Insights into the Mechanism of Action of Helianthus annuus (Sunflower) Seed Essential Oil in the Management of Type-2 Diabetes Mellitus Using Network Pharmacology and Molecular Docking Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
Source of Seeds
2.2. Methods
2.2.1. Seed Preparation, Extraction, and Chromatographic Analysis
2.2.2. Screening of Active Compounds, Drug Therapeutics, and Disease Targets
2.2.3. Protein–Protein Interaction (PPI) Network Construction
2.2.4. Kyoto Encyclopaedia of Genes and Genomes (KEGG) Pathways Enrichment Analyses
2.2.5. Molecular Docking
3. Results
3.1. Compounds Identification and ADME Properties Screening
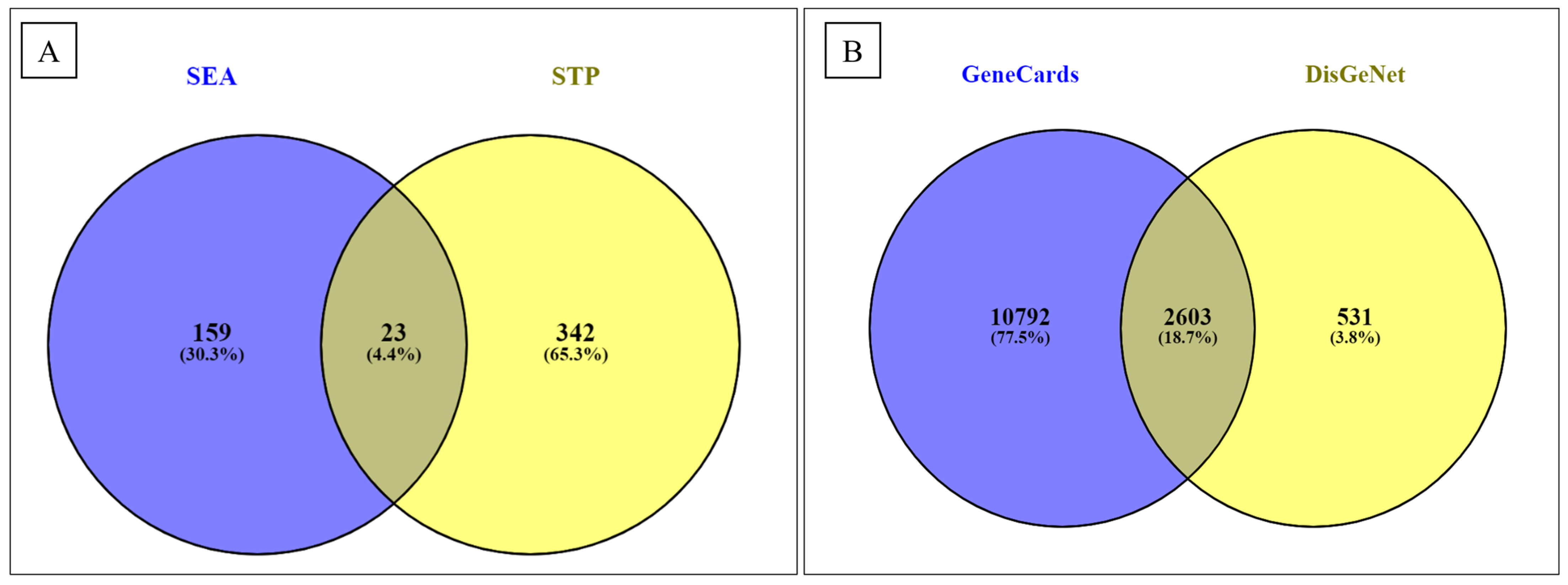
3.2. Screening of Active Compounds and Their Targets
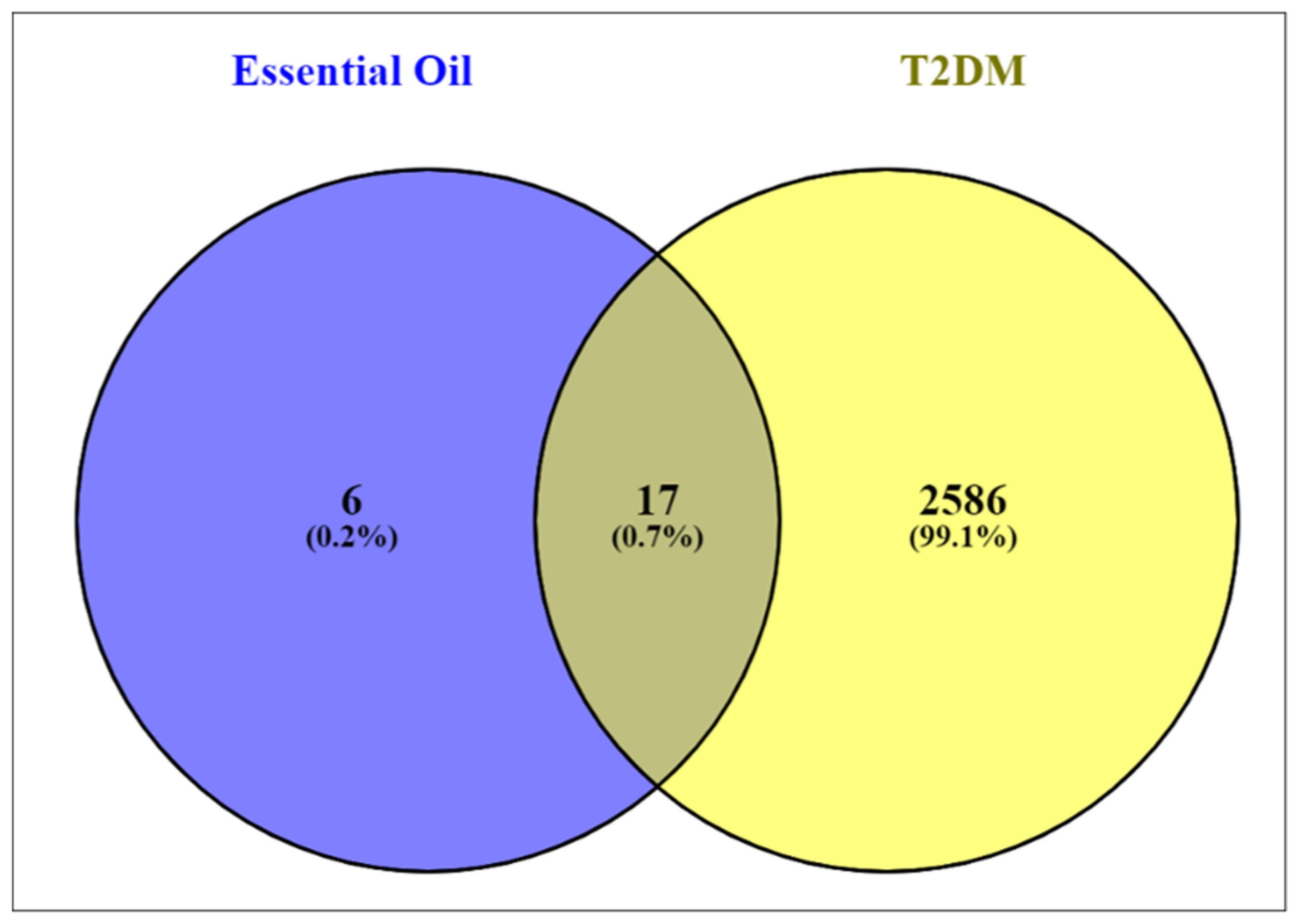
3.3. Screening of T2D Disease Targets, Drugs, and Disease Candidates
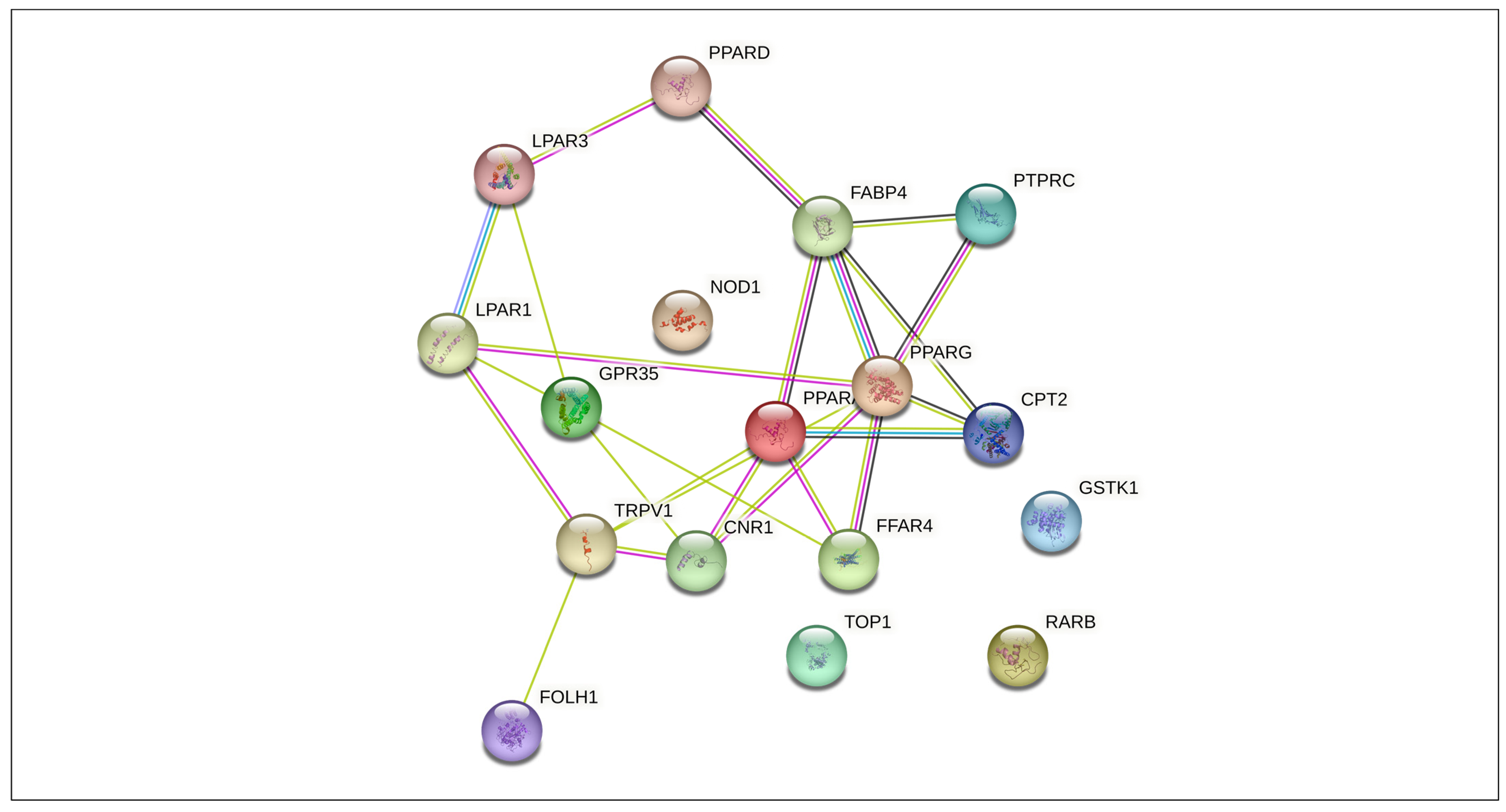
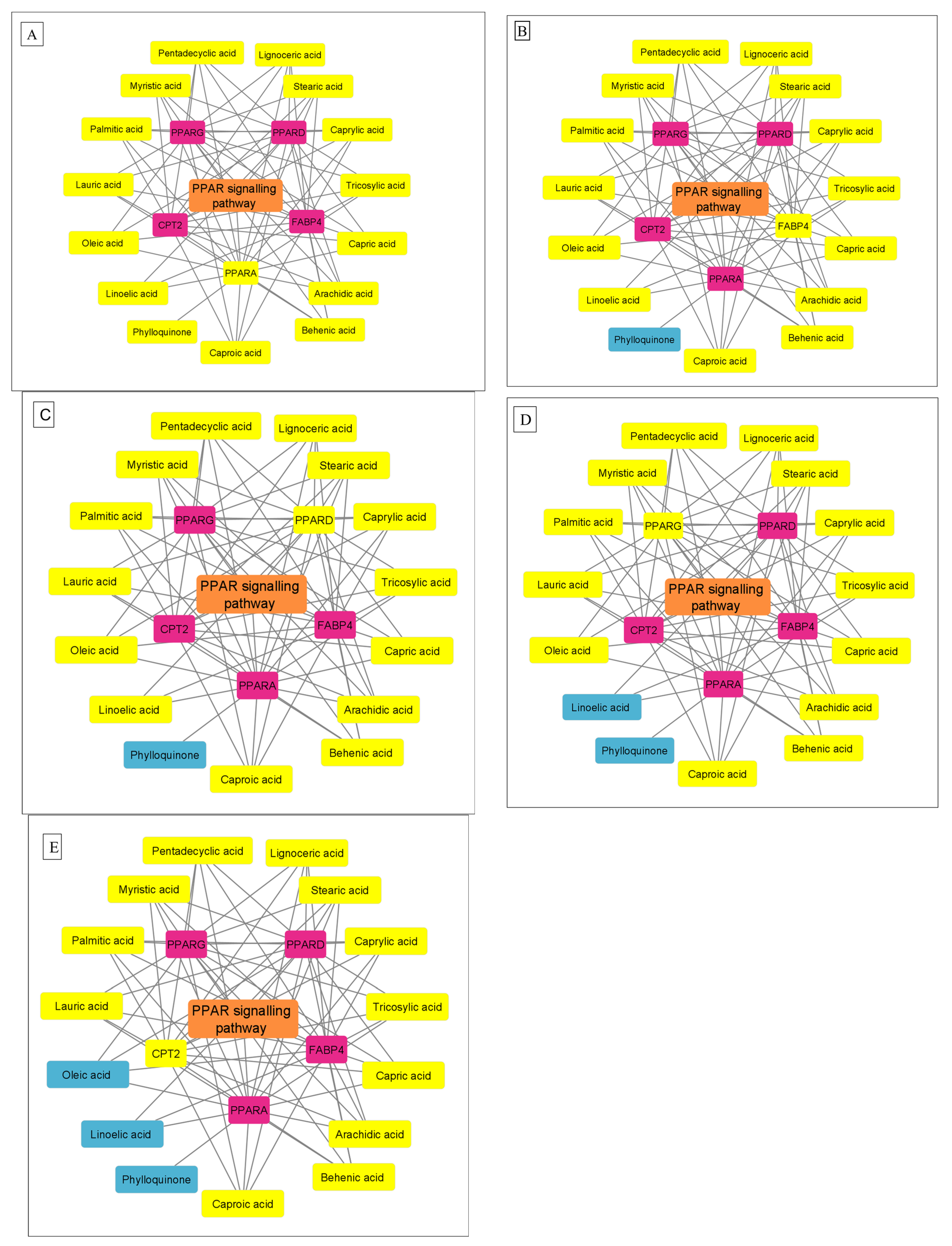
3.4. Protein–Protein Interaction Network
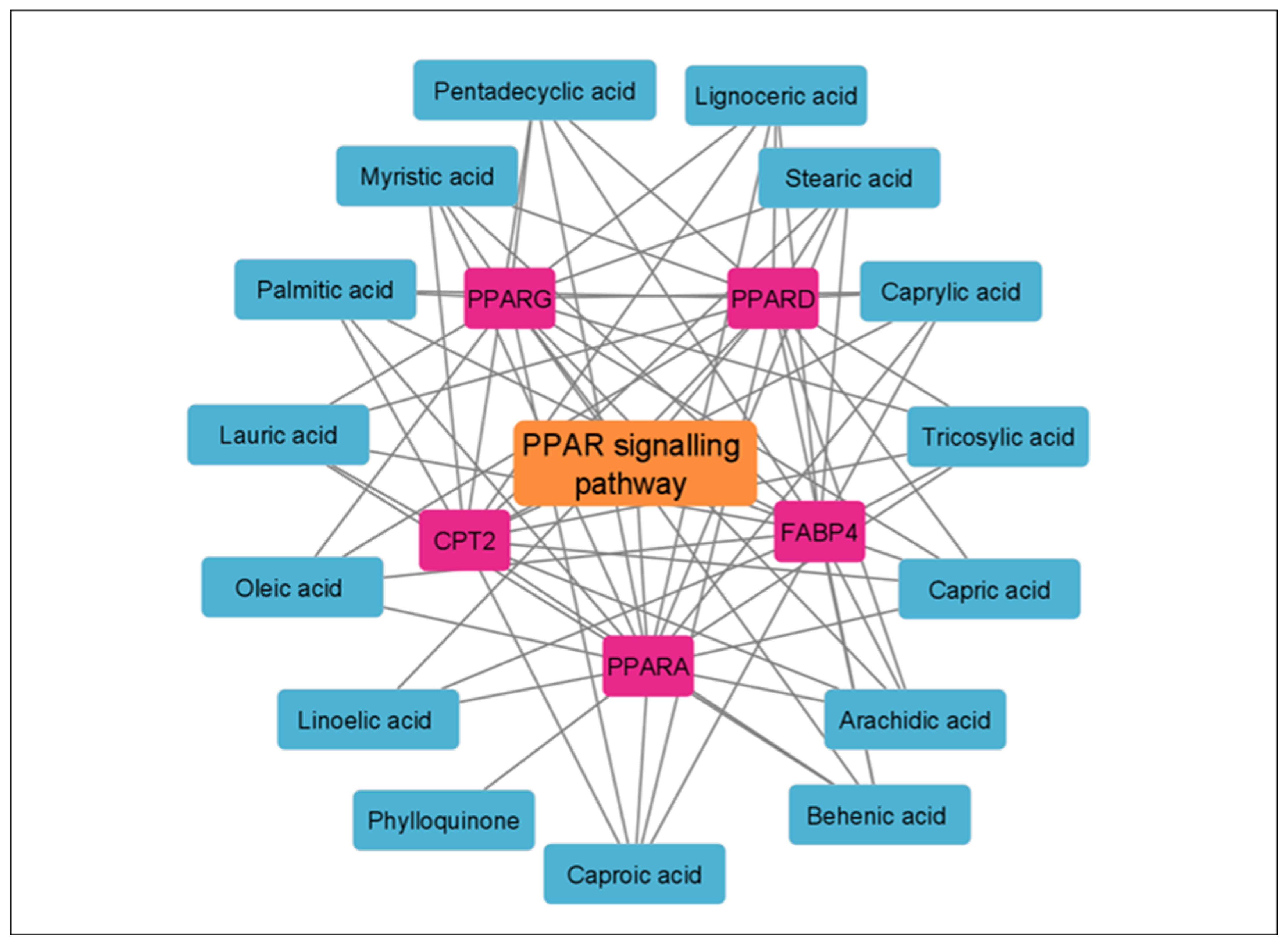
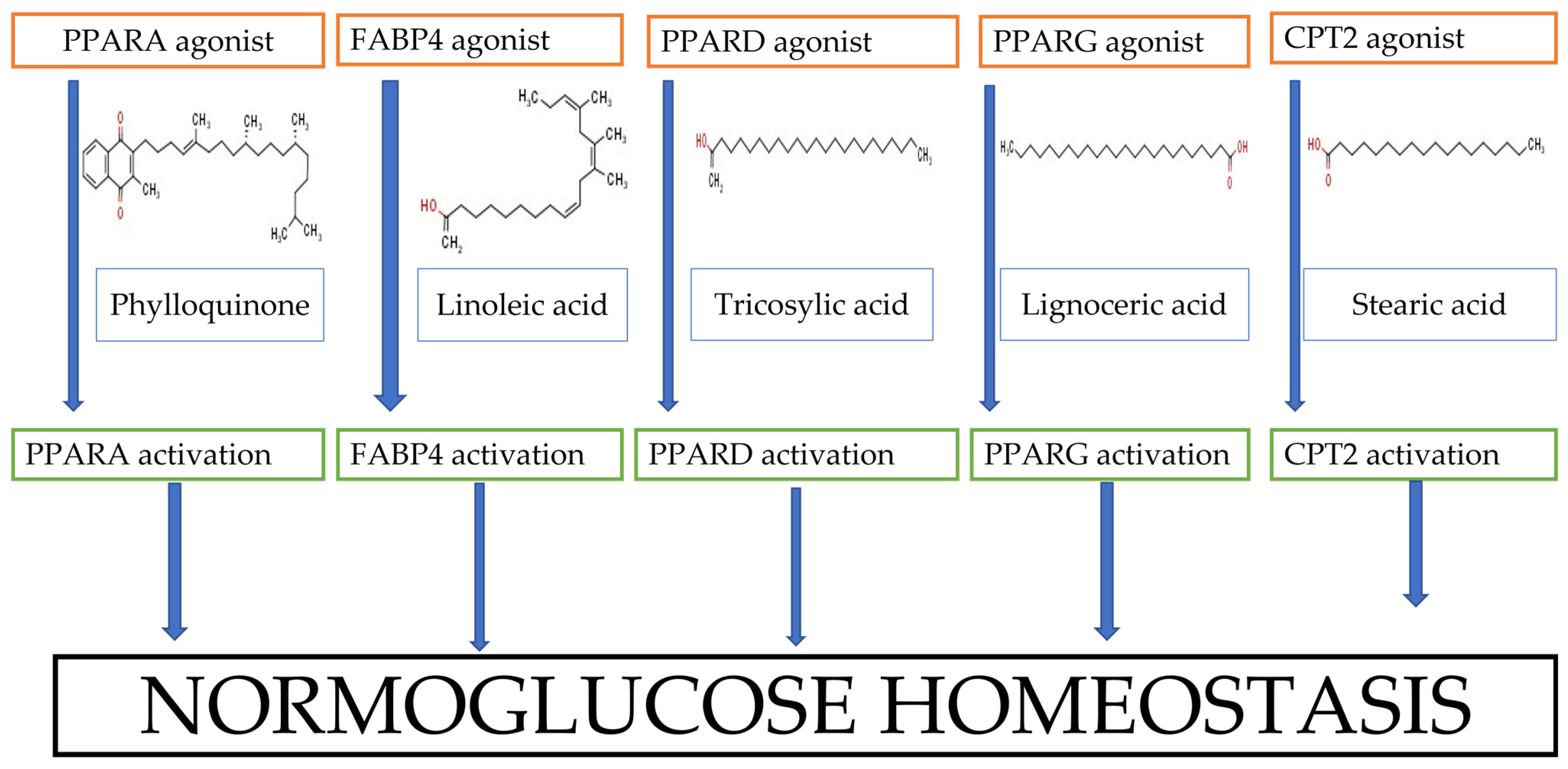
3.5. KEGG Enrichment Analysis
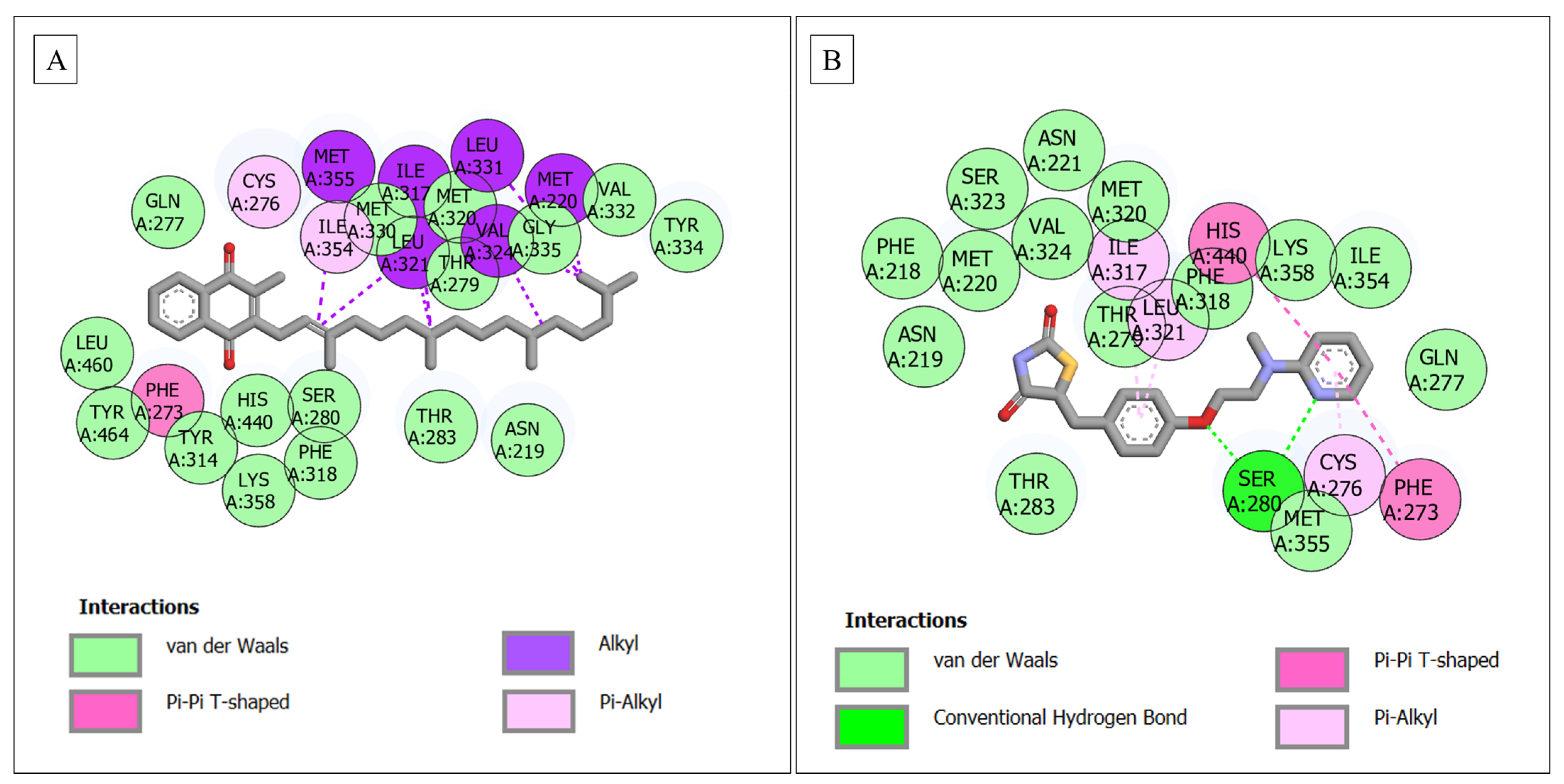
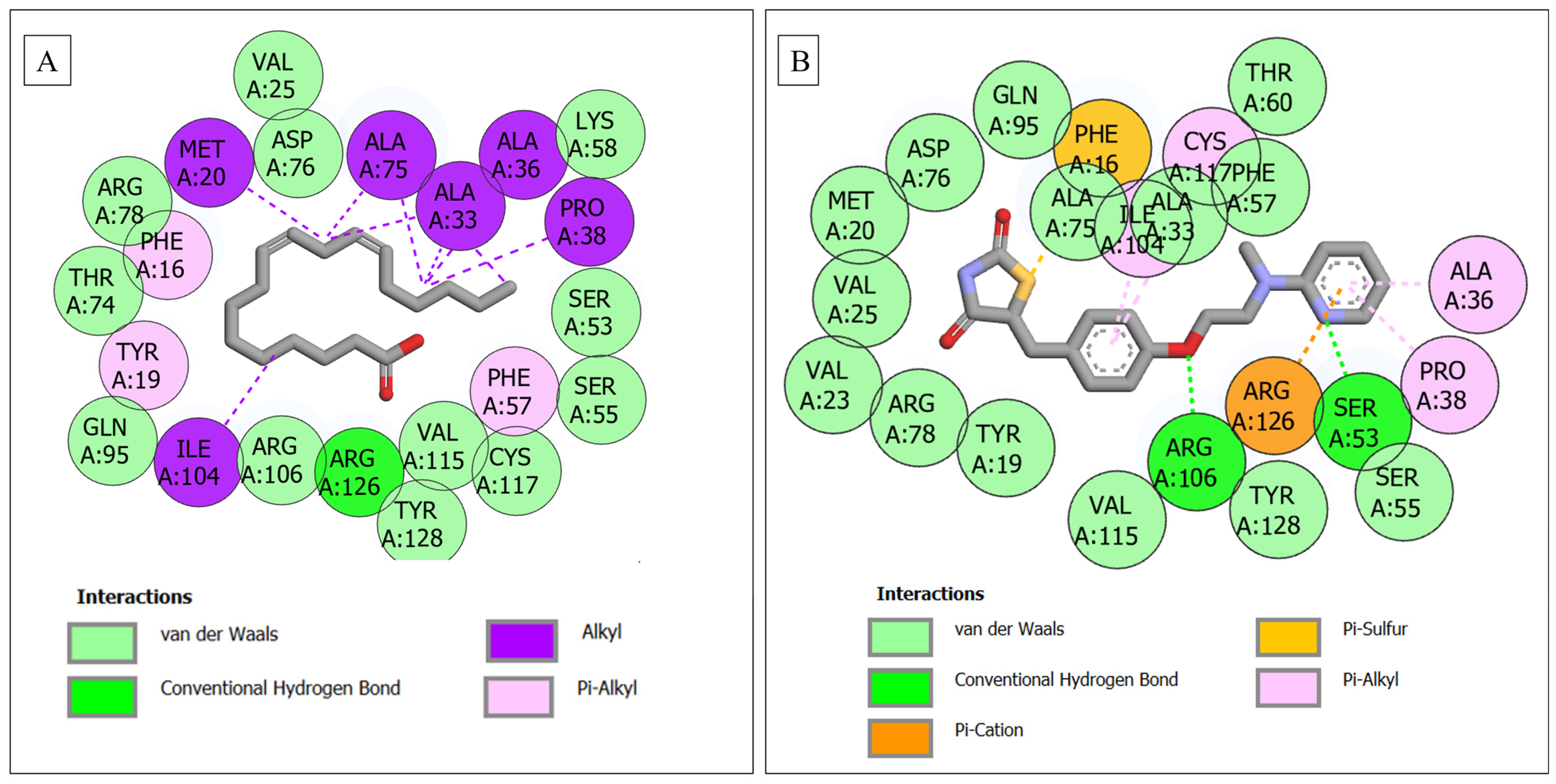
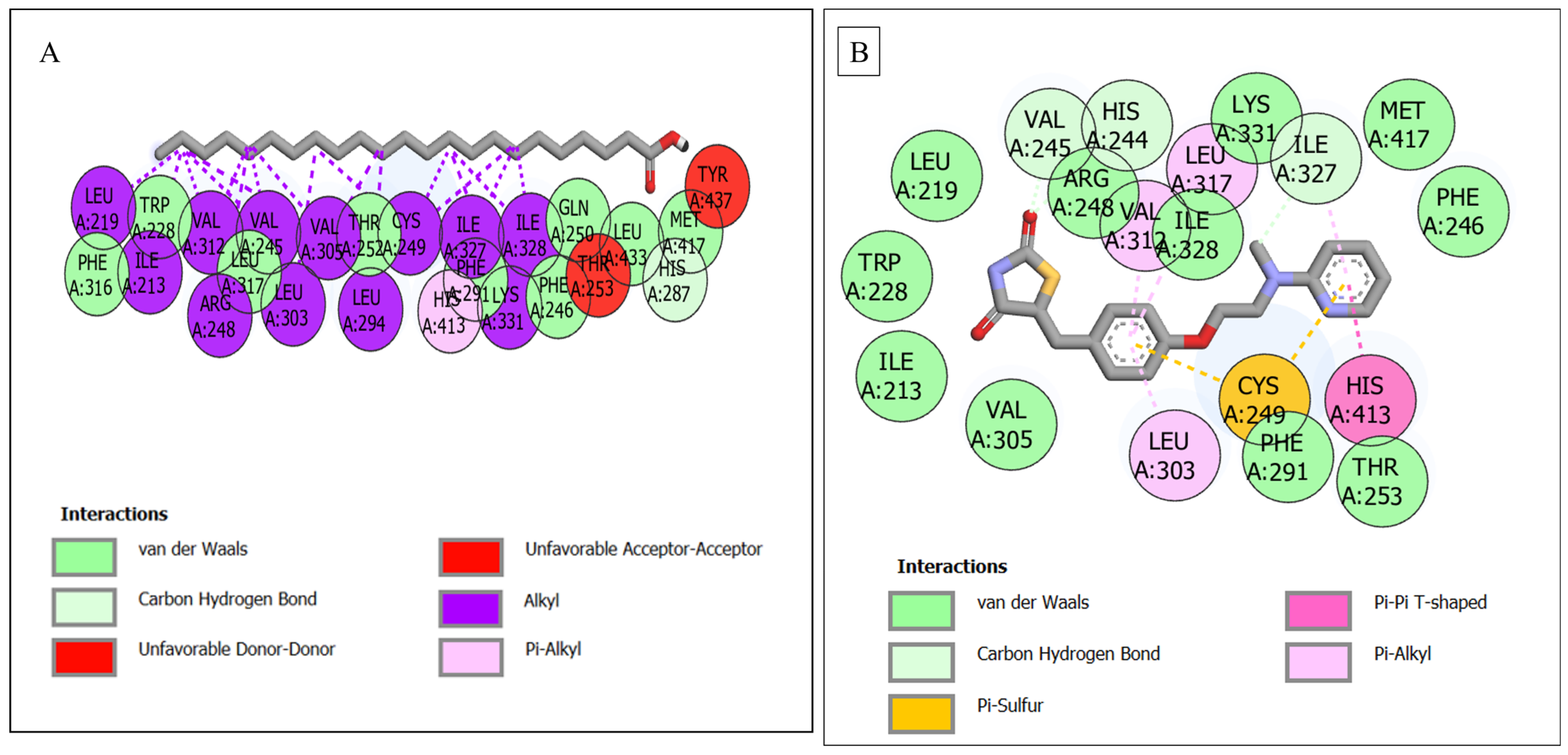
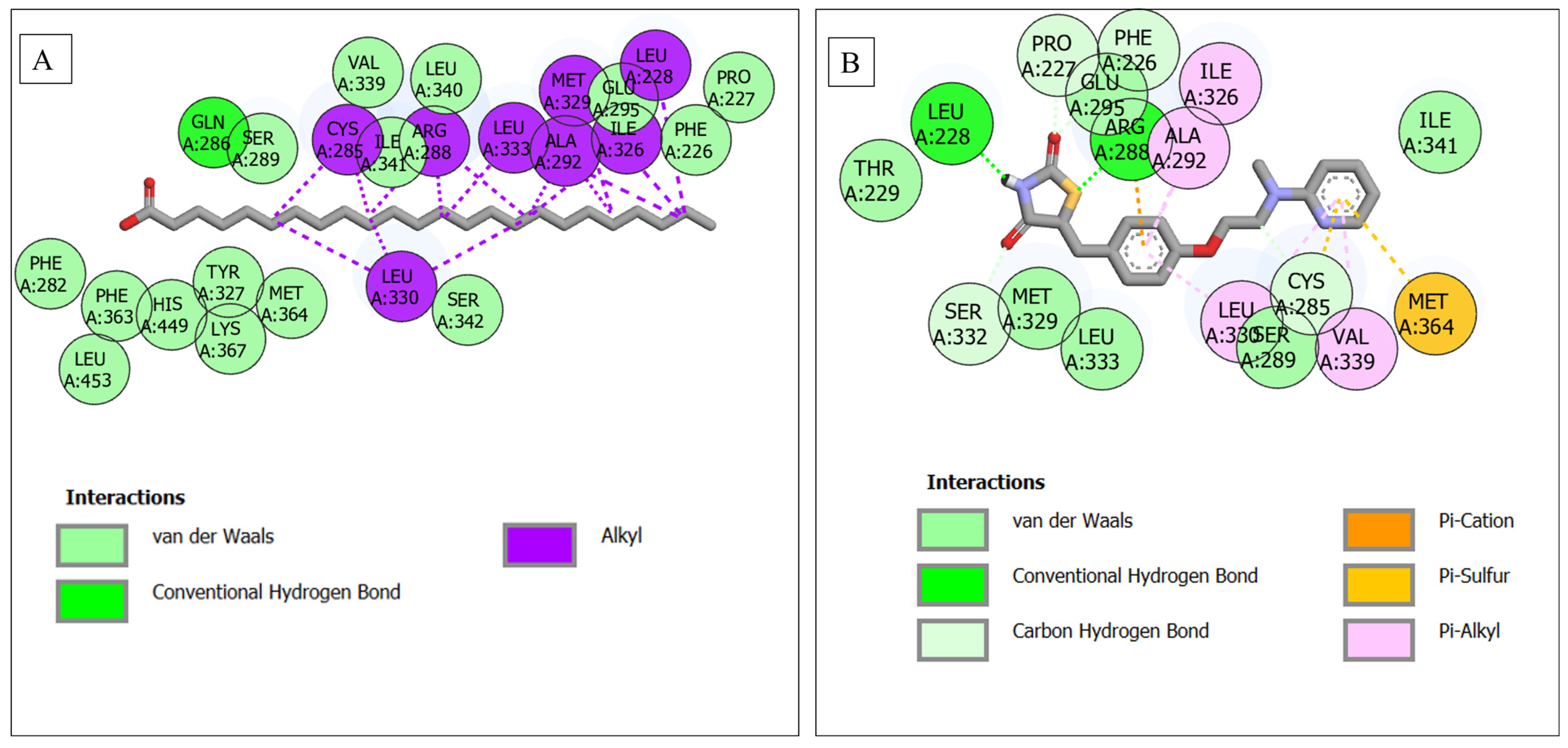
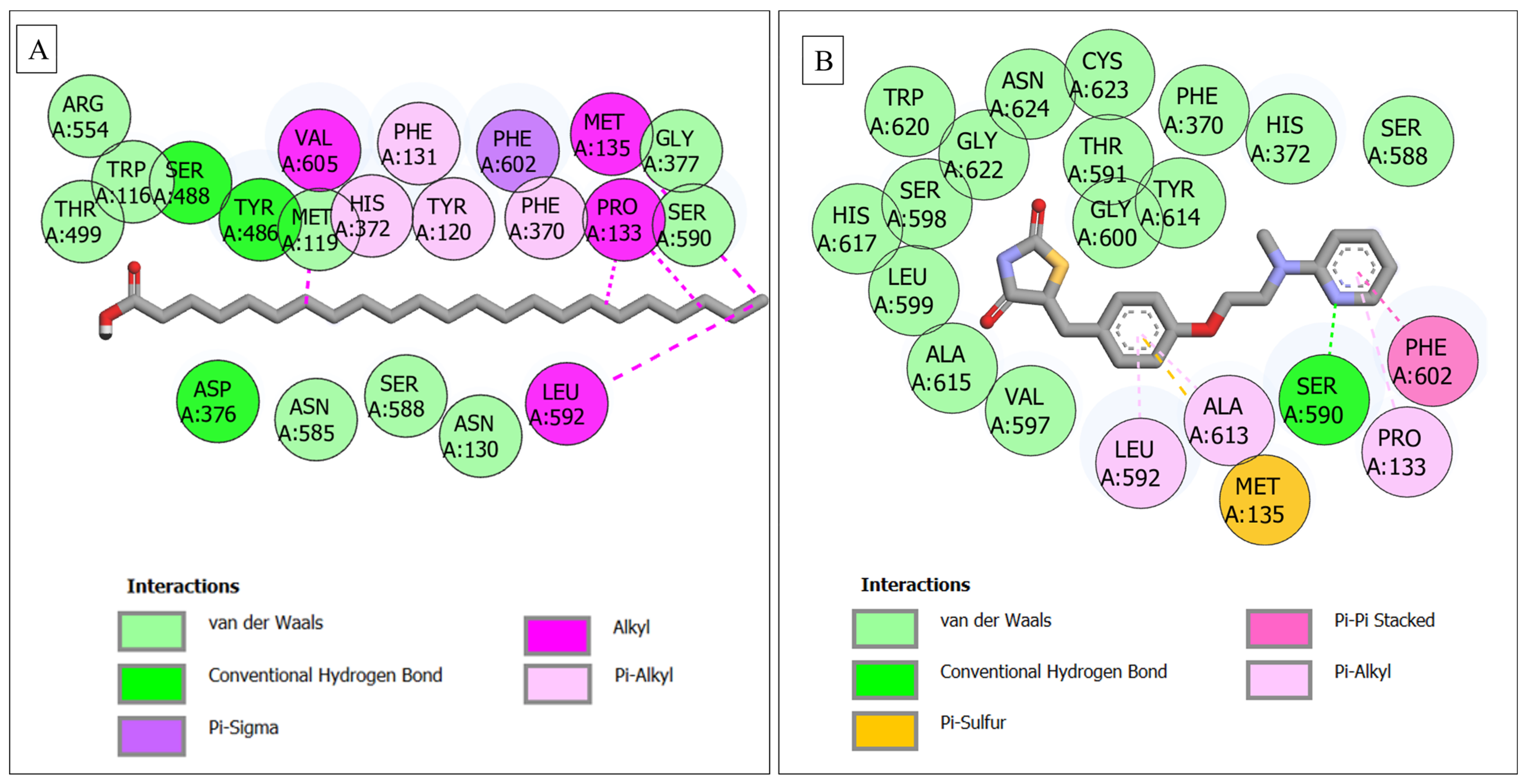
3.6. Molecular Docking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- de Diabetes, A.E. Clasificación y diagnóstico de la diabetes. Diabetes Care 2016, 39, S13–S22. [Google Scholar]
- World Health Organization. Diabetes, Key Facts. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/diabetes (accessed on 24 November 2022).
- Reed, J.; Bain, S.; Kanamarlapudi, V. A review of current trends with type 2 diabetes epidemiology, aetiology, pathogenesis, treatments and future perspectives. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 3567–3602. [Google Scholar] [CrossRef]
- Guariguata, L.; Whiting, D.R.; Hambleton, I.; Beagley, J.; Linnenkamp, U.; Shaw, J.E. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res. Clin. Pract. 2014, 103, 137–149. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021; Available online: https://www.diabetesatlas.org (accessed on 11 April 2023).
- Chennaiah, A.; Bhowmick, S.; Vankar, Y.D. Conversion of glycals into vicinal-1,2-diazides and 1,2-(or 2,1)-azidoacetates using hypervalent iodine reagents and Me3SiN3. Application in the synthesis of N-glycopeptides, pseudo-trisaccharides and an iminosugar. RSC Adv. 2017, 7, 41755–41762. [Google Scholar] [CrossRef]
- Rajasekaran, P.; Ande, C.; Vankar, Y.D. Synthesis of (5,6 & 6,6)-oxa-oxa annulated sugars as glycosidase inhibitors from 2-formyl galactal using iodocyclization as a key step. ARKIVOC 2022, 6, 5–23. [Google Scholar]
- Po-Sen, T.; Chennaiah, A.; Moremen, K.W.; David, C. Influence of Side Chain Conformation on the Activity of Glycosidase Inhibitors. Angew. Chem. 2022, 135, e202217809. [Google Scholar] [CrossRef]
- Yakubu, M.T.; Sunmonu, T.O.; Lewu, F.B.; Ashafa, A.O.; Olorunniji, F.J.; Eddouks, M. Medicinal plants used in the management of diabetes mellitus 2015. Evid. Based Complement. Altern. Med. 2015, 2015, 467196. [Google Scholar] [CrossRef]
- Kooti, W.; Farokhipour, M.; Asadzadeh, Z.; Ashtary-Larky, D.; Asadi-Samani, M. The role of medicinal plants in the treatment of diabetes: A systematic review. Electron. Physician 2016, 8, 1832. [Google Scholar] [CrossRef]
- Panero, J.; Funk, V.A. Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae). Proc. Biol. Soc. Wash. 2002, 115, 760–773. [Google Scholar]
