Process Engineering of the Acetone-Ethanol-Butanol (ABE) Fermentation in a Linear and Feedback Loop Cascade of Continuous Stirred Tank Reactors: Experiments, Modeling and Optimization
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganism and Medium
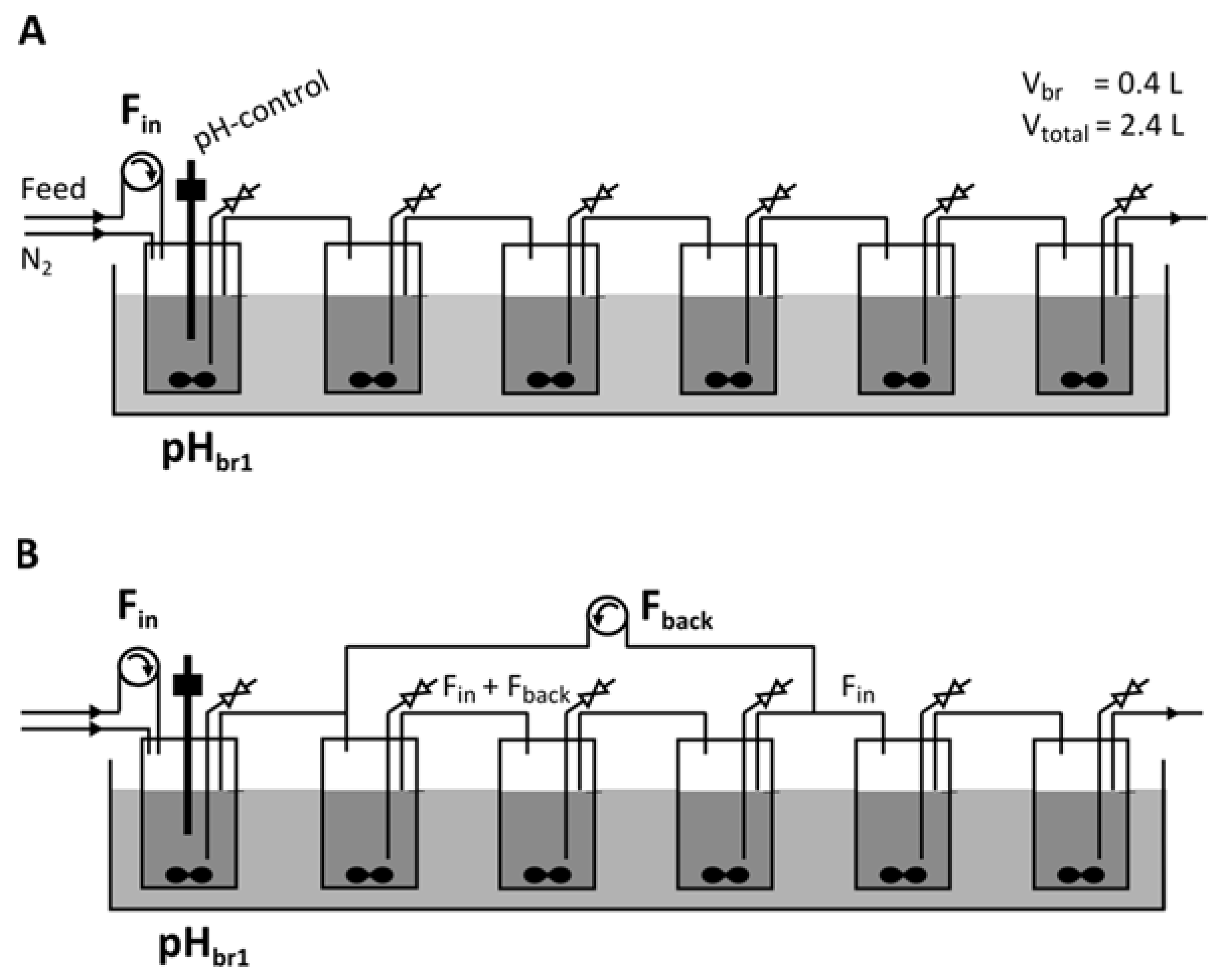
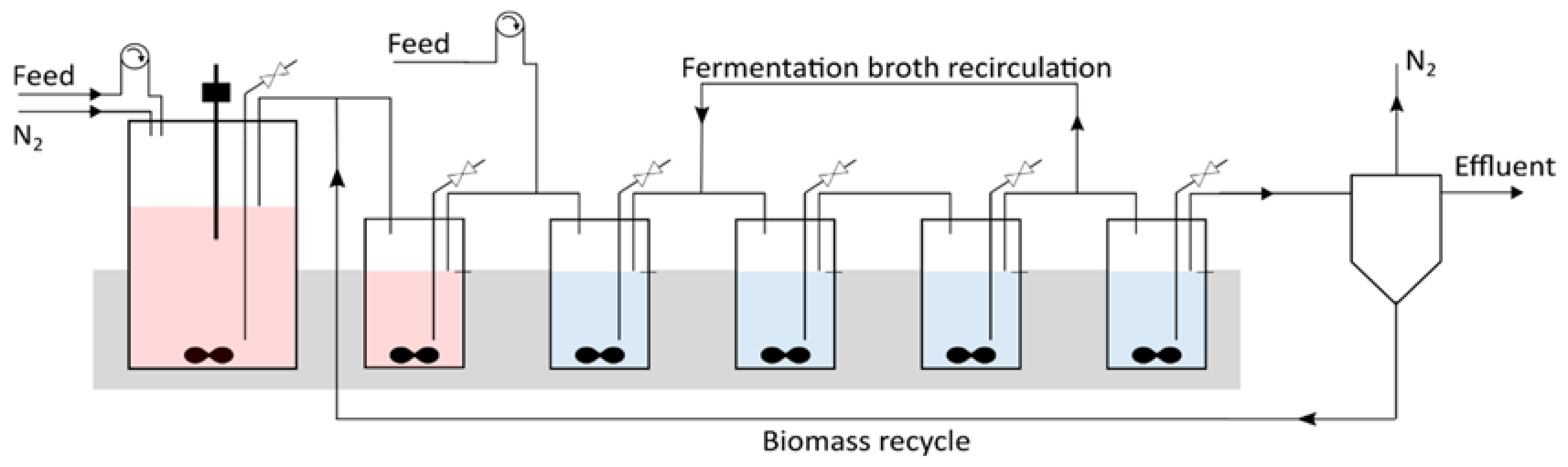
2.2. Continuous Fermentation in a Six-Stage Bioreactor Cascade
- -
- Residence time distribution approximates plug flow behavior, minimizing backmixing.
