Abstract
The evolution of breast cancers results from the emergence of epithelial cell subpopulations containing variant Estrogen Receptor α which is able to bypass conventional treatments aimed at antagonizing the activity of this tumor-promoting receptor. The present investigation concerns a few estradiol derivates bearing substituents in position 11β that might not only contribute to the development of drugs to alleviate this unfortunate issue but that may be also helpful in identifying molecular aspects of resistance to this receptor in order to elaborate other therapeutic approaches. In this regard, AP-1 assisted and ERE-directed ERα transcriptions are demonstrated to be key factors in this area: AP-1 transcriptions are shown to antagonize ERE transcriptions, thereby limiting their tumor-promoting activity. This property results from a conformal change in the receptor, which is induced essentially by estrogenic ligands which, inserted into a cavity of ERα’s ligand-binding pocket, govern this regulatory mechanism. Flexible 11β side-chains favor this insertion, in contrast to their rigid counterparts, which counteract it; these properties give rise to strong estrogenic, SERM or SERD profiles. Suspected extracellular regulatory mechanisms resulting from these ligand-induced transcriptions are elaborated on in the present work in the context of breast cancer development.
1. Introduction
Estrogen Receptor alpha (ERα) is a growth/differentiation modulator pertaining to the class of nuclear receptors implicated in the development of breast cancers. For this reason, multiple studies aimed at discovering its mechanism of action in hopes of curbing its deleterious effects have been conducted. The present study concerns the dynamic character of this mechanism, generated by a high conformational plasticity of the receptor [1,2], which favors rapid transient conformational changes aimed at recruiting a variety of cellular regulators that, under physiological condition, are necessary for survival, while in the present context of cancer must be subjected to selected therapeutic attacks [3,4,5,6,7,8,9]. The present publication overviews this topic in the context of ERα-driven transcriptions induced by its ligands. A set of estradiol (E2) in which 11β hydrogen has been replaced with various hydrophobic substituents, including flexible or inflexible side-chains to reinforce or antagonize the receptor’s activity, is described to illustrate the underlying mechanisms but also to integrate them into a therapeutic perspective. The next sections overview the underlying molecular aspects of this topic.
2. ERα-Mediated Transcriptions
Physiologically, neosynthesized ERα shuttles between targets governing multiple functions; its stabilization on one of these targets, induced by an extracellular stimulus—not necessarily one of estrogenic nature—confers indeed an irreversible ability to accomplish an imposed message [9]. Two peculiar localizations of such ”formatted” ERα pools have largely been identified in publications: the plasma membrane and the nucleus. A palmitoylated receptor form anchored to a specific membrane site induces rapid responses through signal transduction pathways, eventually giving rise to subsequent transcriptions [10,11,12]. An activated receptor, derived at least partly from this pathway, may indeed operate in the nucleus as a co-activator of other transcription factors anchored to the DNA at their own promoter sites [13] (especially Activating Protein-1 (AP-1) consisting of the fos/Jun heterocomplex [14,15]). ERα in a homo-dimeric form which recruits coactivators harboring a LxxLL motif (L = Leucine; x = all other amino acids) completes its global transcription program [3].This homo-dimeric form acts at the level of a specific palindromic sequence of nucleotides (Estrogen Response Element; ERE) localized in the promoter region of the genes it expresses.
These two transcription procedures are under complementary controls exerted by a variety of intra- and extracellular agents which favor or antagonize specific ERα-mediated actions, including also synergistic effects. The selection between these possibilities is related to the flexibility of the receptor, which modulates the exposure of its functional domains (domains present in all nuclear receptors defined by the six first letters of the alphabet) [1,2,3,4,5,6,7,8,9].
The A/B domain, localized at the N-terminal edge of ERα, is mainly unstructured and hydrophilic (rich in positively amino acids), in contrast to the hormone/ligand-binding domain, which is well folded and hydrophobic (E domain). It may associate with an estrogen-binding protein (GPR30) [16] localized on the plasma membrane to satisfy extracellular exigencies not necessarily of estrogenic nature; regulatory peptides targeted in the vicinity of this protein through interaction with the receptor may activate signal transduction pathways as well as gene expression. An activation motif localized within this A/B domain, called Activation Function (AF-1), is implicated in this mechanism.
The E domain contains an AF-2 function, which is essentially estrogen-dependent, in contrast to AF-1. This function regulates the exposure of zinc fingers of the C domain, which interact with EREs for ERα-directed transcriptions; this procedure requires a conformational change at the level of a flexible “hinge” subregion localized in the D/E border, which governs the expression of a third activating site (BF-3) implicated in the recruitment of coregulators [17,18]. A motif of this border hinge subregion (Pro 295-Thr 311) able to interact with these coregulators [19], plays a role of prime importance in this activation mechanism, which coordinates of the actions of AF-1 and AF-2, eventually giving rise to synergistic effects [20,21].
Indeed, this “hinge” subregion coordinates a lot of regulatory requests received at the level of recruitment sites localized along the whole primary structure of the receptor, including also its F domain, the function of which has largely been less well described than that of the other domains [3,4,5,6,7,8]. Nevertheless, its implication in tamoxifen-mediated AF-1 activation [3] as well as in dimerization antagonism [22] is known. ERα conformations resulting from this hinge-mediated action are fixed to avoid incoherent responses potentially generated by incompatible requests [23,24]. This appropriate stabilization is reinforced by irreversible posttranscriptional changes (phosphorylation, methylation, acetylation [25]) which govern the intracellular traffic and turnover rate of ERα implicated in multiple functions(growth/apoptosis, secretion of growth factors for inter-cellular dialogs) [3,4,5,6,7]. The required coordination between these distinct functions is largely enhanced by the presence of some recruitment sites within “hubs” [25]. The Pro295-Thr311 motif is a typical example of this managing structure: it promotes, respectively, the association of ERα with the Hsp 70/90 chaperones that maturates the newly synthesized receptor [19], the GPR30 plasma membrane [16] as well as calmodulin, which stabilizes the receptor within the nucleus in a homo-dimeric form required for ERE-transcriptions [26,27]. Two Lysines (202 and 203) of this motif are directly implicated in these properties as well as its ubiquitination which governs its turnover rate [8,28].
3. Ligand Insertion within the ERα Binding Pocket
The insertion of estrogens as well as their antagonists within the hormone-binding pocket of ERα is logically implicated in the selection of genes whose expression is being requested. This property imposes distinct interactions between the structural chemical elements of these ligands and the residues of this pocket. This topic is addressed in this section in terms of its structure–activity relationship.