- Gai, F.; Karamać, M.; Janiak, M.A.; Amarowicz, R.; Peiretti, P.G. Sunflower (Helianthus annuus L.) Plants at Various Growth Stages Subjected to Extraction-Comparison of the Antioxidant Activity and Phenolic Profile. Antioxidants 2020, 19, 535. [Google Scholar] [CrossRef]
- United States Department of Agriculture. Oilseeds: World Market and Trades; United States Department of Agriculture: Washington, DC, USA, 2023. [Google Scholar]
- Hosni, T.; Abbes, Z.; Abaza, L.; Medimagh, S.; Ben Salah, H.; Kharrat, M. Biochemical Characterization of Seed Oil of Tunisian Sunflower (Helianthus annuus L.) Accessions with Special Reference to Its Fatty Acid Composition and Oil Content. J. Food Qual. 2022, 2022, 2875072. [Google Scholar] [CrossRef]
- Rauf, S.; Jamil, N.; Tariq, S.A.; Khan, M.; Kausar, M.; Kaya, Y. Progress in modification of sunflower oil to expand its industrial value. J. Sci. Food Agric. 2017, 97, 1997–2006. [Google Scholar] [CrossRef] [PubMed]
- Akkaya, M. Fatty acid compositions of sunflowers (Helianthus annuus L.) grown in east Mediterranean region. Riv. Ital. Delle Sostanze Grasse 2018, 95, 239–247. [Google Scholar]
- Gavrilova, V.; Shelenga, T.; Porokhovinova, E.; Dubovskaya, A.; Kon’kova, N.; Grigoryev, S.; Podolnaya, L.; Konarev, A.; Yakusheva, T.; Kishlyan, N. The diversity of fatty acid composition in traditional and rare oil crops cultivated in Russia. Boil. Commun. 2020, 65, 68–81. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. Oilseed crop sunflower (Helianthus annuus) as a source of food: Nutritional and health benefits. Food Sci. Nutr. 2020, 8, 4666–4684. [Google Scholar] [CrossRef]
- Subashini, R.; Rakshitha, S.U. Phytochemical screening, antimicrobial activity and in vitro antioxidant investigation of methanolic extract of seeds from Helianthus annuus L. Chem. Sci. Rev. Lett. 2012, 1, 30–34. [Google Scholar]
- Guo, S.; Ge, Y.; Na Jom, K. A review of phytochemistry, metabolite changes, and medicinal uses of the common sunflower seed and sprouts (Helianthus annuus L.). Chem. Cent. J. 2017, 11, 1–10. [Google Scholar] [CrossRef]
- Al, N.B.M.B.A.; Hossain, M.A. Antimicrobial and cytotoxic potential of seeds and flowers crude extracts of sunflower. Grain Oil Sci. Technol. 2019, 2, 103–108. [Google Scholar]
- Balogun, F.O.; Abdulsalam, R.A.; Ojo, A.O.; Cason, E.; Sabiu, S. Chemical Characterization and Metagenomic Identification of Endophytic Microbiome from South African Sunflower (Helianthus annuus) Seeds. Microorganisms 2023, 11, 988. [Google Scholar] [CrossRef]
- S’thebe, N.W.; Aribisala, J.O.; Sabiu, S. Cheminformatics Bioprospection of Sunflower Seeds’ Oils against Quorum Sensing System of Pseudomonas aeruginosa. Antibiotics 2023, 12, 504. [Google Scholar] [CrossRef]
- Sun, Z.; Chen, J.; Ma, J.; Jiang, Y.; Wang, M.; Ren, G.; Chen, F. Cynarin-rich sunflower (Helianthus annuus) sprouts possess both antiglycative and antioxidant activities. J. Agric. Food Chem. 2012, 60, 3260–3265. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Saeed, A.; Kanwal, R.; Ahmad, S.; Changazi, S.H. Therapeutic effect of sunflower seeds and flax seeds on diabetes. Cureus 2021, 13, e17256. [Google Scholar] [CrossRef]
- Darmstadt, G.L.; Badrawi, N.; Law, P.A.; Ahmed, S.; Bashir, M.; Iskander, I.; Al Said, D.; El Kholy, A.; Husein, M.H.; Alam, A. Topically applied sunflower seed oil prevents invasive bacterial infections in preterm infants in Egypt: A randomized, controlled clinical trial. Pediatr. Infect. Dis. J. 2004, 23, 719–725. [Google Scholar] [CrossRef]