- -
- Total residence time is in the order of the duration of a batch fermentation (up to 36 h [16]).
- -
- Residence time in single reactors allows for resolution of temporal separation of metabolic phases (minimum duration of a metabolic phase approximately 6 h [16]).
2.3. Analytical Procedures
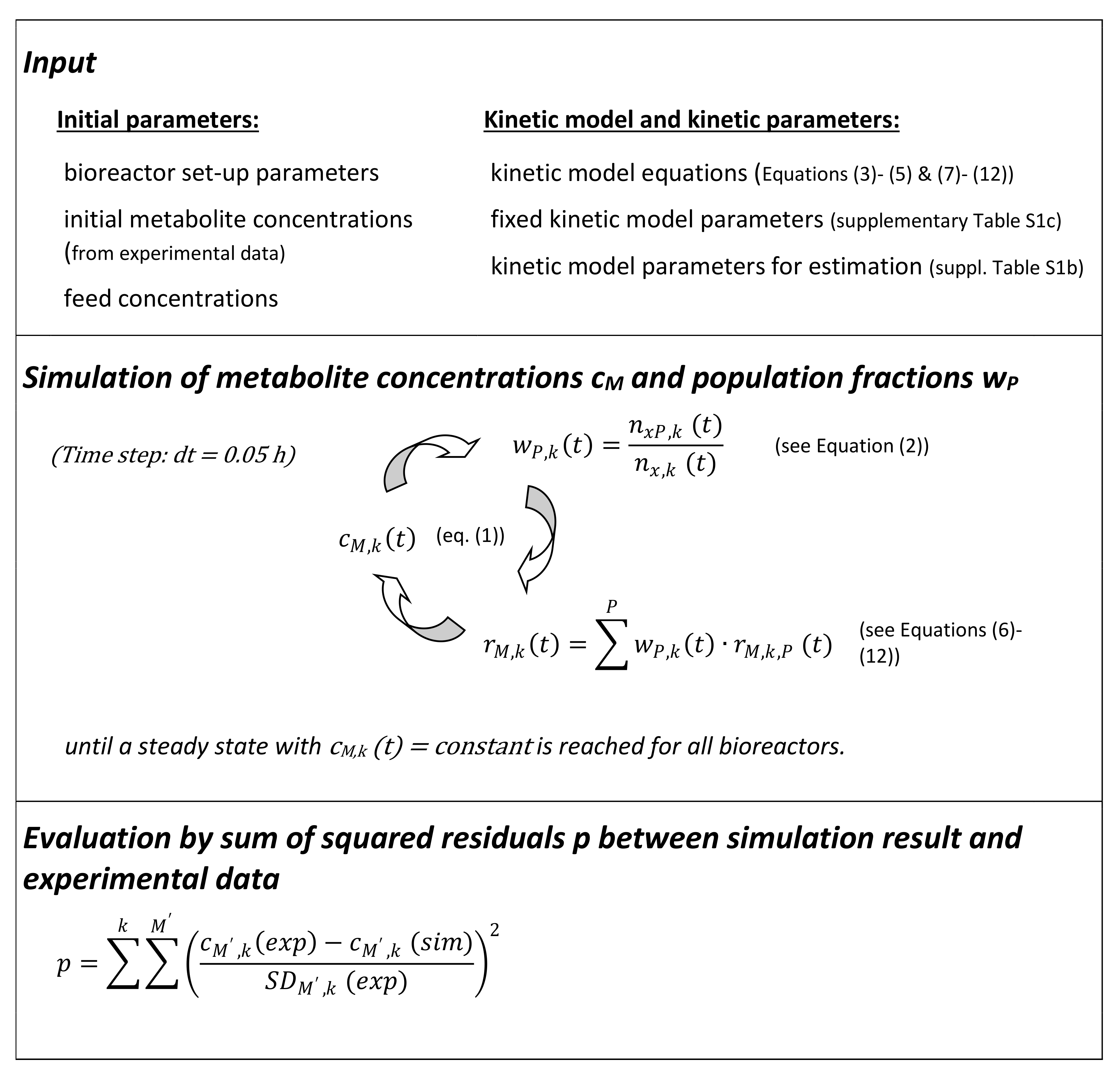
2.4. Mathematical Modeling Approach
3. Results
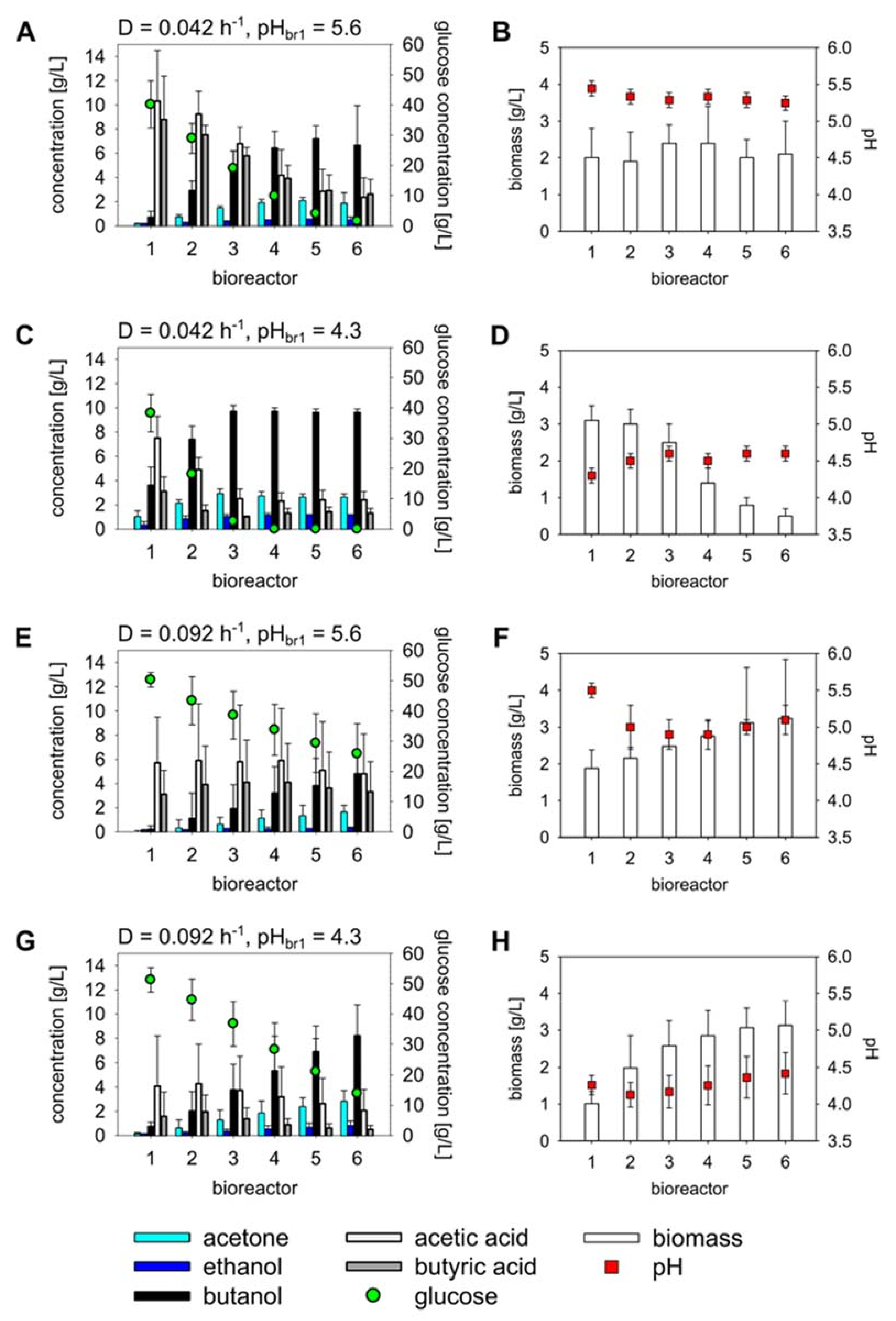
3.1. Continuous Fermentations in a Linear CCSTR under Four Different Operating Conditions
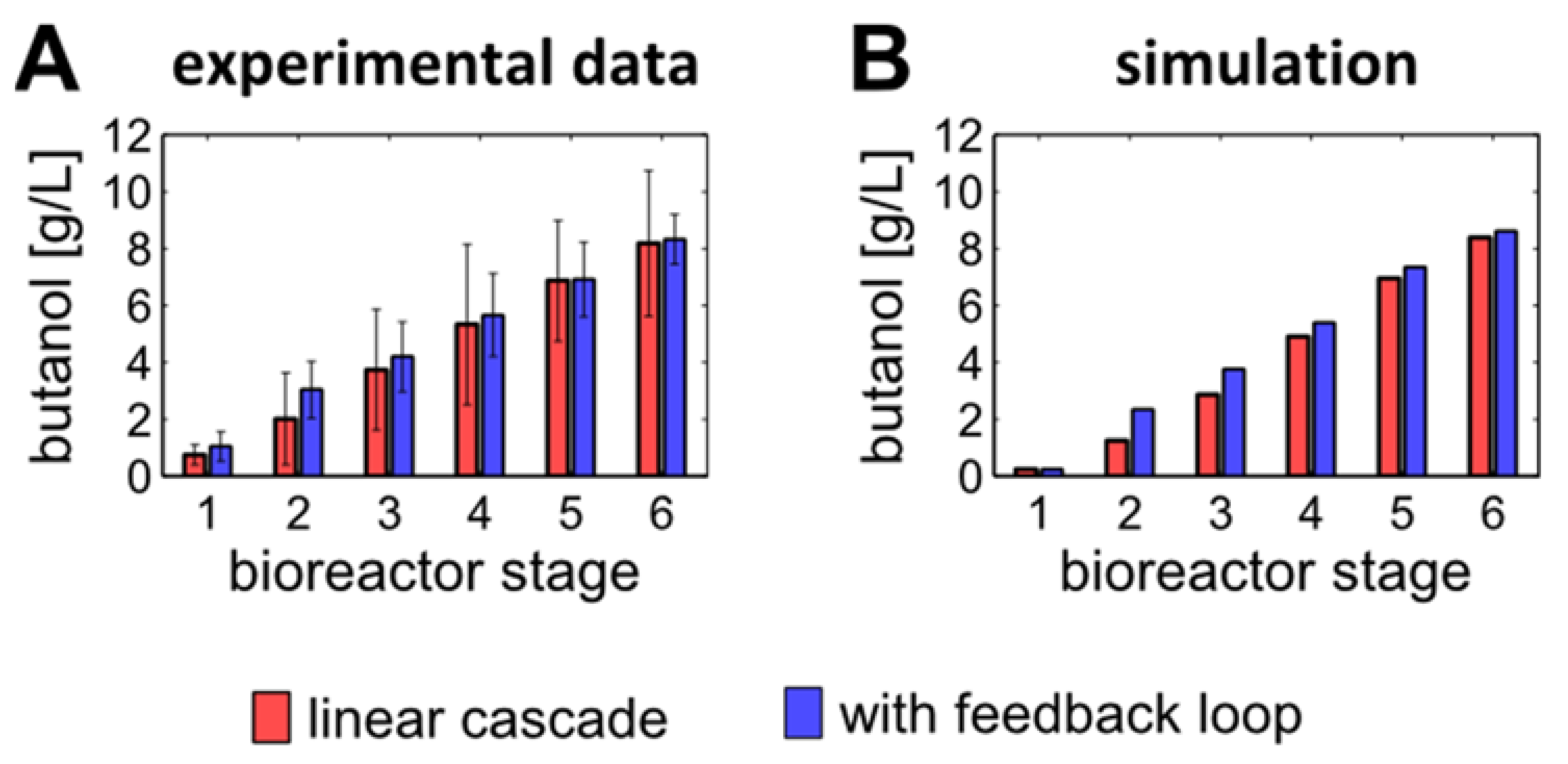
3.2. Estimation and Validation of the Mathematical Model Parameters Based on the Experimental Data from the Linear Cascade
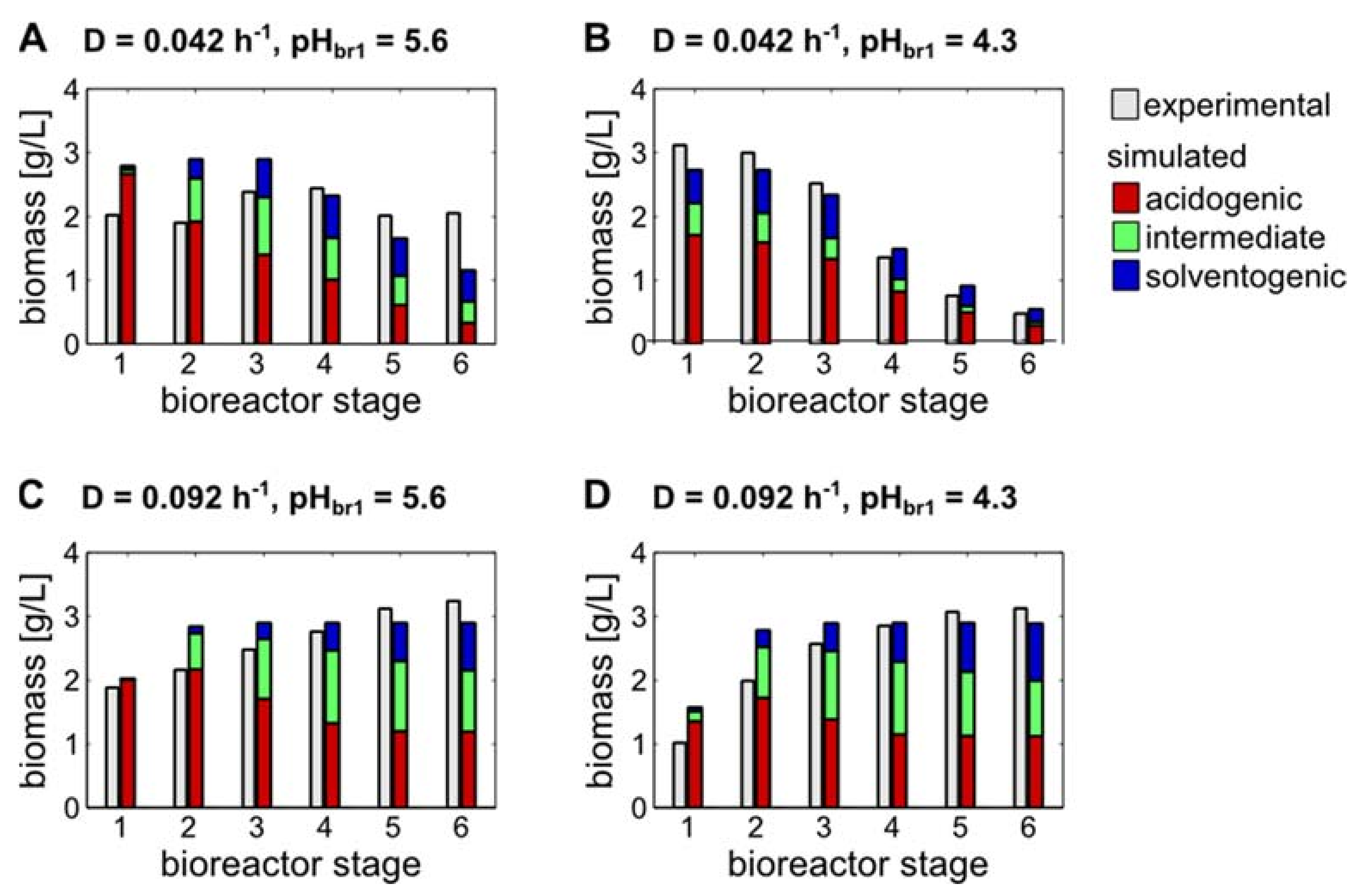
3.3. Model-Based Predictions of Biomass Subpopulations Regarding Acidogenic, Intermediate and Solventogenic States
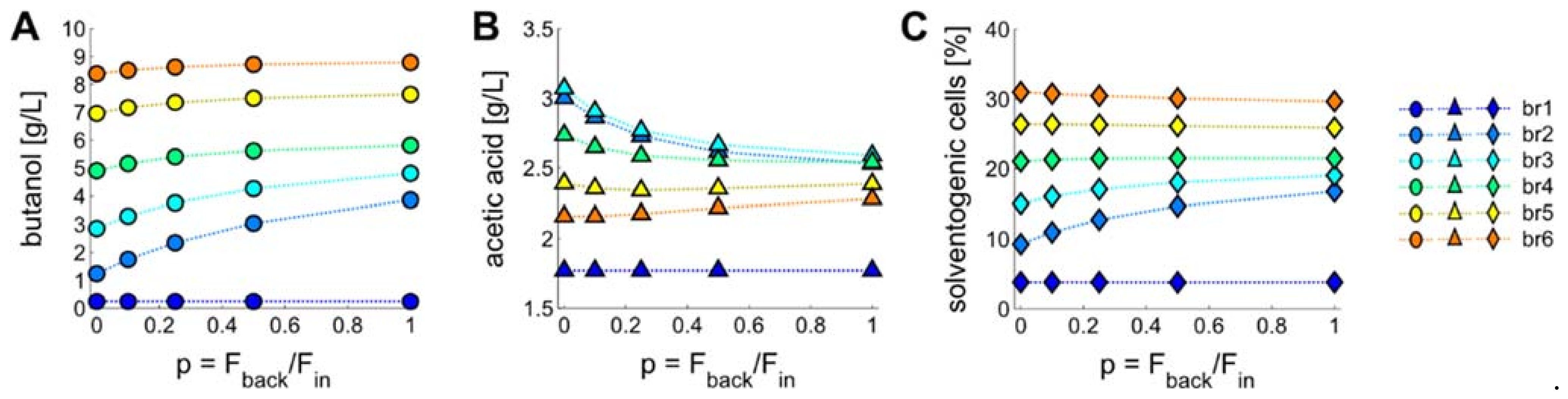
3.4. Introducing a Feedback Loop for Recirculation of Fermentation Broth from Bioreactor 4 to Bioreactor 2
3.5. Product Concentrations, Productivities and Yields of ABE Processes Run in the Continuously Operated, Multi-Stage Bioreactor Cascades
4. Discussion
5. Conclusions
- Reactor system model, depending on the configuration of the reactors, describing residence time distributions and their influence on microbial population fractions and metabolite concentrations;
- kinetic model for microbial metabolism, allowing for populations with different metabolic activities, depending on the bioreactor environment.
- Adapting individual mean residence time in bioreactors by using different working volumes.
- Moving cells and metabolites between bioreactor environments by feed-back and feed-forward loops.
- Feeding additional substrates along the cascade, e.g., organic acids.
- Biomass retention by separation and recycle.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pasteur, L. Quelques résultats nouveaux relatifs aux fermentations acétique et butyrique. Bull Soc. Chim. Paris 1862, 1862, 52–53. [Google Scholar]
- Poehlein, A.; Solano, J.D.M.; Flitsch, S.K.; Krabben, P.; Winzer, K.; Reid, S.J.; Jones, D.T.; Green, E.; Minton, N.P.; Daniel, R.; et al. Microbial solvent formation revisited by comparative genome analysis. Biotechnol. Biofuels 2017, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Dürre, P. Fermentative Butanol Production. Ann. N. Y. Acad. Sci. 2008, 1125, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Weizmann, C. Improvement in the Bacterial Fermentation of Carbohydrates and in Bacterial Cultures for the Same. UK Patent 4845, 6 March 1915. [Google Scholar]
