrossRef]
- Xiao, T.; Xu, T.; Wang, G. Real-Time Detection of Track Fasteners Based on Object Detection and FPGA. Microprocess. Microsyst. 2023, 100, 104863. [Google Scholar] [CrossRef]
- Wang, H.; Xu, S.; Chen, Y.; Su, C. LFD-YOLO: A Lightweight Fall Detection Network with Enhanced Feature Extraction and Fusion. Sci. Rep. 2025, 15, 5069. [Google Scholar] [CrossRef]
- Yoo, J.; Ban, G. Efficient Deep Learning Model Compression for Sensor-Based Vision Systems via Outlier-Aware Quantization. Sensors 2025, 25, 2918. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhong, B.; Jin, G.; Shen, T.; Tan, L.; Li, N.; Zheng, Y. Avltrack: Dynamic Sparse Learning for Aerial Vision-Language Tracking. IEEE Trans. Circuits Syst. Video Technol. 2025, 35, 7554–7567. [Google Scholar] [CrossRef]
- Oks, S.J.; Zöllner, S.; Jalowski, M.; Fuchs, J.; Möslein, K.M. Embedded Vision Device Integration via OPC UA: Design and Evaluation of a Neural Network-Based Monitoring System for Industry 4.0. Procedia CIRP 2021, 100, 43–48. [Google Scholar] [CrossRef]
- Juchem, J.; De Roeck, M.; Loccufier, M. Low-Cost Vision-Based Embedded Control of a 2DOF Robotic Manipulator. IFAC-PapersOnLine 2023, 56, 8833–8838. [Google Scholar] [CrossRef]
- Lemsalu, M.; Bloch, V.; Backman, J.; Pastell, M. Real-Time CNN-Based Computer Vision System for Open-Field Strawberry Harvesting Robot. IFAC-PapersOnLine 2022, 55, 24–29. [Google Scholar] [CrossRef]
- Razali, M.N.; Arbaiy, N.; Lin, P.-C.; Ismail, S. Optimizing Multiclass Classification Using Convolutional Neural Networks with Class Weights and Early Stopping for Imbalanced Datasets. Electronics 2025, 14, 705. [Google Scholar] [CrossRef]
- Macias-Jamaica, R.E.; Castrejón-González, E.O.; Rico-Ramírez, V.; Guillen-Almaraz, X.; Maldonado-Pedroza, C.; Rodríguez-Peña, M.P. Wastewater Treatment Using Constructed Wetlands withForced Flotation: Enhancing Phytoremediation through a Floating Typha latifolia Rhizosphere. Int. J. Environ. Sci. Dev. 2025, 16, 136–145. [Google Scholar] [CrossRef]
- Yadav, D.P.; Jalal, A.S.; Garlapati, D.; Hossain, K.; Goyal, A.; Pant, G. Deep Learning-Based ResNeXt Model in Phycological Studies for Future. Algal Res. 2020, 50, 102018. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, H.; Li, Y.; Wang, D.; Lin, Y.; Li, Z.; Xie, T. Investigation on the Photocatalytic Hydrogen Evolution Properties of Z-Scheme Au NPs/CuInS2/NCN-CNx Composite Photocatalysts. ACS Sustain. Chem. Eng. 2021, 9, 7286–7297. [Google Scholar] [CrossRef]
- Aquino, A.U.; Bautista, M.V.L.; Diaz, C.H.; Valenzuela, I.C.; Dadios, E.P. A Vision-Based Closed Spirulina (A. Platensis) Cultivation System with Growth Monitoring Using Artificial Neural Network. In Proceedings of the 2018 IEEE 10th International Conference on Humanoid, Nanotechnology, Information Technology, Communication and Control, Environment and Management (HNICEM), Baguio City, Philippines, 29 November–2 December 2018. [Google Scholar] [CrossRef]
- Chang, H.-X.; Huang, Y.; Fu, Q.; Liao, Q.; Zhu, X. Kinetic Characteristics and Modeling of Microalgae Chlorella vulgaris Growth and CO2 Biofixation Considering the Coupled Effects of Light Intensity and Dissolved Inorganic Carbon. Bioresour. Technol. 2016, 206, 231–238. [Google Scholar] [CrossRef]
- Ma, Y. Classification of Bacterial Motility Using Machine Learning. Master’s Thesis, University of Tennessee, Knoxville, TN, USA, 2020. Available online: https://trace.tennessee.edu/utk_gradthes/6258 (accessed on 3 June 2025).