- Kumar, A.; Mishra, S.; Singh, S.; Ashraf, S.; Kan, P.; Ghosh, A.K.; Kumar, A.; Krishna, R.; Stevenson, D.K.; Tian, L. Effect of sunflower seed oil emollient therapy on newborn infant survival in Uttar Pradesh, India: A community-based, cluster randomized, open-label controlled trial. PLoS Med. 2021, 18, e1003680. [Google Scholar] [CrossRef]
- Menendez, S.; Falcon, L.; Simon, D.; Landa, N. Efficacy of ozonized sunflower oil in the treatment of tinea pedis. Mycoses 2002, 45, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.A.T., Jr.; Souza, V.R.; Endringer, D.C.; Hendrickson, D.A.; Coelho, C.S. Effects of topical application of sunflower-seed oil on experimentally induced wounds in horses. J. Equine Vet. Sci. 2012, 32, 139–145. [Google Scholar] [CrossRef]
- Kuo, Y.-S.; Hu, M.-H.; Chan, W.-H.; Huang, T.-Y.; Chou, Y.-C.; Huang, G.-S. Evaluation of the Preventive Effects of Fish Oil and Sunflower Seed Oil on the Pathophysiology of Sepsis in Endotoxemic Rats. Front. Nutr. 2022, 9, 857255. [Google Scholar] [CrossRef]
- Wei, X.; Hou, W.; Liang, J.; Fang, P.; Dou, B.; Wang, Z.; Sai, J.; Xu, T.; Ma, C.; Zhang, Q. Network pharmacology-based analysis on the potential biological mechanisms of sinisan against non-alcoholic fatty liver disease. Front. Pharmacol. 2021, 12, 693701. [Google Scholar] [CrossRef]
- He, S.; Wang, T.; Shi, C.; Wang, Z.; Fu, X. Network pharmacology-based approach to understand the effect and mechanism of Danshen against anemia. J. Ethnopharmacol. 2022, 282, 114615. [Google Scholar] [CrossRef] [PubMed]
- Chandran, U.; Mehendale, N.; Patil, S.; Chaguturu, R.; Patwardhan, B. Network Pharmacology. In Innovative Approaches in Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Association of Official Analytical Chemists. Extraction of Oil Content, Standard Method; Association of Official Analytical Chemists: Rockville, MD, USA, 1990. [Google Scholar]
- Fagbemi, K.O.; Aina, D.A.; Olajuyigbe, O.O. Soxhlet extraction versus hydrodistillation using the clevenger apparatus: A comparative study on the extraction of a volatile compound from tamarindus indica seeds. Sci. World J. 2021, 2021, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McCurry, J. GC Analysis of Total Fatty acid Methyl Esters (FAME) and Methyl Linolenate in Biodiesel Using the Revised EN14103: 2011 Method; Agilent Technologies Inc.: Santa Clara, CA, USA, 2012. [Google Scholar]
- Adnan, M.; Jeon, B.-B.; Chowdhury, M.; Uddin, H.; Oh, K.-K.; Das, T.; Chy, M.; Uddin, N.; Cho, D.-H. Network pharmacology study to reveal the potentiality of a methanol extract of Caesalpinia sappan L. Wood against type-2 diabetes mellitus. Life 2022, 12, 277. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H. GeneCards Version 3: The human gene integrator. Database 2010, 2010. [Google Scholar] [CrossRef]
- Blommaert, K.; Steenkamp, J. Tannien-en moontlike kafeieninhoud van rooibostee, Aspalathus (subgen. nortieria) linearis (Burm. Fil.) R. Dahlgr. Agroplantae 1978, 10, 2. [Google Scholar]
- Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Kazuno, S.; Yanagida, M.; Shindo, N.; Murayama, K. Mass spectrometric identification and quantification of glycosyl flavonoids, including dihydrochalcones with neutral loss scan mode. Anal. Biochem. 2005, 347, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Brändén, G.; Sjögren, T.; Schnecke, V.; Xue, Y. Structure-based ligand design to overcome CYP inhibition in drug discovery projects. Drug Discov. Today 2014, 19, 905–911. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function. Effic. Optim. Multithread. 2009, 31, 455–461. [Google Scholar]