- Hijosa-Valsero, M.; Paniagua-García, A.I.; Díez-Antolínez, R. Biobutanol production from apple pomace: The importance of pretreatment methods on the fermentability of lignocellulosic agro-food wastes. Appl. Microbiol. Biotechnol. 2017, 101, 8041–8052. [Google Scholar] [CrossRef]
- Napoli, F.; Olivieri, G.; Russo, M.E.; Marzocchella, A.; Salatino, P. Continuous lactose fermentation by Clostridium acetobutylicum—Assessment of energetics and product yields of the acidogenesis. Enzym. Microb. Technol. 2012, 50, 165–172. [Google Scholar] [CrossRef]
- Sarchami, T.; Munch, G.; Johnson, E.; Kießlich, S.; Rehmann, L. A Review of Process-Design Challenges for Industrial Fermentation of Butanol from Crude Glycerol by Non-Biphasic Clostridium pasteurianum. Fermentation 2016, 2, 13. [Google Scholar] [CrossRef]
- Du, Y.; Zou, W.; Zhang, K.; Ye, G.; Yang, J. Advances and Applications of Clostridium Co-culture Systems in Biotechnology. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Millat, T.; Janssen, H.; Thorn, G.J.; King, J.R.; Bahl, H.; Fischer, R.-J.; Wolkenhauer, O. A shift in the dominant phenotype governs the pH-induced metabolic switch of Clostridium acetobutylicum in phosphate-limited continuous cultures. Appl. Microbiol. Biotechnol. 2013, 97, 6451–6466. [Google Scholar] [CrossRef]
- Engasser, J.-M.; Petitdemange, H. Influence of pH and undissociated butyric acid on the production of acetone and butanol in batch cultures of Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 1984, 19, 422–426. [Google Scholar] [CrossRef]
- Girbal, L.; Vasconcelos, I.; Saint-amans, S.; Soucaille, P. How neutral red modified carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH. FEMS Microbiol. Rev. 1995, 16, 151–162. [Google Scholar] [CrossRef]
- Peguin, S.; Goma, G.; Delorme, P.; Soucaille, P. Metabolic flexibility of Clostridium acetobutylicum in response to methyl viologen addition. Appl. Microbiol. Biotechnol. 1994, 42, 611–616. [Google Scholar] [CrossRef]
- Rao, G.; Mutharasan, R. Alcohol production by Clostridium acetobutylicum induced by methyl viologen. Biotechnol. Lett. 1986, 8, 893–896. [Google Scholar] [CrossRef]
- Junne, S. Stimulus Response Experiments for Modelling Product Formation in Clostridium acetobutylicum Fermentations. Ph.D. Thesis, Technische Universität, Berlin, Germany, 2010. [Google Scholar]