Estrogenic activity is generated by various classes of natural and synthetic compounds (i.e., steroids (E2, E1, E3), phytoestrogens (coumetarol, flavones, isoflavones) and trans-stilbenes (DES, HEX)), all of which share a similar linear hydrophobic structure containing two axial oxygenated functions localized at the ends. These functions play a role of prime importance in attracting estrogens to the hormone-binding pocket of the receptor [29,30] (a property illustrated in 29: Supplementary Materials, Figure S1, with E2 as a reference compound). Thanks to its acidic character (hydrogen donor property) and with the assistance of a water molecule, the phenol in position 3 of the steroid (cycle A) selectively interacts with Glu 353 and Arg 394 of the pocket, thereby favoring its insertion. The oxygen of the hydroxyl in position 17β (cycle D, substituted by a phenol in non-steroidal hormones) stabilizes this anchorage through a complementary attractive action of its oxygen exerted by His 524. Hydrophobic interactions between atoms of the steroid and other residues (note the importance of Phe 404, depicted in Supplementary Materials Figure S1) complete this recruitment procedure, especially for weak estrogens devoid of detectable binding affinity, the chemical structure of which is compatible with an insertion within the pocket [31,32]. In fact, the plasticity of the latter enhances the exposure of a lot of its residues/subdomains and their attraction of a multitude of molecules whose chemical structures are often extremely distinct from steroidal estrogens [33], some of them with antiestrogenic potency stressing a great interest in such molecules and derivatives in a therapeutic context. This interest led V.C. Jordan to propose the regrooming of active compounds within two main classes according to their capacity to generate interactions within a peculiar cavity of the hormone binding pocket [34]: Type I,”linear“ estrogenic structure without any ability of insertion within the pocket and Type II, “angular” structure of which a hydrophobic cluster virtually localized around the central part of the steroid (7α/11β position) favors or decreases interactions in the pocket.
Type II ligands may generate a mixed estrogenic/antiestrogenic profile (SERM) as well as a very strong antiestrogenic activity partly relevant to an ability to degrade ERα (SERD), properties relevant the nature of the cavity inserting agents grafted on the estrogenic substrate, a property recorded in E2 derivatives. Indeed, a substitution of the hydrogen in position 11β of E2 by hydrophobic side-chains may enhance the ability of the hormone to insert itself within this binding pocket [35,36,37], (see [29] for an extended overview of this topic, including several drawings of this cavity). Nevertheless, it should be stressed that an increase in acidity of the 3-phenolic function of E2, related to an electronic displacement within the steroid provoked by the 11β substituent, might also be implicated in the enhancement [38]. Even though this mechanism only concerns some E2 derivatives, it should be taken into account in the interpretation of experimental data in the context of future studies, such as that performed in our laboratory and described in this paper, which focuses especially on the size and the flexibility of these substituents.
The next part of this publication concerns investigations that were initiated some years ago, when the understanding of the molecular aspects of ERα-driven transcriptions was insufficient for the evaluation of our experimental data’s potential therapeutic impact. Given that knowledge on this topic has progressed, I am now able to provide a pertinent global view of our experimentations, including some unpublished data, especially on the subject of the rigidity of fluorinated side-chains in E2 derivatives [37]. This is a topic of interest which has not been addressed in the current literature.
4. Assessing the Activity of 11β Substituted Estradiol Derivatives
4.1. Nature and Origin of the Investigated Compounds
As already mentioned, ERα’s actions are largely dependent on the interrelationships between ERE and AP-1 transcriptions, whose underlying molecular procedures in the context of breast cancers are still largely unknown, especially in terms of AP-1’s role. To contribute to the bridging of this gap, we oriented our study towards the potential impact of 11β E2 substitutions on the transcriptional profile of the hormone in a potential therapeutic optic [39,40,41,42]. In this exploratory phase, a few representative estrogens, namely SERMs and SERDs, were submitted to simple tests to provide complementary information that would direct further investigations (Section 5); this approach was already proposed by other investigators [43]. These compounds were commercially available, except those bearing a perfluorinated substituent side-chain (and non-fluorinated controls), which were synthesized at the Institute Lavoisier de Versailles, Université de Versailles, France (Contact: Prof E. Magnier).
4.2. Experimental Procedures
Our study focused on the binding ability of these compounds to ERα as well as on their induction of receptor-mediated transcriptions in monolayer cultures. The experimental procedures have already been largely reported, which is why only a brief description will be provided here (for experimental details, consult references provided in the text and legends of the figures).
The compounds’ ERα-binding ability was evaluated using a conventional tritiated E2 competitive binding assay performed with a commercial highly purified recombinant receptor (commercially available) adsorbed on a hydroxyl apatite suspension which requires the exposure of its A/B C domains [44]. This experimental approach provided us with an index of AF-1’s exposure, since this site belongs to this region. Moreover, with AF-1 usually being implicated in AP-1 transcriptions, this binding assay appeared extremely adequate to evaluate the potency of a ligand to modulate AP-1 transcriptions. Assays were run at 0 and 25 °C (overnight incubation). These thermodynamic conditions were suspected to modulate conformational changes in ERα induced by the ligand [45].
ERE- and AP-1-mediated transcriptions were assessed with two stably transfected MCF-7 cells, so-called, respectively, MVLN and MTLN cells [46,47], using specific experimental protocols based on the expression of luciferase-reporter genes. Luciferase measurement was performed after 3 days’ exposure of the compounds to the MVLN cells to provide a substantial ERE-dependent response (pVit-TK-Luc) [46]. A similar procedure was used with MTLN cells to activate a TRE with TPA (p(TRE)3-TK-Luc), representative of an ERα-assisted AP-1 transcription, which required 4 days of exposure of the compounds to the cells before the luminometric transcription assessment [47]. The prolonged incubation period with regard to ERα binding and related conformational changes for LxxLL motif recruitment [48] were required for the detection of a substantial response, a property of prominent importance for AP-1 transcription [47].
4.3. Results
4.3.1. Global View of Our Investigation
Table 1 provides the ERα-binding characteristics of the investigated estrogens, SERMs and SERDs. A representative compound of each class was further analyzed to assess their mechanism of action according to the literature data; financial aspects motivated this limited structure–activity approach. E2, tamoxifen and 7α-fulvestrant were included in our study as transcriptional controls. Unfortunately, data relative to AP-1 transcriptions for small-size 11β substituents are partially lacking since MVLN cells were introduced into the laboratory largely before MTLN cells; this handicap failed to affect our conclusions, and the reported data are sufficiently thought-provoking.

Table 1.
11β-estradiol derivatives.
E2, tamoxifen (SERM) and 7α-fulvestrant (SERD) were selected as control ligands. E2 enhances ERE transcription, while tamoxifen, at high concentrations, activates AP-1 transcription [46]. Fulvestrant has mainly been reported as an ERE transcription inhibitor, even though is also inhibits AP-1 transcription, as shown in the present work. In our experiments, the investigated ligands were studied alone or in the presence of one of the control compounds to assess a possibility of synergistic or antagonistic effects. Their inclusion in our study was largely justified by our use of fluorinated ligands (SERMs and SERDs), whose properties in this area are reported here for the first time.