- Balogun, F.O.; Naidoo, K.; Aribisala, J.O.; Pillay, C.; Sabiu, S. Cheminformatics Identification and Validation of Dipeptidyl Peptidase-IV Modulators from Shikimate Pathway-Derived Phenolic Acids towards Interventive Type-2 Diabetes Therapy. Metabolites 2022, 12, 937. [Google Scholar] [CrossRef]
- Li, L.; Dai, W.; Li, W.; Zhang, Y.; Wu, Y.; Guan, C.; Zhang, A.; Huang, H.; Li, Y. Integrated network pharmacology and metabonomics to reveal the myocardial protection effect of Huang-Lian-Jie-Du-Tang on myocardial ischemia. Front. Pharmacol. 2021, 11, 589175. [Google Scholar] [CrossRef] [PubMed]
- Balogun, F.; Ashafa, A. Aqueous root extracts of Dicoma anomala (Sond.) extenuates postprandial hyperglycaemia in vitro and its modulation on the activities of carbohydrate-metabolizing enzymes in streptozotocin-induced diabetic Wistar rats. S. Afr. J. Bot. 2017, 112, 102–111. [Google Scholar] [CrossRef]
- Balogun, F.O.; Ashafa, A.O.T.; Sabiu, S.; Ajao, A.A.-n.; Perumal, P.; Kazeem, M.I.; Adedeji, A.A. Pharmacognosy: Importance and drawbacks. In Pharmacognosy—Medicinal Plants; Intechopen: London, UK, 2019; pp. 1–19. [Google Scholar]
- Ismail, A.I.; Arafat, S.M. Quality characteristics of high-oleic sunflower oil extracted from some hybrids cultivated under Egyptian conditions. J. Food Technol. Res. 2014, 1, 73–83. [Google Scholar] [CrossRef]
- Hernández-Martínez, M.; Gallardo-Velázquez, T.; Osorio-Revilla, G.; Castañeda-Pérez, E.; Uribe-Hernández, K. Characterization of Mexican fishes according to fatty acid profile and fat nutritional indices. Int. J. Food Prop. 2016, 19, 1401–1412. [Google Scholar] [CrossRef]
- Dal Canto, E.; Ceriello, A.; Rydén, L.; Ferrini, M.; Hansen, T.B.; Schnell, O.; Standl, E.; Beulens, J.W. Diabetes as a cardiovascular risk factor: An overview of global trends of macro and micro vascular complications. Eur. J. Prev. Cardiol. 2019, 26, 25–32. [Google Scholar] [CrossRef]
- Das, U.N.; Rao, A.A. Gene expression profile in obesity and type 2 diabetes mellitus. Lipids Health Dis. 2007, 6, 35. [Google Scholar] [CrossRef]
- Oh, K.K.; Adnan, M.; Cho, D.H. Network pharmacology of bioactives from Sorghum bicolor with targets related to diabetes mellitus. PLoS ONE 2020, 15, e0240873. [Google Scholar] [CrossRef]
- Di, S.; Han, L.; An, X.; Kong, R.; Gao, Z.; Yang, Y.; Wang, X.; Zhang, P.; Ding, Q.; Wu, H. In silico network pharmacology and in vivo analysis of berberine-related mechanisms against type 2 diabetes mellitus and its complications. J. Ethnopharmacol. 2021, 276, 114180. [Google Scholar] [CrossRef]
- Noor, F.; Ashfaq, U.A.; Javed, M.R.; Saleem, M.H.; Ahmad, A.; Aslam, M.F.; Aslam, S. Comprehensive computational analysis reveals human respiratory syncytial virus encoded microRNA and host specific target genes associated with antiviral immune responses and protein binding. J. King Saud Univ. Sci. 2021, 33, 101562. [Google Scholar] [CrossRef]
- Noor, F.; Rehman, A.; Ashfaq, U.A.; Saleem, M.H.; Okla, M.K.; Al-Hashimi, A.; AbdElgawad, H.; Aslam, S. Integrating network pharmacology and molecular docking approaches to decipher the multi-target pharmacological mechanism of Abrus precatorius L. acting on diabetes. Pharmaceuticals 2022, 15, 414. [Google Scholar] [CrossRef]
- Roy, S.; Kumar, A.; Baig, M.H.; Masařík, M.; Provazník, I. Virtual screening, ADMET profiling, molecular docking and dynamics approaches to search for potent selective natural molecules based inhibitors against metallothionein-III to study Alzheimer’s disease. Methods 2015, 83, 105–110. [Google Scholar] [CrossRef]