- Grupe, H.; Gottschalk, G. Physiological events in Clostridium acetobutylicum during the shift from acidogenesis to sol-ventogenesis in continuous culture and presentation of a model for shift induction. Appl. Environ. Microbiol. 1992, 58, 3896–3902. [Google Scholar] [CrossRef]
- Jones, S.W.; Paredes, C.J.; Tracy, B.; Cheng, N.; Sillers, R.; Senger, R.S.; Papoutsakis, E.T. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 2008, 9, R114. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Tashiro, Y.; Wang, Q.; Sakai, K.; Sonomoto, K. High acetone–butanol–ethanol production in pH-stat co-feeding of acetate and glucose. J. Biosci. Bioeng. 2016, 122, 176–182. [Google Scholar] [CrossRef]
- Wiesenborn, D.P.; Rudolph, F.B.; Papoutsakis, E.T. Thiolase from Clostridium acetobutylicum ATCC 824 and Its Role in the Synthesis of Acids and Solvents. Appl. Environ. Microbiol. 1988, 54, 2717–2722. [Google Scholar] [CrossRef]
- Junne, S.; Götz, P. Mathematical models for clostridia: From cultivation description to systems biology. In Systems Biology of Clostridium; Dürre, P., Ed.; Imperial College Press: London, UK, 2014; pp. 131–172. [Google Scholar]
- Karstens, K.; Trippel, S.; Görlitz, R.; Niebelschütz, H.; Marzocchella, A.; Götz, P. Modeling Physiological Differences in Cell Populations: Acetone-Butanol-Ethanol (ABE)-Fermentation in a Cascade of Continuous Stirred Tank Reactors. Chem. Eng. Trans. 2016, 49, 271–276. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Volesky, B. Updated model of the batch acetone-butanol fermentation. Biotechnol. Lett. 1990, 12, 693–698. [Google Scholar] [CrossRef]
- Grimmler, C.; Janssen, H.; Krauβe, D.; Fischer, R.-J.; Bahl, H.; Dürre, P.; Liebl, W.; Ehrenreich, A. Genome-Wide Gene Expression Analysis of the Switch between Acidogenesis and Solventogenesis in Continuous Cultures of Clostridium acetobutylicum. J. Mol. Microbiol. Biotechnol. 2011, 20, 1–15. [Google Scholar] [CrossRef]
- Bahl, H.; Andersch, W.; Gotschalk, G. Continuous production of acetone and butanol by Clostridium acetobutylicum in a two-stage phosphate limited chemostat. Appl. Microbiol. Biotechnol. 1983, 17, 73. [Google Scholar] [CrossRef]
- Raganati, F.; Procentese, A.; Olivieri, G.; Russo, M.; Gotz, P.; Salatino, P.; Marzocchella, A. Butanol production by Clostridium acetobutylicum in a series of packed bed biofilm reactors. Chem. Eng. Sci. 2016, 152, 678–688. [Google Scholar] [CrossRef]