Preliminary assays on ERα binding, performed as controls on E2 and a few non-steroidal estrogens (DES, coumestrol, genistein) at 0 and 25 °C, failed to show any influence of this temperature increase on the ability to activate ERE transcriptions and underlying receptor level change. This lack of energy adjunct requirement distinguishes these compounds from estrogens the bear a substituent, such as SERMs and SERDs, as recorded in Table 1. This property largely confirms the importance of these substituents’ flexibility, which is thermodynamically modulated to regulate the stability of the interaction of the steroidal nucleus of these molecules within the ERα binding pocket. This topic is elaborated further in the following paragraphs.
4.3.2. Small-Size 11β Substituents
In -CH3, -CH2Cl, and -CCH E2 derivatives, the temperature jump from 0 to 25 °C provoked a weak increase in binding affinity (RBA vs. E2). This property, which correlates with a 10-fold higher capacity to reach an optimal LxxLL motif recruitment (see Table 1 for molarity) was reflected in a weak ERE-dependent transcription enhancement, suggesting that at 0 °C, these strong agonists had already generated the ERα conformation required for an optimal effect. This property was confirmed by the almost identical ERE transcriptional profile of E2 and these three derivatives, with the hormone being slightly less active than the derivatives (<−10%).
The ethynyl derivative (-CCH), which displayed the highest temperature-dependent increase in ERα-binding affinity, was further analyzed to assess the impact of this property on the receptor turnover rate. Its binding affinity was further analyzed to assess the impact of this property on its turnover rate. After 24 h of incubation, this compound showed a slightly higher transcriptional activity than E2, which was maintained 24 h after the removal of the ligands from the culture medium, confirming that the pursuit of the inductive effect derived from the conformational change in ERα does not require the permanent presence of the ligand (irreversible pulse effect [49] (Figure 1, left)). This property is usually related to a proteasomal downregulation of the receptor [5]. However, under an occasional condition of unknown nature that abrogated this ERα downregulation, the slightly higher transcription potency seen in the ethynyl derivative was not affected (Figure 1, right), stressing that the ligand-induced conformational change in the receptor is independent of the regulation of its turnover rate.
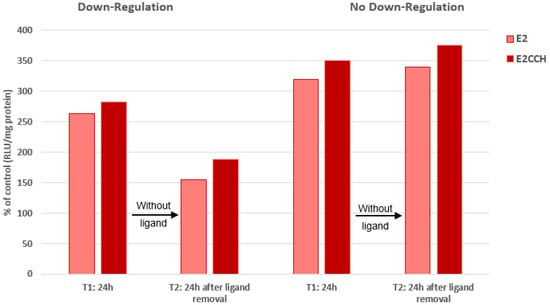
Figure 1.
No requirement of ERα downregulation for estrogen-induced ERE transcription (cells: MVLN, ligands: E2/11β-Ε2CCH at 0.1 nM). Mean of two highly reproducible experiments (SD < 5%). A similar enhancement of transcriptional activity occurred both under conditions that gave rise to the downregulation of the receptor and those that did not. For details of the experiment and related comments, see Section 4.3.2.
This property may be ascribed to the dependence of cells on a complex extracellular network, which governs their vitality as well as their hormonal sensitivity. ERα proteolysis may favor its maintenance through an autocrine/paracrine mechanism, perhaps involving the release of receptor degradation products [9,50]. Under exceptional conditions which liberate the cells from this vital requirement, no message for receptor proteolysis would be issued from this network, explaining our observation. This is of course a hypothesis requiring validation.
On the other hand, the permanent slightly higher transcriptional ability seen in the ethynyl derivative of E2 might reflect a capacity of 11β substituents to bypass the final step of the molecular procedure to reach the optimal activation of ERα, a step that the unsubstituted hormone cannot accomplish.
4.3.3. 11β Side-Chains
SERMs
The E2 derivative bearing a C6H13 flexible side-chain displayed a lower RBA value at 0 than at 25 °C (10–100), while its C6F13 rigid counterpart maintained a low value (3), stressing the possibility that its side-chain’s rigidity maintained ERα in at least a moderately active conformation, a hypothesis that we confirmed (Figure 2).
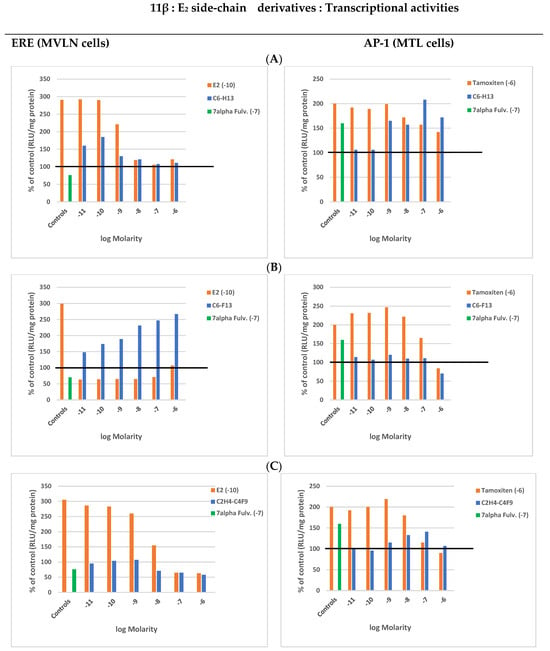
Figure 2.
11β-Ε2 side-chain derivatives: Transcriptional activity induced by increasing concentrations of the compounds alone (blue) or in the presence of a constant concentration of tamoxifen (red). Means of two highly reproducible experiments (SD < 5%). The horizontal line helps differentiate very weak stimulations from those that would produce at least a substantial effect. (A) 11β-C6H13. (B) 11β-C6F13. (C) 11β-C2H4-C4F9.
Indeed, the C6H13 compound showed a concentration-dependent progressive decrease in its relatively weak ERE-dependent transcription (with regards to E2), while a marked inverse increase in AP-1 transcription, reaching the level induced by tamoxifen at 1 μM (control induction), was observed in parallel (Figure 2A). In contrast, its C6F13 counterpart induced a very weak ERE-dependent activity without an almost total absence of AP-1 activity (Figure 2B). Of note, the smaller fluoride side-chain (C2H4-C4F9) mainly decreased its ERE activity at high concentrations (intermediate profile) (Figure 2C).