- Aronson, J.K. Meyler’s Side Effects of Drugs. In Tesaglitarzar and Other PPAR Dual Agonist; Elsevier: Amsterdam, The Netherlands, 2016; pp. 762–763. [Google Scholar]
- Hashemzadeh, A.A.; Nasoohi, N.; Raygan, F.; Aghadavod, E.; Akbari, E.; Taghizadeh, M.; Memarzadeh, M.R.; Asemi, Z. Flaxseed oil supplementation improve gene expression levels of PPAR-γ, LP (a), IL-1 and TNF-α in type 2 diabetic patients with coronary heart disease. Lipids 2017, 52, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Laugerette, F.; Vors, C.; Peretti, N.; Michalski, M.-C. Complex links between dietary lipids, endogenous endotoxins and metabolic inflammation. Biochimie 2011, 93, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Masi, L.N.; Martins, A.R.; Neto, J.C.R.; Amaral, C.L.D.; Crisma, A.R.; Vinolo, M.A.R.; de Lima Junior, E.A.; Hirabara, S.M.; Curi, R. Sunflower oil supplementation has proinflammatory effects and does not reverse insulin resistance in obesity induced by high-fat diet in C57BL/6 mice. J. Biomed. Biotechnol. 2012, 2012, 945131. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, S. Effect of berberine on cyclin dependent kinase 9 and cyclin T1 expressions in type 2 diabetic rat kidney. Chin. J. Pharmacol. Toxicol. 2008, 6, 81–87. [Google Scholar]
- Wang, M.; Wang, J.; Tan, R.; Wu, Q.; Qiu, H.; Yang, J.; Jiang, Q. Effect of berberine on PPARα/NO activation in high glucose-and insulin-induced cardiomyocyte hypertrophy. Evid. Based Complement. Altern. Med. 2013, 2013, 285489. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhou, S. Berberine regulates peroxisome proliferator-activated receptors and positive transcription elongation factor b expression in diabetic adipocytes. Eur. J. Pharmacol. 2010, 649, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front Pharm. 2018, 9, 923. [Google Scholar] [CrossRef]
- Sabiu, S.; Balogun, F.O.; Amoo, S.O. Phenolics profiling of Carpobrotus edulis (L.) NE Br. and insights into molecular dynamics of their significance in type 2 diabetes therapy and its retinopathy complication. Molecules 2021, 26, 4867. [Google Scholar] [CrossRef]
- Akoonjee, A.; Rampadarath, A.; Aruwa, C.E.; Ajiboye, T.A.; Ajao, A.A.-n.; Sabiu, S. Network Pharmacology-and Molecular Dynamics Simulation-Based Bioprospection of Aspalathus linearis for Type-2 Diabetes Care. Metabolites 2022, 12, 1013. [Google Scholar] [CrossRef]
- Vergara, R.; Romero-Romero, S.; Velázquez-López, I.; Espinoza-Pérez, G.; Rodríguez-Hernández, A.; Pulido, N.O.; Sosa-Peinado, A.; Rodríguez-Romero, A.; Fernández-Velasco, D.A. The interplay of protein–ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein. FEBS J. 2020, 287, 763–782. [Google Scholar] [CrossRef]
- Hernández-Santoyo, A.; Tenorio-Barajas, A.Y.; Altuzar, V.; Vivanco-Cid, H.; Mendoza-Barrera, C. Protein-Protein and Protein-Ligand Docking. In Protein Engineering—Technology and Application (Ogawa, T); InTech Open: London, UK, 2013. [Google Scholar]
- Khan, M.S.; Husain, F.M.; Alhumaydhi, F.A.; Alwashmi, A.S.; Rehman, M.T.; Alruwetei, A.M.; Hassan, M.I.; Islam, A.; Shamsi, A. Exploring the molecular interactions of Galantamine with human Transferrin: In-silico and in vitro insight. J. Mol. Liq. 2021, 335, 116227. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.-L.; Ai, S.-M.; Liang, J.; Sang, P.; Ji, X.-L.; Liu, S.-Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Jay, M.A.; Ren, J. Peroxisome proliferator-activated receptor (PPAR) in metabolic syndrome and type 2 diabetes mellitus. Curr. Diabetes Rev. 2007, 3, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Gross, B.; Staels, B. PPAR agonists: Multimodal drugs for the treatment of type-2 diabetes. Best Pract. Res. Clin. Endocrinol. Metab. 2007, 21, 687–710. [Google Scholar] [CrossRef] [PubMed]