- Xu, L.; Chen, Y.; Zhang, Y.; Xu, L.; Yang, J. Accurate Classification of Algae Using Deep Convolutional Neural Network with a Small Database. ACS EST Water 2022, 2, 1921–1928. [Google Scholar] [CrossRef]
- Correa, I.; Drews, P., Jr.; Botelho, S.; de Souza, M.S.; Tavano, V.M. Deep Learning for Microalgae Classification. In Proceedings of the 2017 16th IEEE International Conference on Machine Learning and Applications, Cancun, Mexico, 18–21 December 2017. [Google Scholar] [CrossRef]
- Pardeshi, R.; Deshmukh, P.D. Classification of Microscopic Algae: An Observational Study with AlexNet. In Soft Computing and Signal Processing; Reddy, V., Prasad, V., Wang, J., Reddy, K., Eds.; Springer: Singapore, 2020; Volume 1118, pp. 309–316. [Google Scholar] [CrossRef]
- Meenatchi Sundaram, K.; Sravan Kumar, S.; Deshpande, A.; Chinnadurai, S.; Rajendran, K. Machine Learning Assisted Image Analysis for Microalgae Prediction. ACS EST Eng. 2025, 5, 541–550. [Google Scholar] [CrossRef]
- Kanwal, S.; Khan, F.; Alamri, S.A. A Multimodal Deep Learning Infused with Artificial Algae Algorithm—An Architecture of Advanced E-Health System for Cancer Prognosis Prediction. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 2707–2719. [Google Scholar] [CrossRef]
- Kim, S.; Sosnowski, K.; Hwang, D.S.; Yoon, J.-Y. Smartphone-Based Microalgae Monitoring Platform Using Machine Learning. ACS EST Eng. 2023, 3, 186–195. [Google Scholar] [CrossRef]