Interestingly, AP-1 transcription induced by tamoxifen at 1 μM (control) failed to display any synergistic effect in the presence of the C6H13 compound, while this effect was visible with its C6F13 counterpart over a large range of its lower concentrations (maximal effect at 1 nM); at this molarity, a slight synergy with the C2H4-C4F9 compound was recorded, confirming its intermediary status. Moreover, the AP-1 transcription induced by these three compounds decreased at their highest concentrations, C6F13 being the most efficient antagonist. Confronting this information with ERE transcription data clearly indicated that AP-1 and ERE transcriptions operated in opposition to each other. This conclusion is in agreement with the finding that the C6H13 SERM profile correlated with a loss of LxxLL motif recruitment ability conjugated with a nuclear upregulation of ERα, while its C6F13 weak estrogenic counterpart correlated with a weak LxxLL recruitment ability conjugated with an induced receptor downregulation (Table 1). This was most probably derived from an easier insertion of the flexible C6H13 side-chain into the sub pocket of the ERα ligand-binding site, which is where the side-chain of tamoxifen also enters [51], allowing for its regulatory function. This would be hindered by the rigidity of the C6F13 side-chain.
SERDs
The side-chain of fulvestrant (grafted in position 7α of E2) is known to eliminate ERα to abrogate ERE-dependent transcriptions through a complex procedure implicating SUMOylation [52,53,54]. Grafting of this chain in the 11β position maintains these properties [55], which is logical since 7α and 11β substituents are symmetrically positioned with regard to the 3/17β hydroxyl axis of E2, allowing for identical displacements during the molecule’s rotation along this axis to allow for an optimal insertion within the ERα binding pocket [29]. Hence, it is normal that 7α- and 11β-fulvestrants share a similar binding temperature dependence for receptor binding (α: 10–100, β: 6–80), a property reflected in their almost identical ability to abrogate the recruitment of co-activators harboring a LxxLL motif, which consequently provoke a similar ERE transcription inhibition.
These antagonistic properties of 7α- and 11β-fulvestrants were conjugated with a marked AP-1 transcription in MTLN cells, reaching the control value of tamoxifen, at a 100-fold lower concentration for its 11β isomer (Figure 3, upper panel). This property is reminiscent of the observation that the substitution of the amino alkyl side-chain of SERM RU 394 411 with a quasi-identical 7α-fulvestrant side-chain generates a SERD (RU 58 668; Supplementary Materials Figure S2) which abrogates its Progesterone Receptor-inducing ability [56], stressing the supremacy of the side-chain of SERDs over that of SERMs. This supremacy supplants the prolonged SERM-induced stabilization of ERα within the cell nucleus, favoring its release from the DNA, coupled with a compaction of chromatin at the level of E2 target genes, which generates multiple disturbances at the level of vitally important transcriptions implicating members of the AP-1 family. This would explain the strong growth-antagonistic activity of 7α-fulvestrant [51,52,53,54,55,57,58,59] underlying its drastic receptor elimination. The enhanced AP-1 transcriptional antagonism against ERE transcriptions that we recorded in the residual cell population after its exposure to both isomers of fulvestrant would most probably be related to this state.
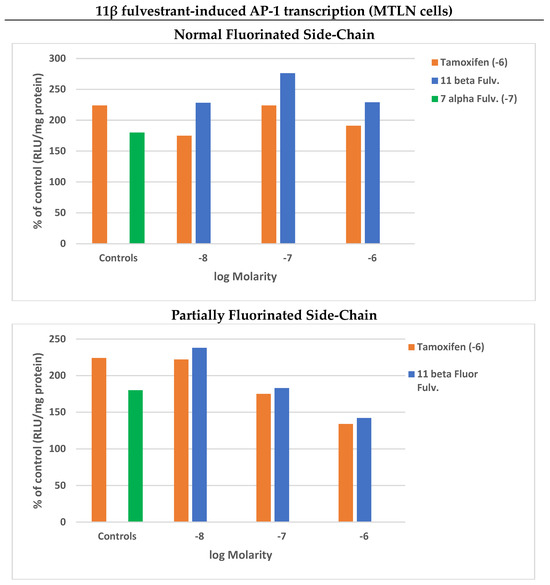
Figure 3.
Concentration-dependent 11β-fulvestrant-induced AP-1 transcription in the absence (blue) or presence of a constant concentration of tamoxifen (red; 1 μM). Partial decrease in side-chain fluorination of the compound limits its transcriptional potency. Mean of two reproducible experiments (SD < 5%, two values < 10%).
Of note, SUMOylation, which is implicated in these mechanisms, was found to occur even in the absence of an accelerated turnover rate [54]. This was also seen in a 11β-fulvestrant derivative, the flexibility of whose side-chain had been decreased by a partial fluorination [55], consequently displaying a lack of temperature dependence in its ERα-binding affinity, in contrast to its parent (2–10 vs. 6–80). This estrogenic property was logically found to be combined with a decreased AP-1 expression (Figure 3, lower panel), validating the importance of side-chain flexibility of all types of ligands (estrogens, SERMs and SERDs) in the regulation of AP-1/ERE transcription balance as well as a very strong loss of ERα elimination (undetectable at 1 μM).
5. Concluding Remarks
The present study reveals that ERα conformal changes reflected in a temperature dependence of ligand-binding affinity are an indication of the ligand’s (estrogen, SERM or SERD) ability to modulate ERE and AP-1 transcriptions. This indication seems to be devoid of any information relative to the influence of these ligands on the turnover rate of the receptor, which, consequently, appears independent of this conformational change. Our conclusion introduces the concept that ligand-induced conformational changes in ERα (and, logically, in other nuclear receptors as well) provoke intracellular events that favor the insertion of epithelial breast cancer cells into an extracellular network governing their growth and differentiation. If this insertion imposes a prompt decrease in the receptor level (which is apparently a usual condition in estrogen-free cell cultures), this network forces the cells to satisfy this requirement.
In fact, this temperature dependence of ERα conformational changes, always seen in the case of AP-1 transcriptions, may reflect a two-step mechanism: the first step corresponds to ERα intracellular trafficking, eventually reflected in weak ERE transcriptions, and the second is relevant to the association of this formatted receptor with the AP1 complex to active a TRE with TPA. A physiological/pathological thermodynamic regulation is logically related to energy production changes [60], represented by the temperature dependence of binding affinity. Hence, according to our data, AP-1/ERE antagonism would govern the development of breast cancers according to energy-regulating cycles in which the mitochondria are implicated [61].
The evolution of breast cancer, which can initially be maintained in a relatively stable state with the use of the appropriate endocrine therapeutic approach (antiestrogens, aromatase inhibitors), unfortunately often becomes resistant to these treatments, a phenomenon related to the emergence of receptor variants. Our study justifies the use of fulvestrant as a curative agent against this disastrous issue. Indeed, resistant cells become progressively subjected to estrogen-induced apoptosis related to an abnormally overactivated ERα transcription of proteins (especially AP-1 family members) which provoke various lethal stress responses at the level of the endoplasmic reticulum [62]. One may postulate that the drastic elimination of these receptor variants using fulvestrant may favor a selective maintenance of a minor residual ERα-activatable cell population, perhaps through the auto-/paracrine external network, which regulates receptor activation [9]. Such a hypothetical perspective finds some complementary support in the observation that E2 inhibits the metastatic ability of ERα-negative cells following their transfection with the receptor [63]. Hence, the scope of the present investigation may contribute to generating new therapeutic modalities.