- Setlhaku, M.; Heitmann, S.; Górak, A.; Wichmann, R.D. Investigation of gas stripping and pervaporation for improved feasibility of two-stage butanol production process. Bioresour. Technol. 2013, 136, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Ezeji, T.; Qureshi, N.; Blaschek, H. Production of acetone, butanol and ethanol by Clostridium beijerinckii BA101 and in situ recovery by gas stripping. World J. Microbiol. Biotechnol. 2003, 19, 595–603. [Google Scholar] [CrossRef]
- Qureshi, N.; Maddox, I.S.; Friedl, A. Application of continuous substrate feeding to the ABE fermentation: Relief of product inhibition using extraction, perstraction, stripping, and pervaporation. Biotechnol. Prog. 1992, 8, 382–390. [Google Scholar] [CrossRef]
- Xue, C.; Yang, D.; Du, G.; Chen, L.; Ren, J.; Bai, F. Evaluation of hydrophobic micro-zeolite-mixed matrix membrane and integrated with acetone–butanol–ethanol fermentation for enhanced butanol production. Biotechnol. Biofuels 2015, 8, 1–9. [Google Scholar] [CrossRef]
- Xue, C.; Zhao, J.; Lu, C.; Yang, S.-T.; Bai, F.; Tang, I.-C. High-titern-butanol production by Clostridium acetobutylicum JB200 in fed-batch fermentation with intermittent gas stripping. Biotechnol. Bioeng. 2012, 109, 2746–2756. [Google Scholar] [CrossRef] [PubMed]
- Napoli, F.; Olivieri, G.; Russo, M.E.; Marzocchella, A.; Salatino, P. Butanol production by Clostridium acetobutylicum in a continuous packed bed reactor. J. Ind. Microbiol. Biotechnol. 2010, 37, 603–608. [Google Scholar] [CrossRef]
- Survase, S.A.; Van Heiningen, A.; Granström, T. Continuous bio-catalytic conversion of sugar mixture to acetone–butanol–ethanol by immobilized Clostridium acetobutylicum DSM 792. Appl. Microbiol. Biotechnol. 2011, 93, 2309–2316. [Google Scholar] [CrossRef]
- Tashiro, Y.; Takeda, K.; Kobayashi, G.; Sonomoto, K. High production of acetone–butanol–ethanol with high cell density culture by cell-recycling and bleeding. J. Biotechnol. 2005, 120, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ma, Y.; Yang, F.; Zhang, C. Continuous acetone–butanol–ethanol production by corn stalk immobilized cells. J. Ind. Microbiol. Biotechnol. 2009, 36, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- González-Peñas, H.; Lu-Chau, T.A.; Moreira, M.T.; Lema, J.M. Assessment of morphological changes of Clostridium acetobutylicum by flow cytometry during acetone/butanol/ethanol extractive fermentation. Biotechnol. Lett. 2014, 37, 577–584. [Google Scholar] [CrossRef]