- Mazibuko-Mbeje, S.E.; Dludla, P.V.; Roux, C.; Johnson, R.; Ghoor, S.; Joubert, E.; Louw, J.; Opoku, A.R.; Muller, C.J. Aspalathin-enriched green rooibos extract reduces hepatic insulin resistance by modulating PI3K/AKT and AMPK pathways. Int. J. Mol. Sci. 2019, 20, 633. [Google Scholar] [CrossRef] [PubMed]
| S/N | Compound Names | Lipinski’s Rule Remarks | No of Violations |
|---|---|---|---|
| 1 | Capric acid | Yes | None |
| 2 | Caproic acid | Yes | None |
| 3 | Caprylic acid | Yes | None |
| 4 | Lauric acid | Yes | None |
| 5 | Myristic acid | Yes | None |
| 6 | Palmitic acid | Yes | None |
| 7 | Pentadecyclic acid | Yes | None |
| 8 | Stearic acid | Yes | None |
| 9 | Oleic acid | Yes | None |
| 10 | Linoleic acid | Yes | None |
| 11 | Arachidic acid | Yes | None |
| 12 | Behenic acid | Yes | None |
| 13 | Tricosylic acid | Yes | None |
| 14 | Lignoceric acid | Yes | None |
| 15 | Phylloquinone | Yes | None |
| Term ID | Pathways | No of Genes | False Discovery Rate | Genes |
|---|---|---|---|---|
| hsa03320 | PPAR signaling pathway | 5 | 2.01 × 10−6 | FABP4, PPARG, PPARD, CPT2, PPARA |
| hsa04080 | Neuroactive ligand-receptor interaction | 5 | 1.20 × 10−3 | CNR1, LPAR1, LPAR3, GPR35, TRPV1 |
| hsa05200 | Pathways in cancer | 5 | 7.00 × 10−3 | PPARG, PPARD, RARB, LPAR1, LPAR3 |
| S/N | Compounds/Standards | Docking Scores (kcal/mol) | ||||
|---|---|---|---|---|---|---|
| PPARA | FABP4 | PPARD | PPARG | CPT2 | ||
| 1 | Linoleic acid | −6.3 | −6.3 | NA | NA | NA |
| 2 | Lignoceric acid | −6.7 | −6.0 | NA | −6.1 | −5.7 |
| 3 | Behenic acid | −6.5 | −5.9 | NA | NA | −6.1 |
| 4 | Phylloquinone | −9.2 | NA | NA | NA | NA |
| 5 | Tricosylic acid | −6.6 | −6.0 | −7.1 | −5.7 | −5.7 |
| 6 | Arachidic acid | NA | −5.9 | −6.1 | −5.8 | −6.2 |
| 7 | Pentadecyclic acid | NA | NA | −6.3 | NA | NA |
| 8 | Stearic acid | NA | NA | −6.3 | −5.8 | −6.3 |
| 9 | Oleic acid | NA | NA | −6.4 | NA | NA |
| 10 | Palmitic acid | NA | NA | NA | −5.8 | NA |
| 11 | Rosiglitazone | −8.6 | −8.3 | −8.7 | −8.2 | −9.4 |
| 12 | Metformin | −5.2 | −4.5 | −5.2 | −4.9 | −5.3 |
| Complex | Number of Interactions | Number of H-Bonds and Interaction Residues | Number of van der Waal Forces and Interaction Residues | Other Important Interactions and Residues |
|---|---|---|---|---|
| PPARA–phylloquinone | 25 | - | 16 (Gln277, Met330, Met320, Thr279, Gly335, Val332, Tyr334, Leu460, Tyr464, His440, Tyr314, Ser280, Thr283, Asn219, Phe318, Lys358) | 9 (Cys276, Met355, Ile354, Ile317, Leu321, Val324, Met220, Leu331, Phe273) |
| PPARA–lignoceric acid | 22 | 2 (His440, Tyr464) | 14 (Tyr314, Lys358, Met355, Cys276, Gln277, Ser280, Thr283, Met320, Asn219, Thr279, Tyr334, Gly335, Val332, Leu331) | 6 (Phe318, Ile217, Leu321, Met220, Met330, Val324) |
| PPARA–behenic acid | 21 | - | 12 (Phe218, Met320, Val324, Thr283, Thr279, Tyr314, Phe318, Ile354, Gln277, Asn219, Met220, Ser280) | 9 (Leu321, Ile317, His440, Cys226, Phe273, Leu458, Leu460, Val444, Tyr464) |
| PPARA–tricosylic acid | 21 | 3 (Gln277, Ser280, Tyr464) | 9 (Val444, Phe318, Thr279, Val332, Tyr334, Leu331, Thr283, Tyr314, Leu460) | 9 (Ile354, Phe273, Lys276, His440, Leu321, Ile317, Met320, Val324, Met220) |