In this regard, the extension of our transcriptional assessments to the proliferation of MCF-7 cells (Supplementary Materials Table S1) indicates their perfect relevance to this crucially important property. This assessment would justify the elaboration of a model referring to all the elements of this study, including some growth aspects that we did not explore (Figure 4). The growth of epithelial breast cancer cells is indeed dependent on a variety of extracellular factors present in serum, probably including peptides resulting from its ligand-induced proteolysis, whose survival is modulated by its interaction with the cells (auto-/paracrine procedures) [9]. Relationships between ERα and the cell cycle that governs proliferation [64,65] including Ki-67 as commentary marker of tumor evolution [66] may also be taken into account. Whether these suspected regulatory procedures were implicated in the generation of our data, especially in AP-1 transcriptions, merits confirmation through experiments conducted on extracts from conditioned culture media and mass spectrometry analyses, comparison of data from cultures with our non-fluorinated and fluorinated compounds.
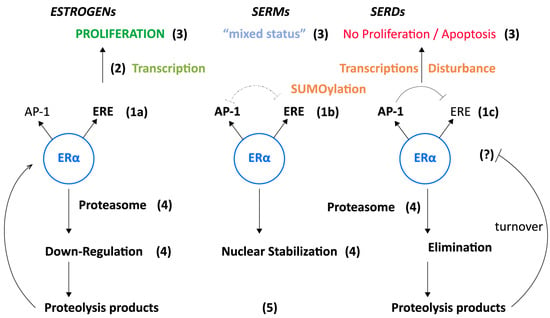
Figure 4.
Model of ligand-induced AP-1/ERE transcription antagonism’s impact on breast cancer development. (1) Ligand-induced ERα conformational changes favor ERE-directed transcriptions (estrogens, 1a), AP-1 assisted transcriptions (SERDs, 1c) or an intermediary transcriptional profile (depending on the chemical nature of the SERM, 1b). Side-chain rigidity in position 11βΕ2 orients ERα towards ERE, while flexibility orients it towards AP-1. (2) Influence of transcription on epithelial cell proliferation/viability: ERE operates on selected genes implicated in proliferation enhancement while AP-1 operates extensively on DNA/chromatin (SUMOylation), provoking disturbances in the action of multiple transcription factors. (3) Consequence of (2): ERE: breast cancer evolution, AP-1: inverse status. (4) Consequence of (3) on ERα turnover rate: proteasomal degradation eventually conjugated with an arrest of receptor synthesis (estrogens, SERDs), maintenance of receptor synthesis and shuttling, provoking its nuclear accumulation (SERMs). (5) Extracellular release of ERα proteolysis products potentially implicated in receptor restoration through an autocrine/paracrine mechanism acting on specific site(s) localized on the plasma membrane, including GPR30. A mechanism that may (?) regulate the AP-1/ ERE transcription balance.
Regarding future investigations, it should be stressed that the temperature/energy aspect of our work was not addressed in our structural molecular analyses concerning ERα conformational changes [2,67,68,69,70]. I suggest its introduction into future investigations, even if this would be quite difficult to achieve. I am aware that our data are rather simplistic compared with those provided by such analyses; however, the suggestion of a potential implication of electronic displacement within steroids induced by a substituent may open new avenues of investigation, which would not be specifically restricted to a hydrophobic aspect [71]. In fact, as already mentioned, our goal was to evaluate the validity of tests to determine the selection of appropriate ligands for such future structural approaches, in an original perspective which progressively leads to a concept overviewed in Figure 4. The incorporation of multiple regulatory facets of ERα’s mechanism of action, as reported in the first sections of this publication, into this model may also be of interest for the elaboration of new experimentations.
Finally, the peculiar fluorinated fulvestrant which does not eliminate ERα may potentially be used for the in vivo identification of “receptor-positive tumors” using tomography with a F18 analog, as was already indicated in the case of another 11 β derivative [72]. In this therapeutic perspective, one may logically consider that perfluoro side- chains linked in another position of the hormone might also present an interest, as demonstrated with a set of 17 α derivatives displaying agonistic/antagonistic profiles related to the nature (α/β) of the receptor [73]. Hence, I believe that this publication might be of interest to a relatively wide group of scientific/medical investigators with distinct objectives.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/endocrines5010007/s1. Figure S1: Implication of Glu-353/Arg-394 and His-524 of ERα in the binding of estradiol; Figure S2: Importance of the nature of a side-chain in postion 11β of Estradiol to generate a SERM of a SERD; Table S1: Influence of fluorinated of 11β E2 derivatives on MCF-7 cell proliferation.
Funding
This publication was not supported by any funding, whether academic and private. Moreover, the writing of this manuscript was performed without any administrative assistance (Honorary Professor status).
Acknowledgments
I am extremely grateful to E. Magnier and J.-C. Blazejewski (retired) of the Institut Lavoisier of the Université of Versailles, St Quentin en Yveline (France), who produced the fluorinated compounds and their non-fluorinated analogs used in the present investigation. Thanks also to J.-C. Tabet (retired) of Université Pierre et Marie Curie, Paris 6, who introduced the concept of a potential contribution of an electronic movement within the investigated estrogen derivatives to the induction of the reported transcriptions. Thanks also to my colleagues at the laboratory and associated students who performed the reported experiments and whose names are specified in publications issued some years ago [19,24,27,44,55,56]. I also greatly appreciate the contribution of my friend P. Thoul, who produced the illustrations for this article.
Conflicts of Interest
The author declares no conflicts of interest.