- Chong, J.W.R.; Khoo, K.S.; Chew, K.W.; Ting, H.-Y.; Show, P.L. Trends in Digital Image Processing of Isolated Microalgae by Incorporating Classification Algorithm. Biotechnol. Adv. 2023, 63, 108095. [Google Scholar] [CrossRef]
- Madkour, D.M.; Shapiai, M.I.; Mohamad, S.E.; Aly, H.H.; Ismail, Z.H.; Ibrahim, M.Z. A Systematic Review of Deep Learning Microalgae Classification and Detection. IEEE Access 2023, 11, 57529–57555. [Google Scholar] [CrossRef]
- Ning, H.; Li, R.; Zhou, T. Machine Learning for Microalgae Detection and Utilization. Front. Mar. Sci. 2022, 9, 947394. [Google Scholar] [CrossRef]
- Pyo, J.; Duan, H.; Baek, S.; Kim, M.S.; Jeon, T.; Kwon, Y.S.; Lee, H.; Cho, K.H. A Convolutional Neural Network Regression for Quantifying Cyanobacteria Using Hyperspectral Imagery. Remote Sens. Environ. 2019, 233, 111350. [Google Scholar] [CrossRef]
- Macias-Jamaica, R.E.; Castrejón-González, E.O.; González-Alatorre, G.; Alvarado, J.F.J.; Díaz-Ovalle, C.O. Molecular models for sodium dodecyl sulphate in aqueous solution to reduce the micelle time formation in molecular simulation. J. Mol. Liq. 2019, 274, 90–97. [Google Scholar] [CrossRef]



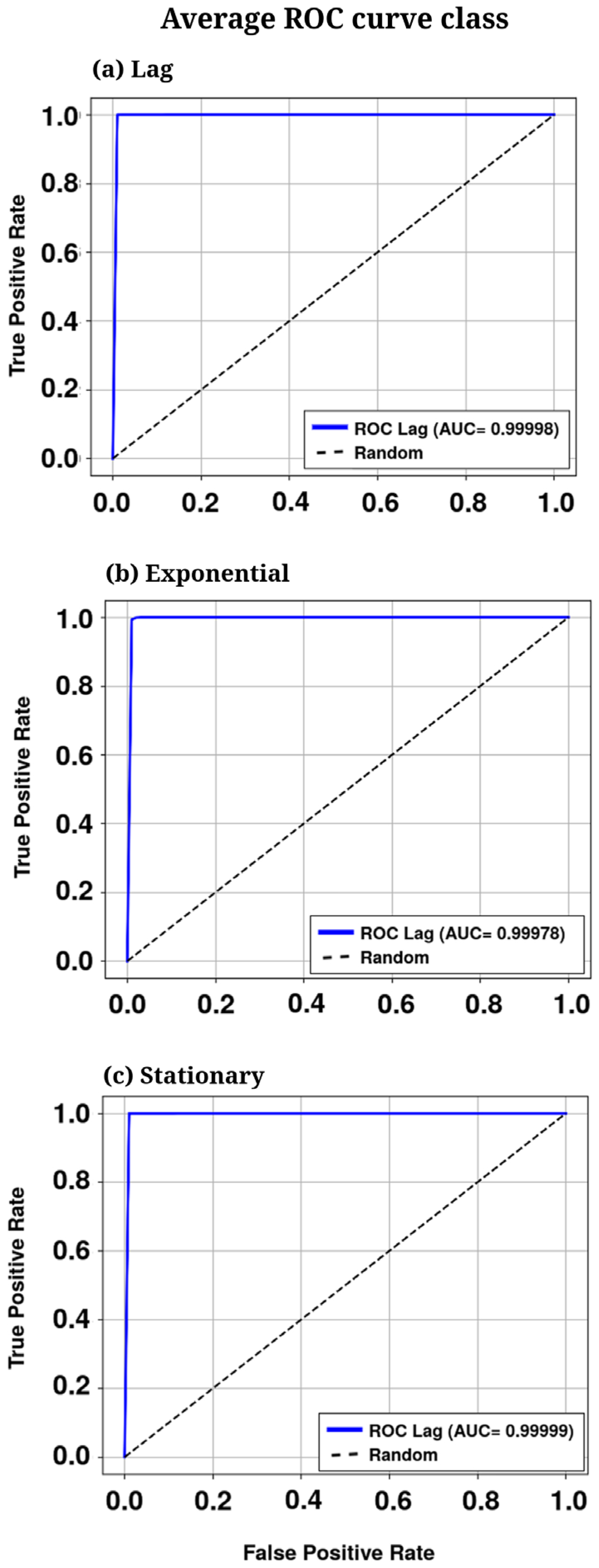
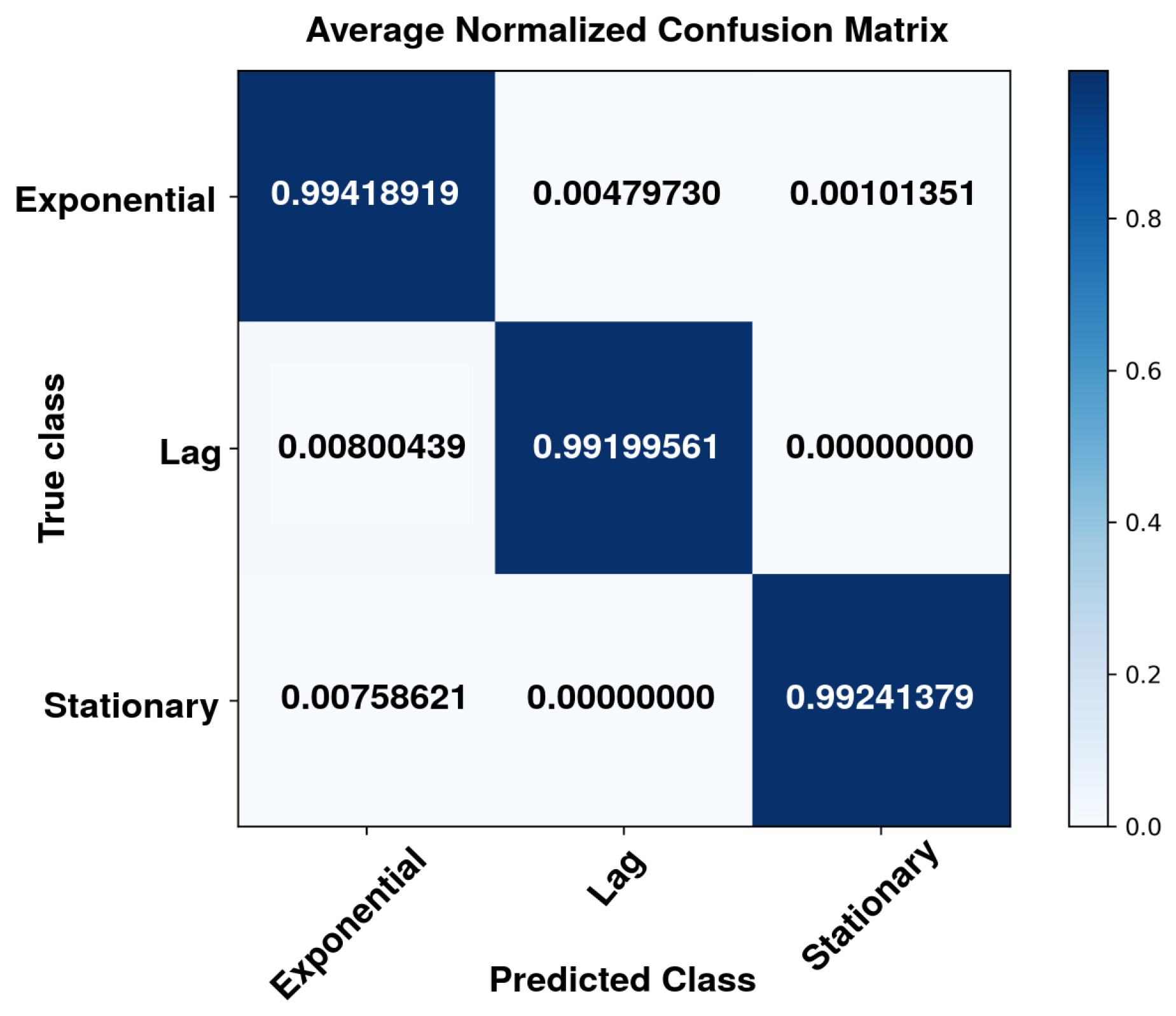
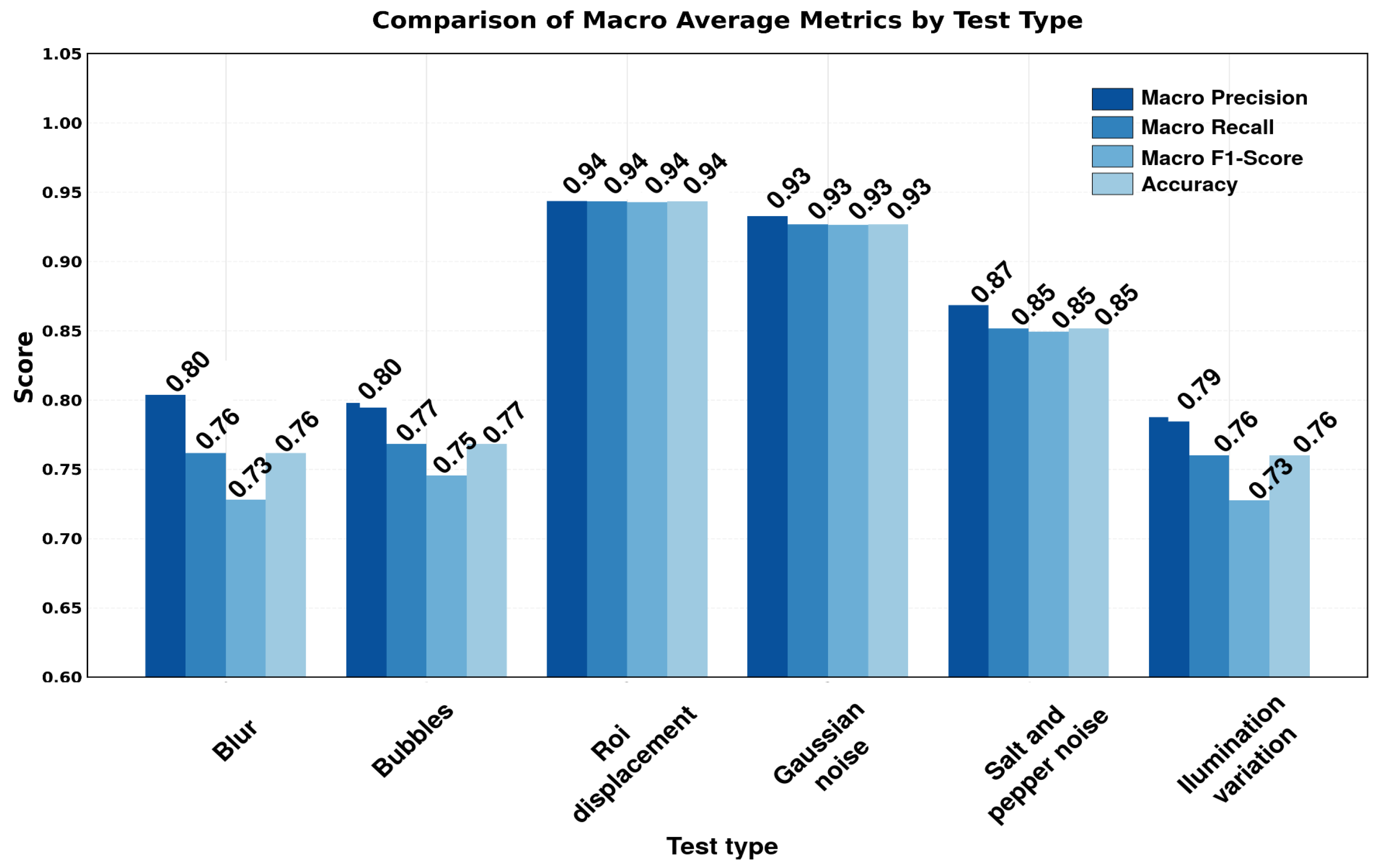

| No. | Feature | Symbol | Observed Range* | Normalization Method | Normalized Range |
|---|---|---|---|---|---|
| 1 | Mean red intensity | 1.8–249.9 (8-bit) | Z-score (μ = 32.2, σ = 62.9) | −0.4–+3.4 | |
| 2 | Mean green intensity | 1.8–252.9 (8-bit) | Z-score (μ = 64.8, σ = 84.4) | −0.7–+2.2 | |
| 3 | Mean blue intensity | 1.6–251.7 (8-bit) | Z-score (μ = 45.6, σ = 69.7) | −0.6–+2.9 | |