- Patakova, P.; Linhova, M.; Vykydalova, P.; Branská, B.; Rychtera, M.; Melzoch, K. Use of fluorescent staining and flow cytometry for monitoring physiological changes in solventogenic clostridia. Anaerobe 2014, 29, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Branska, B.; Pechacova, Z.; Kolek, J.; Vasylkivska, M.; Patakova, P. Flow cytometry analysis of Clostridium beijerinckii NRRL B-598 populations exhibiting different phenotypes induced by changes in cultivation conditions. Biotechnol. Biofuels 2018, 11, 99. [Google Scholar] [CrossRef]
- Tracy, B.P.; Gaida, S.M.; Papoutsakis, E.T. Development and Application of Flow-Cytometric Techniques for Analyzing and Sorting Endospore-Forming Clostridia. Appl. Environ. Microbiol. 2008, 74, 7497–7506. [Google Scholar] [CrossRef] [PubMed]
- Junne, S.; Klein, E.; Angersbach, A.; Goetz, P. Electrooptical measurements for monitoring metabolite fluxes in acetone–butanol–ethanol fermentations. Biotechnol. Bioeng. 2008, 99, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Lönnberg, T. Global and targeted approaches to single-cell transcriptome characterization. Brief. Funct. Genom. 2018, 17, 209–219. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L.; Chen, Z.; Zhang, W. RNA-seq based transcriptomic analysis of single bacterial cells. Integr. Biol. 2015, 7, 1466–1476. [Google Scholar] [CrossRef]
- Janssen, H.; Döring, C.; Ehrenreich, A.; Voigt, B.; Hecker, M.; Bahl, H.; Fischer, R.-J. A proteomic and transcriptional view of acidogenic and solventogenic steady-state cells of Clostridium acetobutylicum in a chemostat culture. Appl. Microbiol. Biotechnol. 2010, 87, 2209–2226. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bauer, R.; Hekmat, D. Development of a Transient Segregated Mathematical Model of the Semicontinuous Microbial Production Process of Dihydroxyacetone. Biotechnol. Prog. 2006, 22, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Casadesús, J.; Low, D.A. Programmed Heterogeneity: Epigenetic Mechanisms in Bacteria. J. Biol. Chem. 2013, 288, 13929–13935. [Google Scholar] [CrossRef] [PubMed]
- Lidstrom, M.E.; Meldrum, D.R. Life-on-a-chip. Nat. Rev. Genet. 2003, 1, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Veening, J.-W.; Smits, W.K.; Kuipers, O.P. Bistability, Epigenetics, and Bet-Hedging in Bacteria. Annu. Rev. Microbiol. 2008, 62, 193–210. [Google Scholar] [CrossRef] [PubMed]