References
- Katzenellenbogen, J.A. Stringing along the estrogen receptor to engage DNA. Proc. Natl. Acad. Sci. USA 2023, 120, e230060812. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Jin, S.; Chen, M.; Bueno, C.; Wolynes, P.G. The marionette mechanism of domain-domain communication in the antagonist, agonist, and coactivator response of the estrogen receptor. Proc. Natl. Acad. Sci. USA 2023, 120, e2216906120. [Google Scholar] [CrossRef] [PubMed]
- Arao, Y.; Korach, K.S. The physiological role of estrogen receptor functional domains. Essays Biochem. 2021, 65, 867–875. [Google Scholar] [CrossRef]
- Perissi, V.; Rosenfeld, M.G. Controlling nuclear receptors: The circular logic of cofactor cycles. Nat. Rev. Mol. Cell. Biol. 2005, 6, 542–554. [Google Scholar] [CrossRef]
- Tecalco-Cruz, A.C.; Ramirez-Jarquin, J.O.; Cruz-Ramos, E. Estrogen Receptor Alpha and its Ubiquitination in Breast Cancer. Curr. Drug Targets 2019, 20, 690–704. [Google Scholar] [CrossRef]
- Habara, M.; Shimala, M. Estrogen receptor a revised: Expression, function and stability. BioEssays 2022, 44, e2200148. [Google Scholar] [CrossRef] [PubMed]
- Arnal, J.-F.; Lenfant, F.; Metivierr, R.; Flouriot, G.; Henrion, D.; Adlanmerini, M.; Gourdy, P.; Chambon, P.; Katzenellenbogen, B.; Katzenellenbogen, J.A.; et al. Membrane and nuclear estrogen receptor alpha actions: From tissue specificity to medical implications. Physiol. Rev. 2017, 17, 1045–1087. [Google Scholar] [CrossRef]
- Winkeldfeld, S.R.; Lin, F.D.E. Communication between genomic and non-genomic signaling events coordinate steroid hormone actions. Steroids 2018, 133, 2–7. [Google Scholar]
- Leclercq, G. Pathological Maintenance and Evolution of Breast Cancer: The Convergence of Irreversible Biological Actions of ER Alpha. Endocrines 2021, 2, 1–14. [Google Scholar] [CrossRef]
- Marino, M.; Ascenzi, P.; Acconcia, F. S-palmitoylation modulates estrogen receptor alpha localisation and function. Steroids 2006, 71, 298–303. [Google Scholar] [CrossRef]
- Razandi, M.; Pedram, A.; Levin, E.R. Heat shock protein 27 is required for sex steroid receptor trafficking to and functioning at the plasma membrane. Mol. Cell. Biol. 2010, 30, 3249–3261. [Google Scholar] [CrossRef]
- Acconcia, A.; Fiocchetti, M.; Busnero, C.; Fernandez, V.S.; Montalesi, E.; Cipoletti, M.; Pallottini, V.; Marino, M. The extra-nuclear interactome of the estrogen receptors: Implications for the physiological functions. Molec. Cell. Endocrinol. 2021, 538, 111452. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Kim, K. Non Classical ER/Sp and AR/AP-1 signaling Pathways. J. Mol. Endocrinol. 2008, 41, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.; Nguyen, P.; Valentine, C.; Lopez, G.N.; Kwok, R.; Mclnerney, E.; Katzenellenbogen, B.S.; Enmark, E.; Gustafsson, A.; Nilsson, S.; et al. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol. Endocrinol. 1999, 13, 1672–1685. [Google Scholar] [CrossRef]
- Kushner, P.J.; Agard, D.A.; Greene, G.L.; Scanlan, T.S.; Shiau, A.K.; Uth, R.M.; Webb, P. Estrogen receptor pathways to A-P-1. J. Steroid Biochem. Mol. Biol. 2000, 74, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Acramel, A.; Jacquot, Y. Deciphering of a Putative GPER Recognition Domain in ERα and ERα36. Front. Endocrinol. 2022, 13, 943343. [Google Scholar] [CrossRef]
- Buzon, V.; Carbo, L.R.; Estruch, S.B.; Fletterick, R.J.; Estebanez-Perpina, E.A. A conserved surface on the ligand binding domain of nuclear receptors for allosteric controls. Mol. Cell. Endocrinol. 2012, 348, 394–402. [Google Scholar] [CrossRef]
- Fischer, A.; Smiesko, M. Allosteric Binding Sites On Nuclear Receptors: Focus on Drug Efficacy and Selectivity. Int. J. Mol. Sci. 2020, 21, 534. [Google Scholar] [CrossRef]
- Gallo, D.; Haddad, I.; Duvillier, H.; Jacquemotte, F.; Laïos, I.; Laurent, G.; Jacquot, I.; Vinh, H.; Leclercq, G. Trophic effect in MCF-7cells of ERalpha17p, a peptide corresponding to a platform regulatory motif of the estrogen receptor alpha—Underlying mechanism. J. Steroid Mol. Biol. 2008, 109, 138–149. [Google Scholar] [CrossRef]
- Métivier, R.; Penot, G.; Flouriot, G.; Pakdel, P. Synergism between ER alpha transactivation function 1 (AF-1) and AF-2 mediated by steroid receptor coactivator protein-1: Requirement for AF-I alpha-helical core and for a direct interaction between the N- and C- terminal domains. Mol. Endocrinol. 2001, 15, 1953–1970. [Google Scholar]
- Zwart, W.; de Leeuw, R.; Rondaij, M.; Neefjes, J.; Mancini, M.A.; Michalides, R. The hinge region of the human estrogen receptor determines functional energy between AF-1 and AF-2 in the quantitative response to estradiol and tamoxifen. J. Cell Sci. 2010, 123, 1253–1261. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Singleton, D.W.; Shaughnessy, S.A.; Khan, S.A. The F-domain of estrogen receptor-alpha inhibits ligand induced receptor dimerization. Mol. Cell Endocrinol. 2008, 295, 94–100. [Google Scholar] [CrossRef]
- Carlson, K.E.; Choi, I.; Gee, G.; Katzenellenbogen, B.S.; Katzenellenbogen, J.A. Altered Ligand binding properties and enhanced stability of a constitutively active estrogen receptor: Evidence that an open pocket conformation is required for ligand interaction. Biochemistry 1997, 36, 14897–14905. [Google Scholar] [CrossRef]
- El Khisiin, A.; Leclercq, G. Exchange of bound estrogens and antiestrogens In MCF-7 cell: Evidence for ligand-induced stable configurations of the estrogen receptor. Steroids 1998, 63, 565–574. [Google Scholar] [CrossRef]
- Le Romancer, M.; Poulard, C.; Cohen, P.; Sentis, S.; Renoir, J.-M.; Corbo, L. Cracking the estrogen receptors posttranscriptional code in breast tumors. Endocr. Rev. 2011, 32, 597–622. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Hedman, A.C.; Ames, J.B.; Sacks, D.B. Calmodulin lobes facilitate dimerization and activation of estrogen receptor-α. J. Biol. Chem. 2017, 292, 4614–4622. [Google Scholar] [CrossRef] [PubMed]