| 4 | Mean perceptual luminance | 1.8–251.2 (8-bit) | Z-score (μ = 55.3, σ = 76.6) | −0.7–+2.5 | |
| 5 | Red channel std. dev. | σR | 0.3–37.5 (8-bit) | Min–max | 0–1 |
| 6 | Green channel std. dev. | σG | 0.3–39.3 (8-bit) | Min–max | 0–1 |
| 7 | Blue channel std. dev. | σB | 0.2–43.0 (8-bit) | Min–max | 0–1 |
| 8 | Luminance std. dev. | σI | 0.2–35.9 (8-bit) | Min–max | 0–1 |
| 9 | Mean pH | pHmean | 7.20–10.40 | Min–max | 0–1 |
| 10 | Mean temperature (°C) | Tmean | 21.69–31.31 | Min–max | 0–1 |
| Metric | Lag Phase | Exponential Phase | Stationary Phase | Macro Average |
|---|---|---|---|---|
| Accuracy | 0.9923 | 0.9665 | 0.9997 | 0.9862 |
| Recall | 0.9920 | 0.9941 | 0.9924 | 0.9928 |
| F1-score | 0.9920 | 0.9799 | 0.9960 | 0.9893 |
| AUC-ROC | 0.9999 | 0.9997 | 0.9999 | 0.9999 |
| Metric | Mean Value | Standard Deviation |
|---|---|---|
| CPU Usage (%) | 22.4 | 1.2 |
| RAM Consumption (MB) | 2450 | 950 |
| Training Time per Epoch (s) | 9.8 | 1.5 |
| Inference Latency (ms) | 134 | 5 |
| Metric | Mean Value | Description |
|---|---|---|
| Model size | 13.48 KB | 8-bit quantized model (TFLite Micro format) |
| Processor frequency | 240 MHz | ESP32 dual-core (Xtensa LX6, no operating system) |
| Inference latency | 4.8 ms | Average time per sample (measured directly on the embedded environment) |
| Speed-up vs. PC environment | ×28 | Compared to single-sample validation inference (134 ms on PC) |
| Tensor arena usage | 2 KB | Memory allocated for internal model tensors |