- Meeks, J.C.; Campbell, E.L.; Summers, M.L.; Wong, F.C. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 2002, 178, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.; Fischer, R.J.; Bahl, H.; (Universität Rostock, Rostock, Germany). Personal communication, 2010.

| Fin (L h−1) | Dbr1 (h−1) | D (h−1) | td (h) |
|---|---|---|---|
| 0.1 | 0.25 | 0.042 | 24 |
| 0.22 | 0.55 | 0.092 | 10.9 |
| Kinetic Model Parameter | Unit | Acidogenic Cells | Intermediate Cells | Solventogenic Cells | All |
|---|---|---|---|---|---|
| biomass evolution | |||||
| mu_max_ (P) | h−1 | 0.7273 | 0.4641 | 0.4204 | |
| KsPO4 | gKH2PO4 L−1 | 0.005 | |||
| KsGLU | gGLU L−1 | 6.5 | |||
| KiB | gBUT L−1 | 5 | |||
| niB | - | 3 | |||
| Kd | L gBUT−1 h−1 | 0.02 | |||
| n_dGLU | - | 1.7332 | |||
| KdGLU | gGLU L−1 | 1.0353 | |||
| acid production and uptake | |||||
| r_aa_max_ (P) | gAA gX−1 h−1 | 0.5097 | 0.0296 | 0 | |
| Y_aax_ (P) | gAA gX−1 | 0.5224 | 0.5 | 0 | |
| k_AA_up_ (P) | gAA gX−1 h−1 | 0 | 0.4224 | 0.9485 | |
| KsAA | gAA L−1 | 0.6 | |||
| r_ba_max_ (P) | gBA gX−1 h−1 | 0.5248 | 0 | 0 | |
| Y_bax_ (P) | gBA gX−1 | 0.5 | 0 | 0 | |
| k_BA_up_ (P) | gBA gX−1 h−1 | 0 | 0.5 | 1.8594 | |
| KsBA | gBA L−1 | 0.734 | |||
| solvent production | |||||
| r_eth_max_ (P) | gETH gX−1 h−1 | 0 | 0 | 0.2458 | |
| r_act_max_ (P) | gACT gX−1 h−1 | 0 | 0.0866 | 0.6213 | |
| r_but_max_ (P) | gBUT gX−1 h−1 | 0 | 0.0203 | 2.4485 | |
| phosphate and glucose uptake | |||||
| Y_xp | gX gKH2PO4−1 | 29 | |||
| sk | molC molGLU−1 | 1.1020 | |||
| r_CO2_ (P) | molC molGLU−1 | 0.0019 | 0 | 2.4904 | |
| n_C_GLU | molC molGLU−1 | 6 | |||
| Y_xs | gX gGLU−1 | 0.475 | |||
| Y_aas | gAA gGLU−1 | 0.9091 | |||
| Y_bas | gBA gGLU−1 | 0.6667 | |||
| Y_eths | gETH gGLU−1 | 0.6970 | |||
| Y_acts | gACT gGLU−1 | 0.5859 | |||
| Y_buts | gBUT gGLU−1 | 0.5606 | |||
| differentiation | |||||
| mu_d_ (P) | h−1 | 0.1681 | 0.1681 | 0 | |
| K_UDA_ (P) | g L−1 | 3.0203 | 0 | 0 | |
| n_iUDA_ (P) | - | 4.3175 | 0 | 0 | |
| p | Bioreactor Stage | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | Σ | |
| D = 0.042 h−1/ pHbr1= 5.6 | 4.11 | 10.36 | 11.89 | 8.07 | 10.70 | 12.33 | 57.46 |
| D = 0.042 h−1/ pHbr1= 4.3 | 15.07 | 9.80 | 1.83 | 19.96 | 5.56 | 3.19 | 55.40 |
| D = 0.092 h−1/ pHbr1= 5.6 | 3.45 | 5.62 | 3.33 | 2.32 | 9.97 | 15.97 | 40.66 |
| D = 0.092 h−1/ pHbr1= 4.3 | 5.60 | 2.36 | 2.33 | 2.21 | 1.88 | 1.89 | 16.29 |
| Σ | 28.23 | 28.14 | 19.38 | 32.56 | 28.10 | 33.38 | 169.80 |