- Bourgouin-Voillard, S.; Fournier, F.; Alfonso, C.; Jacquot, Y.; Leclercq, G.; Tabet, J.-C. Calmodulin association with the synthetic ERα 17p peptide investigated by Mass Spectrometry. Int. J. Mass Spectrom. 2011, 305, 87–94. [Google Scholar] [CrossRef]
- Berry, N.B.; Fan, M.; Nephew, K.P. Estrogen Recceptor-α Hinge- Region Lysines 302 and 303 RegulateReceptor Degadation by the prteasome. Mol. Endocrinol. 2008, 22, 1535–1551. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, J.A. The 2010 Philip S. Portoghese Medicinal Chemistry Lecture: Addressing the 3 core Issue in the Design of Estrogen Receptor Ligands. J. Med. Chem. 2011, 54, 5271–5282. [Google Scholar] [CrossRef]
- Amstead, G.M.; Carson, K.M.; Katzenellenbogen, J.A. The estradiol pharmacophore: Ligand structure-estrogen receptor binding affinity relationships and a model for the receptor binding site. Steroids 1997, 62, 268–303. [Google Scholar] [CrossRef] [PubMed]
- Kekenes-Huskey, P.M.; Megge, I.; von Rauch, M.; Gust, R. Knapp. A molecular Docking study of Estrrogenically active componds with 1,2-diarylethane and 1,é-diarylethenePharmacophore. Biorg. Med. Chem. 2004, 12, 6527–6537. [Google Scholar] [CrossRef]
- Ogawa, T.; Otha, K.; Ijima, T.; Suzuki, T.; Otha, S.; Endo, Y. Synthesis and biological evaluation of p-carborane bispheols and their derivatives: Structure-activity relationship for estrogenic activity. Biorg. Med. Chem. 2009, 17, 1109–1117. [Google Scholar] [CrossRef]
- Lorand, T.; Vigh, E.; Garai, J. Hormonal action of plant derived and anthropogenic non-steroidal estrogenic compounds: Phytoestrogens and xenoestrogens. Curr. Med. Chem. 2010, 17, 2632–2653. [Google Scholar] [CrossRef]
- Jordan, V.C.; Schafer, J.M.; Levenson, A.S.; Liu, H.; Pease, K.M.; Simons, L.A.; Zapf, J.W. Molecular classificatin of estrogens. Cancer Res. 2001, 61, 6619–6623. [Google Scholar]
- Zhang, J.-X.; Labaree, D.C.; Hochberg, R.B. Nonpolar and short side chain groups at C-11beta of estradiol result in antiestrogens. J. Med. Chem. 2005, 48, 1428–1447. [Google Scholar] [CrossRef] [PubMed]
- Hanson, R.N.; Hua, E.; Hendricks, A.; Labaree, D.; Hochberg, R.B. Synthesis and evaluation of 11β-(4-substituted phenyl) estradiol analogs: Transition from estrogen receptor agonist to antagonist. Biorg. Med. Chem. 2012, 20, 3768–3780. [Google Scholar] [CrossRef] [PubMed]
- Agouridas, V.; Blazejewski, J.-C.; Cleeren, A.; Laïos, I.; Leclercq, G.; Magnier, E. Fluorous tolerance of the estrogen receptor alpha as probed by 11-fluoroalkylestradiol derivatives. Steroids 2008, 73, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Bourgouin-Voillard, S.; Fournier, F.; Alfonso, C.; Zins, E.-L.; Jacquot, Y.; Pèpe, C.; Leclercq, G.; Tabet, J.-C. Electronic effect of 11βsubstituted 17β-estradiol and instrumental effects on the relative gas phase acidity. J. Am. Soc. Mass Spectrom. 2012, 23, 2167–2177. [Google Scholar] [CrossRef]
- Jakaka, M.; Ito, M.; Weis, J.; Chien, P.; Gehem, B.D.; Jameson, J.L. Estrogen receptor binding to DNA is not required for its activity through-h the nonclassical AP1 pathway. J. Biol. Chem. 2001, 276, 13615–13621. [Google Scholar] [CrossRef] [PubMed]
- Heldring, N.; Isaacs, G.D.; Diehl, A.G.; Sun, M.; Cheung, E.; Ranish, J.A.; Lee Kraus, W. Multiple sequence-specific DNA-binding proteins mediate estrogen receptor signaling through a tethering pathway. Mol. Endocrinol. 2011, 25, 564–574. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Sinha, I.; Fan, R.; Haldosen, L.-A.; Zhao, C.; Dalman-Wright, K. c-Jun/AP-1 overexpression reprograms ERα signaling related to tamoxifen response in in ERα -positive breast cancer. Oncogene 2018, 19, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Lian, Y.; Zhang, L. The potential activator protein 1 (AP-1)in breast cancer targeted therapy. Front. Immunol. 2023, 14, 1224892. [Google Scholar] [CrossRef] [PubMed]
- Cheung, E.; Acevedo, M.L.; Cole, P.A.; Lee Kraus, W. Altered pharmacology and distinct coactivators usage for estrogen receptor-dependent transcription through activating protein -1. Proc. Natl. Acad. Sci. USA 2005, 102, 559–564. [Google Scholar] [CrossRef]
- Maaroufi, Y.; Leclercq, G. Importance of AB and C Domains of the Estrogen Receptor for its Adsorption to Hydroxylapatite. J. Steroid Biochem. Mol. Biol. 1994, 48, 155–1631. [Google Scholar] [CrossRef]
- Spera, D.; Cabrera, G.; Fiashi, R.; Carlson, K.E.; Katzenellenbogen, J.A.; Napolitano, E. Estradiol derivatives bearing sulfur-containing substituents at the 11β or7α positions: Versatile reagents for the preparation of estrogen conjugates. Biorg. Med. Chem. 2004, 12, 4393–4401. [Google Scholar] [CrossRef]
- Pons, M.; Gagne, D.; Nicolas, J.-C.; Methalli, M.A. A new cellular model of response to estrogens: A bioluminescent test to characterize (anti)estrogen molecules. Biotechniques 1990, 9, 450–459. [Google Scholar]
- Astruc, M.E.; Chabret, C.; Ball, P. Prolonged Treatment of breast cancer cells with antiestrogens increases the activating propein-1-mediated response: Involvement of the estrogen receptor. Endocrinology 1995, 136, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Bourgoin-Voillard, S.; Gallo, D.; Laïos, I.; Cleeren, A.; El Bali, L.; Jacquot, Y.; Nonclercq, D.; Laurent, G.; Tabet, J.-C.; Leclercq, G. Capacity of Type I and II to confer to estrogen receptor alpha an appropriate conformation for the recruitment of coactivators containing a LxxLL motif—Relationship with the regulation of receptor level and ERE-dependent transcription in MCF-7 cells. Biochem. Pharmacol. 2010, 79, 746–757. [Google Scholar] [CrossRef][Green Version]
- Otto, A.M. A one minute pulse of estradiol to MCF-7 breast cancer cells changes estrogen receptor binding properties and commits cells to induce estrogenic responses. J. Steroid Biochem. Mol. Biol. 1995, 54, 39–46. [Google Scholar] [CrossRef]