| Estimated total RAM usage | <25 KB | Includes model, tensors, stack, and auxiliary buffers |
| Available memory (SRAM) | ~520 KB | On standard ESP32 versions (without external PSRAM) |
| Estimated CPU usage | ~24% | Assuming 50 inferences per second (continuous operation) |
| Model accuracy | 98% | Maintained after quantization and deployment on embedded device |
| Study | Domain/Task | Model and Platform | Accuracy/mAP | Embedded Inference | Inference Latency | Δ Accuracy vs. Ours |
|---|---|---|---|---|---|---|
| This study | Microalgae growth-phase classification | Dense network—ESP32 (TFL-Micro) | 0.9862 | Yes | 4.8 ms | — |
| [41] | Optical density prediction from images | DNN—PC | 0.9600* | No | NA | +2.6 pp |
| [44] | Morphological classification via microscopy | CNN/SVM—PC | 0.9500* | No | NA | +3.6 pp |
| [45] | Systematic review: algae detection | Various (CNN, SVM)—PC | 0.9997* (máximo) | No | NA | –1.3 pp |
| [42] | Cancer prognosis via algae algorithm | CNN-XGBoost—PC | 0.9900 | No | NA | –0.4 pp |
| [38] | Algae species classification (13 classes) | CNN + SENet—PC | 0.9390 | No | NA | +4.7 pp |
| [46] | Detection/growth/utilization | CNN, RF, SVM—PC | 0.9200* | No | NA | +6.2 pp |
| [33] | Algal taxonomy (16 genera) | ResNeXt—GPU | 0.9997 | No | NA | –1.3 pp |
| [35] | Growth monitoring (Spirulina) | ANN—Arduino | 0.9522* | No | NA | +3.4 pp |
| [40] | Morphological classification | AlexNet—PC | 0.9600 | No | NA | +2.6 pp |
| [47] | Cyanobacteria quantification (PC/Chl-a) | PRCNN—PC (hyperspectral) | 0.8600* | No | NA | +12.6 pp |