| PPARA–linoleic acid | 14 | 1 (Cys276) | 9 (Val332, Leu331, Asn219, Glu286, Phe218, Thr283, Thr279, Ile317, Ser280) | 4 (Met220, Met320, Val324, Leu321) |
| PPARA–rosiglitazone | 20 | 1 (Ser280) | 14 (Asn219, Phe218, Met220, Ser323, Val324, Asn221, Met320, Thr279, Phe318, Lys358, Ile354, Gln277, Thr283, Met355) | 5 (Ile317, Leu321, His440, Cys276, Phe273) |
| PPARA–metformin | 13 | 2 (Tyr214, Thr283) | 9 (Lys 222, Thr279, Val324, Ser323, Asn221, Met220, Met220, Asn219, Phe218) | 2 (Glu286, Asp372) |
| FABP4–linoleic acid | 22 | 1 (Arg126) | 12 (Gln95, Thr74, Arg78, Val25, Asp76, Lys58, Ser53, Ser55, Cys117, Val115, Tyr128, Arg106) | 9 (Tyr19, Phe16, Met20, Ala75, Ala33, Ala36, Pro38, Phe57, Ile104) |
| FABP4–lignoceric acid | 8 | 1 (Thr29) | 4 (Met35, Phe27, Phe57, Lys58) | 3 (Lys31, Val32, Ala28) |
| FABP4–aradichic acid | 10 | 2 (Ala75, Asp76) | 4 (Asp77, Thr29, Lys58, Phe27) | 4 (Val32, Phe57, Ala28, Lys31) |
| FABP4–behenic acid | 22 | 1 (Arg106) | 11 (Arg78, Asp76, Ser55, Lys58, Ser53, Arg126, Thr60, Ile104, Met40, Val115, Tyr128) | 10 (Val25, Val23, Tyr19, Met20, Phe57, Ala33, Ala36, Ala75, Phe16, Pro38) |
| FABP4–tricosylic acid | 24 | 2 (Ala75, Thr24) | 11 (Glu72, Thr60, Asp76, Arg78, Arg126, Ser53, Lys58, Ser55, Val25, Tyr19, Arg106) | 11 (Met20, Ala36, Pro38, Phe51, Ala33, The16, Tyr128, Cys117, Ile104, Met40, Val115) |
| FABP4–rosiglitazone | 22 | 2 (Arg106, Ser53) | 14 (Val23, Arg78, Tyr19, Val115, Tyr128, Ser55, Val25, Met20, Asp76, Gln95, Ala75, Ala33, Phe57, Thr60) | 6 (Phe16, Cys117, Ile104, Arg126, Pro38, Ala36) |
| FABP4–metformin | 10 | 1 (Tyr128) | 7 (Met40, Pro38, Ala75, Ala36, Phe57, Phe16, Ala33) | 2 (Arg126, Ser53) |
| PPARD–tricosylic acid | 25 | 1 (His287) | 9 (Phe316, Trp228, Leu317, Thr252, Phe291, Phe246, Gln250, Leu433, Met417) | 15 (Leu219, Ile213, Val312, Arg218, Val245, Leu323, Val305, Leu294, Cys249, Ile322, Ile328, Tyr437, Thr253, Lys331, His413) |
| PPARD–oleic acid | 19 | 1 (Thr252) | 4 (Trp228, Thr253, Ile290, Leu317) | 13 (Phe316, Arg248, Val305, Leu294, Leu303, Cys249, Ile328, Lys331, Phe291, Val312, Val245, Leu219, Ile213) |
| PPARD–stearic acid | 20 | 6 (Thr253, Gln250, Phe246, Phe291, His244, Phe316), 1 (His413) | 1 (His413) | 13 (Leu219, Arg248, Val245, Val312, Leu303, Cys249, Lys331, Ile328, Leu317, Leu294, Ile213, Val305, Trp228) |
| PPARD–arachidic acid | 23 | 2 (His413, Thr253) | 8 (Trp228, Phe316, Ile290, Tyr437, His287, Met417, Leu433, Gln250) | 13 (Lys331, Phe291, Leu294, Ile328, Val305, Val312, Ile213, Cys249, Arg248, Leu219, Val245, Leu317, Leu303) |
| PPARD—pentadecylic acid | 19 | - | 7 (His287, His413, Ile290, Thr253, Phe291, Phe316, Trp228) | 12 (Ile328, Val305, Val245, Leu219, Leu294, Leu303, Arg248, Lys331, Leu317, Val312, Ile213, Cys249) |
| PPARD—rosiglitazone | 19 | 3 (Val245, His244, Ile327) | 11 (Leu219, Arg248, Ile328, Lys331, Met417, Phe246, Thr253, Phe291, Val305, Ile213, Trp228) | 5 (Val312, Leu317, Leu303, Cys249, His413) |
| PPARD–metformin | 9 | 2 (Met293, Thr256) | 5 (Tyr186, Asn191, Met192, Ile297, Ser296) | 2 (Glu259, Phe190) |
| PPARG–lignoceric acid | 24 | 1 (Gln286) | 15 (Ser289, Val339, Leu340, Ile341, Glu295, Phe226, Pro227, Phe287, Phe363, His449, Tyr327, Met364, Ser342, Lys367, Leu453) | 8 (Cys285, Arg288, Leu333, Ala292, Met329, Ile326, Leu228, Leu330) |
| PPARG–palmitic acid | 20 | 1 (Gln286) | 12 (Leu228, Glu295, Ile296, Ser289, Tyr322, His323, Tyr473, His449, Leu453, Lys367, Cys285, Met364) | 7 (Pro227, Phe226, Met329, Arg288, Ile326, Ala292, Leu330) |