- Lippman, M.E.; Dickson, R.B.; Kasid, A.; Gelmann, E.; Davidson, N.; McManaway, M.; Huff, K.; Bronzert, D.; Bates, S.; Swain, S.J. Autocrine and pararacrine growth regulation of breast cancer. J. Steroid Biochem. 1986, 24, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Levenson, S.A.; Mac Gregor Schafer, J.I.; Bentrem, D.J.; Pease, K.M.; Jordan, V.C. Control of the estrogen-like actions of the tamoxifen-estrogen receptor complex by the surface amino acid at position 351. J. Steroid Biochem. Mol. Bol. 2001, 76, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Vallet, A.; El Ezzy, M.; Diennet, M.; Haidar, S.; Bouvier, M.; Mader, S. The AF-2 cofactor binding region is a key for the selective SUMOylation of estrogen receptor alpha by antiestrogens. J. Biol. Chem. 2023, 229, 102757. [Google Scholar] [CrossRef] [PubMed]
- Traboulsi, T.; El Ezzy, M.; Dumeaux, V.; Audemard, E.; Mader, S. Role of SUMOylation in differential ERα transcriptional repression by tamoxifen and fulvestrant in breast cancer cells. Oncogene 2019, 38, 1019–1037. [Google Scholar] [CrossRef]
- Himi, K.; Hussein, N.; Mendoza-Sanchez, R.; El-Ezzy, M.; Ismail, H.; Durette, C.; Bail, M.; Rozendaal, M.J.; Bouvier, M.; Thibaut, P.; et al. Role of SUMOylation in Full Antiestrogenic. Mol. Cell. Biol. 2012, 32, 3823–3837. [Google Scholar]
- Agouridas, V.; Magnier, E.; Blazejewski, J.-C.; Laïos, I.; Cleeren, A.; Nonclercq, D.; Laurent, G.; Leclercq, G. Effect of Fluorination on the Pharmacological Profile of Fulvestrant in Breast Carcinoma Cells. J. Med. Chem. 2009, 52, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Borras, M.; Lacroix, M.; Legros, N.; Leclercq, G. Antiestrogenic activity of two 11β-estradiol derivatives on MCF-7 breast cancer cells. Steroids 1995, 60, 512–518. [Google Scholar] [CrossRef]
- Tempé, D.; Vives, E.; Brocky, F.; Brooks, H.S.; De Rossi, S.; PLechaczyk, M.; Bossis, G. SUMOylation of the inductible (c-Fois:c-Jun)/AP-1 transcription complex occurs on target promoters to limit Transcriptional activation. Oncogene 2014, 33, 921–927. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, N.; Yang, X.; Feng, S.; Wang, F.; Zhang, W.; He, Z. ERα promotes SUMO1- mediated protein SUMOylation in breast cancer. Gland Surg. 2023, 12, 963–973. [Google Scholar] [CrossRef]
- Guan, J.; Zhou, W.; Hafner, M.; Blake, R.A.; Chalouni, C.; Chen, I.P.; De Bruyn, T.; Giltnane, J.M.; Hartman, S.J.; Heidersbach, A.; et al. Therapeutic Ligands Antagonize Estrogen Receptor Function By Impairing Its Mobility. Cell 2019, 178, 949–963. [Google Scholar] [CrossRef]
- Scherbacov, A.M.; Sorokin, D.V.; Omelchuk, O.A.; Shchekotikhin, A.E.; Krasil’nikov, M.A. Glucose starvation greatly enhance antiproliferative and antiestrogenic potency of oligomycin A in MCF-7 breast cancer. Biochimie 2021, 186, 51–58. [Google Scholar] [CrossRef]
- Klinge, C.M. Estrogens regulates life and death in mitochondria. J. Bioenerg. 2017, 49, 307–324. [Google Scholar] [CrossRef] [PubMed]
- Maximov, P.Y.; Curpan, P.; Jordan, V.C. Estrogen Receptor Complex to Trigger or Delay Estrogen-Induced Apoptosis in Long Term Estrogen Deprived Breast Cancer. Front. Endocrinol. 2022, 13, 869562. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.; Derocq, D.; Freiss, G.; Rochefort, H. Activation of estrogen receptor transfected into a receptor-negative breast cancer cell line decreases the metastatic and invasive potential of the cells. Proc. Natl. Acad. Sci. USA 1992, 89, 11518–11542. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Murphy, L.C.L. Regulation of steroid hormones receptors and co regulators during cycle highlights potential novel functions in addition to transcription factors. Nuclear Recept. Signal 2016, 14, e001. [Google Scholar]
- JavanModoghadam, S.; Weihua, Z.; Hunt, K.K.; Keyomarsi, K. Estrogen receptor alpha is cell cycle-regulated and regulates cell cycle in a ligand-dependent fashion. Cell Cycle 2016, 15, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Kaufman, P.D. Ki-67:More than a prokiferation marker. Chromosoma 2018, 127, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Nettles, K.W.; Bruning, J.B.; Gil, G.; O’Neil, E.E.; Nowak, J.; Guo, J.; Kim, Y.; De Sombre, R.; Dilis, R.; Hanson, R.N.; et al. Structural plasticity in the estrogen receptor ligand-binding domain. Look. Nucl. Recept. New Rep. 2007, 8, 563–568. [Google Scholar]
- Khan, S.H.; Braet, S.M.; Koehler, S.J.; Elacua, E.; Anand, G.S.; Okafor, C.D. Ligand-induced shifs in conformational ensembles that describe transcriptioal activation. Elife 2022, 11, e80140. [Google Scholar] [CrossRef]
- Helsen, C.; Claessens, F. Looking at nuclear receptors from a new angle. Mol. Cell. Endocrinol. 2014, 382, 97–106. [Google Scholar] [CrossRef]
- WeiKum, E.R.; Liu, X.; Ortlund, E.A. The nuclear receptor superfamily: A structural Perspective. Protein Sci. 2018, 27, 1876–1892. [Google Scholar] [CrossRef]
- Bourgoin-Voillard, S.A.; Zins, E.-E.; Fournier, F.; Jacquot, Y.; Alfonso, C.; Pèpe, C.; Leclercq, G.; Tabet, J.-C. Stereochemical Effects During [M-H]-Dissociations of Epimeric 11-OH-17β Estradiols at Distant Electronic Effects of substituents at Position C(11) on Gas Phase Acidity. J. Am. Soc. Mass Spectrom. 2009, 20, 3318–3333. [Google Scholar] [CrossRef] [PubMed][Green Version]
- French, A.N.; Napolitano, E.; VanBroklin, R.N.; Hanson, R.N.; Welch, R.J.; Katenellenbogen, J.A. Synthesis, radiolabeling and tissue distribution of 11 beta- fluoroalkyl- and 11 beta fluoroalkoxy- substituted estrogens: Target tissue uptake activity and defluorination of a homologous series of fluorine-18-labeled estrogens. Nucl. Med. Biol. 1993, 20, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Eignerova, B.; Sedlak, D.; Drcinsky, M.; Bartunek, P.; Kotora, M. Synthesis and Biochemical Characterization of 17α-Perfluoro alkylated Estradiols as Selective Ligands for Estrogen Receptor α. J. Med. Chem. 2010, 53, 6947–6953. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).