| [39] | FlowCAM-based algae classification | CNN—PC | 0.8859 | No | NA | +10.3 pp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez-Ramírez, J.J.; Macias-Jamaica, R.E.; Zamudio-Rodríguez, V.M.; Sotelo, H.A.; Velázquez-Vázquez, D.A.; de Anda-Suárez, J.; Gutiérrez-Hernández, D.A. A Modular Framework for RGB Image Processing and Real-Time Neural Inference: A Case Study in Microalgae Culture Monitoring. Eng 2025, 6, 221. https://doi.org/10.3390/eng6090221
Gutiérrez-Ramírez JJ, Macias-Jamaica RE, Zamudio-Rodríguez VM, Sotelo HA, Velázquez-Vázquez DA, de Anda-Suárez J, Gutiérrez-Hernández DA. A Modular Framework for RGB Image Processing and Real-Time Neural Inference: A Case Study in Microalgae Culture Monitoring. Eng. 2025; 6(9):221. https://doi.org/10.3390/eng6090221
Chicago/Turabian StyleGutiérrez-Ramírez, José Javier, Ricardo Enrique Macias-Jamaica, Víctor Manuel Zamudio-Rodríguez, Héctor Arellano Sotelo, Dulce Aurora Velázquez-Vázquez, Juan de Anda-Suárez, and David Asael Gutiérrez-Hernández. 2025. "A Modular Framework for RGB Image Processing and Real-Time Neural Inference: A Case Study in Microalgae Culture Monitoring" Eng 6, no. 9: 221. https://doi.org/10.3390/eng6090221
APA StyleGutiérrez-Ramírez, J. J., Macias-Jamaica, R. E., Zamudio-Rodríguez, V. M., Sotelo, H. A., Velázquez-Vázquez, D. A., de Anda-Suárez, J., & Gutiérrez-Hernández, D. A. (2025). A Modular Framework for RGB Image Processing and Real-Time Neural Inference: A Case Study in Microalgae Culture Monitoring. Eng, 6(9), 221. https://doi.org/10.3390/eng6090221