| PPARG–stearic acid | 18 | 2 (Gln286, His449) | 6 (Glu295, Lys367, Phe363, Ser280, Tyr322, Ile296) | 10 (Phe226, Met329, Ala297, Ile326, Arg288, Glu330, Pro227, Leu228, Cys285, Met364) |
| PPARG–arachidic acid | 16 | 1 (Glu343) | 9 (Leu228, Leu340, Ser342, Val339, Met364, Leu333, Ser289, Glu295, Tyr327) | 6 (Cys285, Ile341, Leu330, Ile326, Ala292, Met329) |
| PPARG–tricosylic acid | 18 | 1 (Glu295) | 7 (Glu343, Ser342, Pro227, Phe226, Leu340, Ser289, Ile325) | 10 (Leu228, Leu333, Arg288, Leu330, Ala292, Met329, Ile326, Ile296, Cys285, Ile341) |
| PPARG–rosiglitazone | 17 | 6 (Leu228, Arg288, Pro227, Phe226, Ser332, Cys285) | 6 (Glu295, Ile341, Thr229, Met329, Leu333, Ser289, Ile341) | 5 (Ile326, Ala292, Leu330, Val339, Met364) |
| PPARG–metformin | 12 | 2 (Leu228, Ile326) | 9 (Pro227, Phe226, Arg288, Met329, Leu333, Ala292, Ser332, Ile296, Leu330) | 1 (Glu295) |
| CPT2–stearic acid | 16 | 3 (Thr499, Ser488, Tyr120) | 7 (Asp376, Trp116, Arg554, Ser588, Asn585, Met119, Asn130), | 6 (Tyr486, Val605, His372, Phe131, Phe602, Pro133) |
| CPT2–tricosylic acid | 22 | 3 (Ser488, Tyr486, Asp376) | 9 (Arg554, Trp116, Thr499, Met119, Gly377, Ser590, Asn585, Ser588, Asn130) | 9 (Val605, Phe131, Phe602, Met135, His372, Tyr120, Phe370, Pro133, Leu592) |
| CPT2–lignoceric acid | 15 | 2 (Val175, Glu174) | 3 (Ser490, Val378, Arg382) | 10 (Phe176, Leu212, Ala547, Tyr210, Ala493, Phe494, Met548, Pro50, Trp201, Tyr205) |
| CPT2–arachidic acid | 16 | 2 (Ser498, Tyr486) | 9 (Trp116, Thr499, Arg554, Met119, Asn130, Ser590, Phe370, Ty120, Ser588) | 5 (Val605, Phe131, Phe602, His372, Pro133) |
| CPT2–behenic acid | 17 | 3 (Tyr486, Ser488, Asp376), | 8 (Asn585, Ser588, Ser590, Trp116, Asn130, Tyr120, Thr499, Arg554) | 6 (Val605, Phe131, Met119, Phe602, His372, Pro133) |
| CPT2–rosiglitazone | 21 | 1 (Ser590) | 15 (Val597, Ala615, Leu599, His617, Ser598, Trp620, Gly622, Asn624, Cys623, Thr591, Gly600, Tyr614, Phe370, His372, Ser588) | 4 (Ala613, Phe602, Pro133, Met135, Leu592) |
| CPT2–metformin | 12 | 3 (Phe131, Ala603, Leu129) | 8 (Asn132, Pro133, Asn130, His372, Leu343, Pro604, Val605, Ser588) | 1 (Phe602) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rampadarath, A.; Balogun, F.O.; Sabiu, S. Insights into the Mechanism of Action of Helianthus annuus (Sunflower) Seed Essential Oil in the Management of Type-2 Diabetes Mellitus Using Network Pharmacology and Molecular Docking Approaches. Endocrines 2023, 4, 327-349. https://doi.org/10.3390/endocrines4020026
Rampadarath A, Balogun FO, Sabiu S. Insights into the Mechanism of Action of Helianthus annuus (Sunflower) Seed Essential Oil in the Management of Type-2 Diabetes Mellitus Using Network Pharmacology and Molecular Docking Approaches. Endocrines. 2023; 4(2):327-349. https://doi.org/10.3390/endocrines4020026
Chicago/Turabian StyleRampadarath, Athika, Fatai Oladunni Balogun, and Saheed Sabiu. 2023. "Insights into the Mechanism of Action of Helianthus annuus (Sunflower) Seed Essential Oil in the Management of Type-2 Diabetes Mellitus Using Network Pharmacology and Molecular Docking Approaches" Endocrines 4, no. 2: 327-349. https://doi.org/10.3390/endocrines4020026
APA StyleRampadarath, A., Balogun, F. O., & Sabiu, S. (2023). Insights into the Mechanism of Action of Helianthus annuus (Sunflower) Seed Essential Oil in the Management of Type-2 Diabetes Mellitus Using Network Pharmacology and Molecular Docking Approaches. Endocrines, 4(2), 327-349. https://doi.org/10.3390/endocrines4020026