| Operating Condition | Product Concentration | Volumetric Productivity | Yield | ||||
|---|---|---|---|---|---|---|---|
| (g L−1) | (g L−1 h−1) | (g gGLU−1) | |||||
| cbutanol | cABE | rbutanol | rABE | YBUT/GLU | YABE/GLU | ||
| D = 0.042 h−1/ pHbr1 5.6 | f | 6.7 ± 3.3 | 8.9 ± 4.5 | 0.28 ± 0.14 | 0.37 ± 0.19 | 0.11 ± 0.06 | 0.15 ± 0.09 |
| m | 7.2 ± 1.1 br5 | 9.7 ± 1.5 br5 | 0.42 ± 0.10 br3 | 0.56 ± 0.13 br3 | 0.13 ± 0.04 br4 | 0.17 ± 0.06 br4 | |
| D = 0.042 h−1/ pHbr1 4.3 | f | 9.6 ± 0.3 | 13.3 ± 0.7 | 0.40 ± 0.01 | 0.55 ± 0.03 | 0.16 ± 0.01 | 0.22 ± 0.01 |
| m | 9.7 ± 0.5 br3 | 13.6 ± 1.1 br3 | 0.93 ± 0.14 br2 | 1.29 ± 0.21 br2 | 0.18 ± 0.05 br2 | 0.25 ± 0.07 br2 | |
| D = 0.092 h−1/ pHbr1 5.6 | f | 4.8 ± 1.9 | 6.7 ± 2.7 | 0.44 ± 0.18 | 0.62 ± 0.25 | 0.14 ± 0.10 | 0.20 ± 0.14 |
| m | ~ | ~ | ~ | ~ | ~ | ~ | |
| D = 0.092 h−1/ pHbr1 4.3 | f | 8.2 ± 2.6 | 11.8 ± 3.9 | 0.75 ± 0.23 | 1.08 ± 0.36 | 0.18 ± 0.09 | 0.26 ± 0.13 |
| m | ~ | ~ | 0.76 ± 0.23 br5 | 1.08 ± 0.36 br5 | ~ | ~ | |
| D = 0.092 h−1/ pHbr1 4.3 | f | 8.3 ± 0.9 | 11.9 ± 1.5 | 0.76 ± 0.08 | 1.09 ± 0.14 | 0.18 ± 0.06 | 0.26 ± 0.09 |
| feedback loop | m | ~ | ~ | 0.78 ± 0.20 br4* | 1.10 ± 0.28 br4* | ~*,§ | ~*,§ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karstens, K.; Trippel, S.; Götz, P. Process Engineering of the Acetone-Ethanol-Butanol (ABE) Fermentation in a Linear and Feedback Loop Cascade of Continuous Stirred Tank Reactors: Experiments, Modeling and Optimization. Fuels 2021, 2, 108-129. https://doi.org/10.3390/fuels2020007
Karstens K, Trippel S, Götz P. Process Engineering of the Acetone-Ethanol-Butanol (ABE) Fermentation in a Linear and Feedback Loop Cascade of Continuous Stirred Tank Reactors: Experiments, Modeling and Optimization. Fuels. 2021; 2(2):108-129. https://doi.org/10.3390/fuels2020007
Chicago/Turabian StyleKarstens, Katja, Sergej Trippel, and Peter Götz. 2021. "Process Engineering of the Acetone-Ethanol-Butanol (ABE) Fermentation in a Linear and Feedback Loop Cascade of Continuous Stirred Tank Reactors: Experiments, Modeling and Optimization" Fuels 2, no. 2: 108-129. https://doi.org/10.3390/fuels2020007
APA StyleKarstens, K., Trippel, S., & Götz, P. (2021). Process Engineering of the Acetone-Ethanol-Butanol (ABE) Fermentation in a Linear and Feedback Loop Cascade of Continuous Stirred Tank Reactors: Experiments, Modeling and Optimization. Fuels, 2(2), 108-129. https://doi.org/10.3390/fuels2020007






