ChatCVD: A Retrieval-Augmented Chatbot for Personalized Cardiovascular Risk Assessment with a Comparison of Medical-Specific and General-Purpose LLMs
Abstract
1. Introduction
- Comparative Evaluation of LLMs: We present a direct comparison of eight fine-tuned LLMs (medical-specific and general-purpose) for CVD risk classification, with an emphasis on recall for identifying high-risk individuals.
- extualization of Health Data: We propose a method to convert structured numerical health data into narrative-style textual profiles to enhance LLM interpretability.
- RAG-Based Personalized Recommendations: We develop a RAG framework to generate actionable recommendations aligned with authoritative medical guidelines.
- ChatCVD—A User-Centered Interface: We introduce ChatCVD, a chatbot that makes complex risk assessments and personalized health advice accessible to users in natural language.
- From Research to Application: This study demonstrates a pathway to translate LLM-based AI advances into practical, accessible tools for real-world healthcare use.
2. Related Work
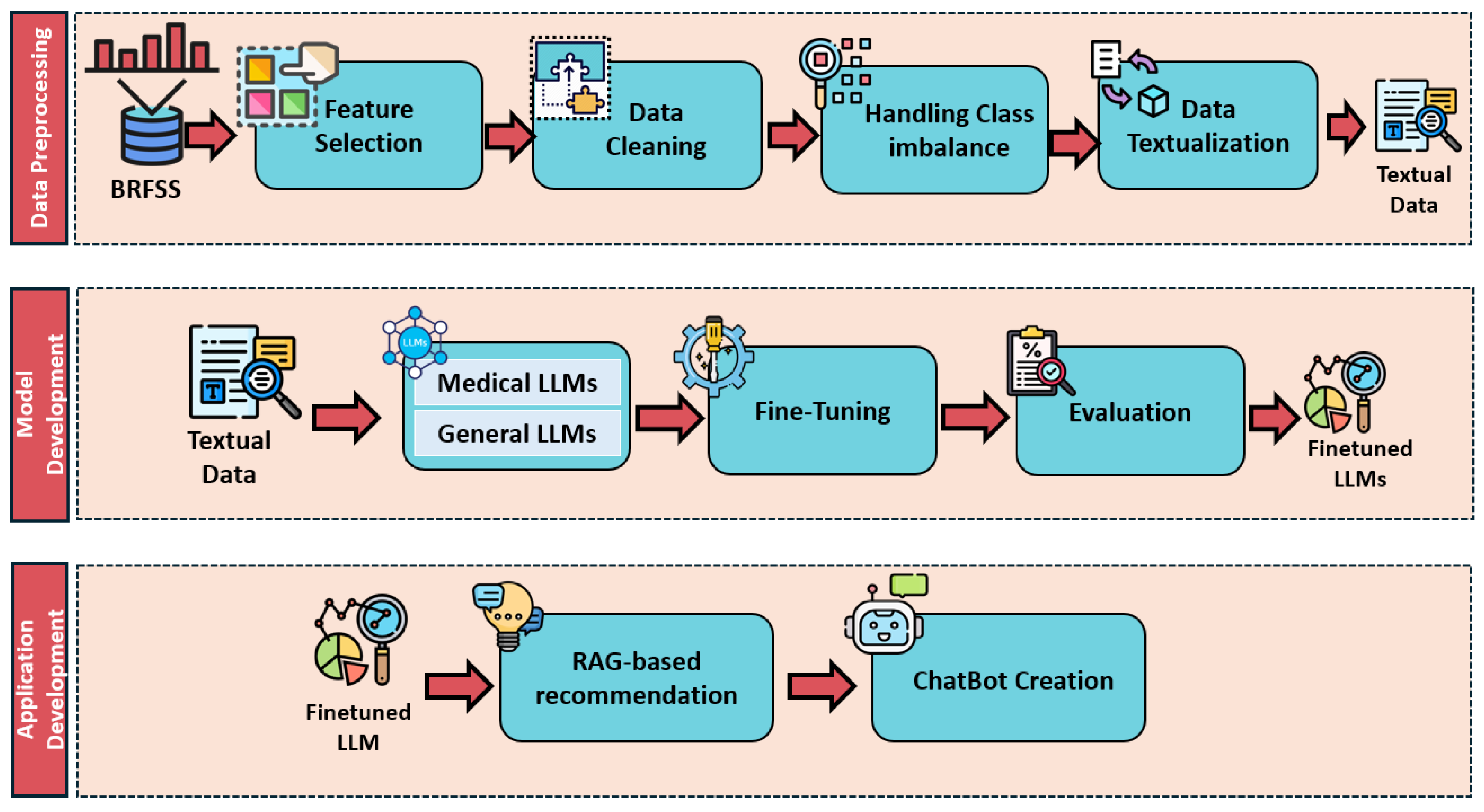
3. Methods
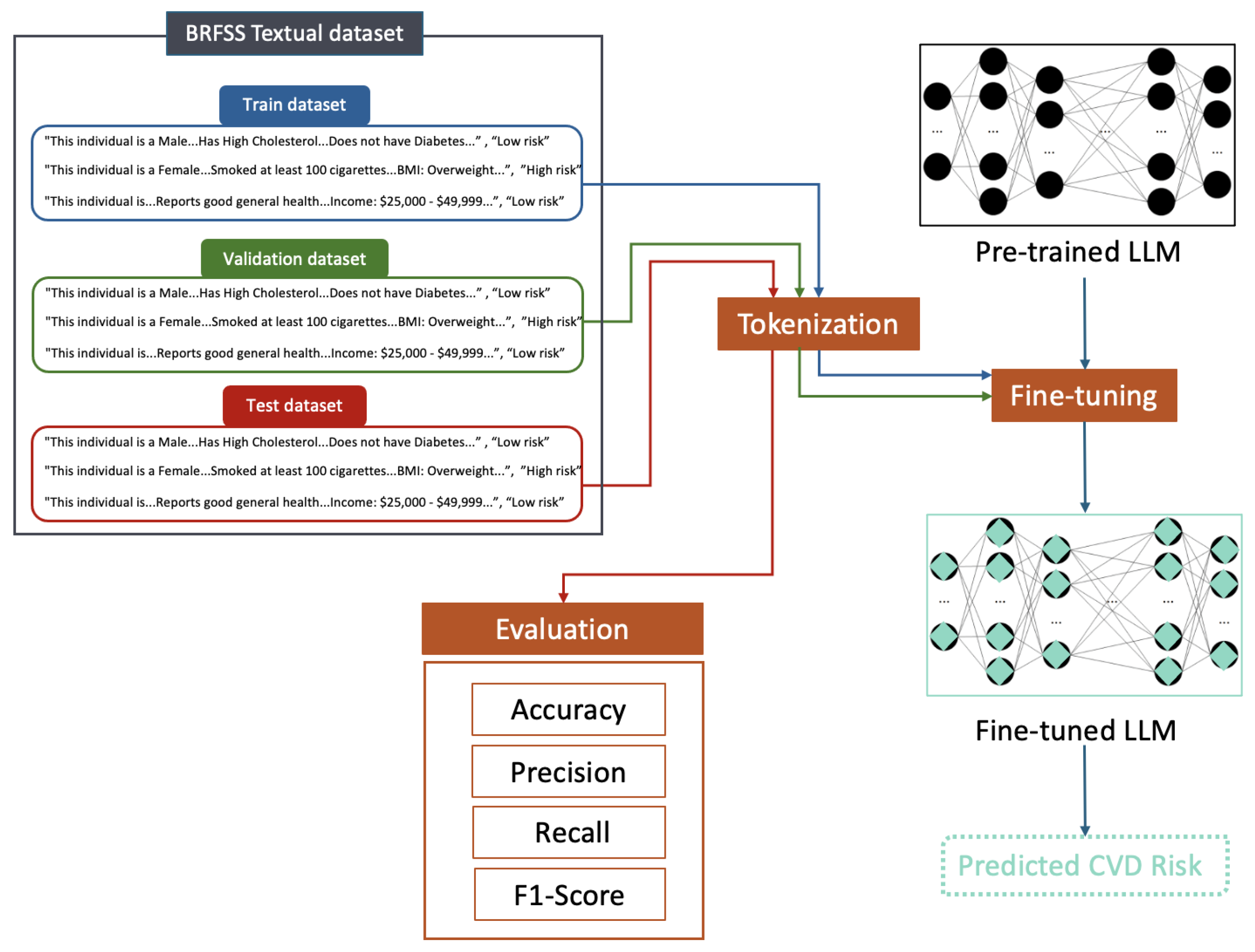
3.1. Dataset
3.2. Data Preprocessing
3.2.1. Feature Selection
3.2.2. Data Cleaning
3.2.3. Class Imbalance Handling
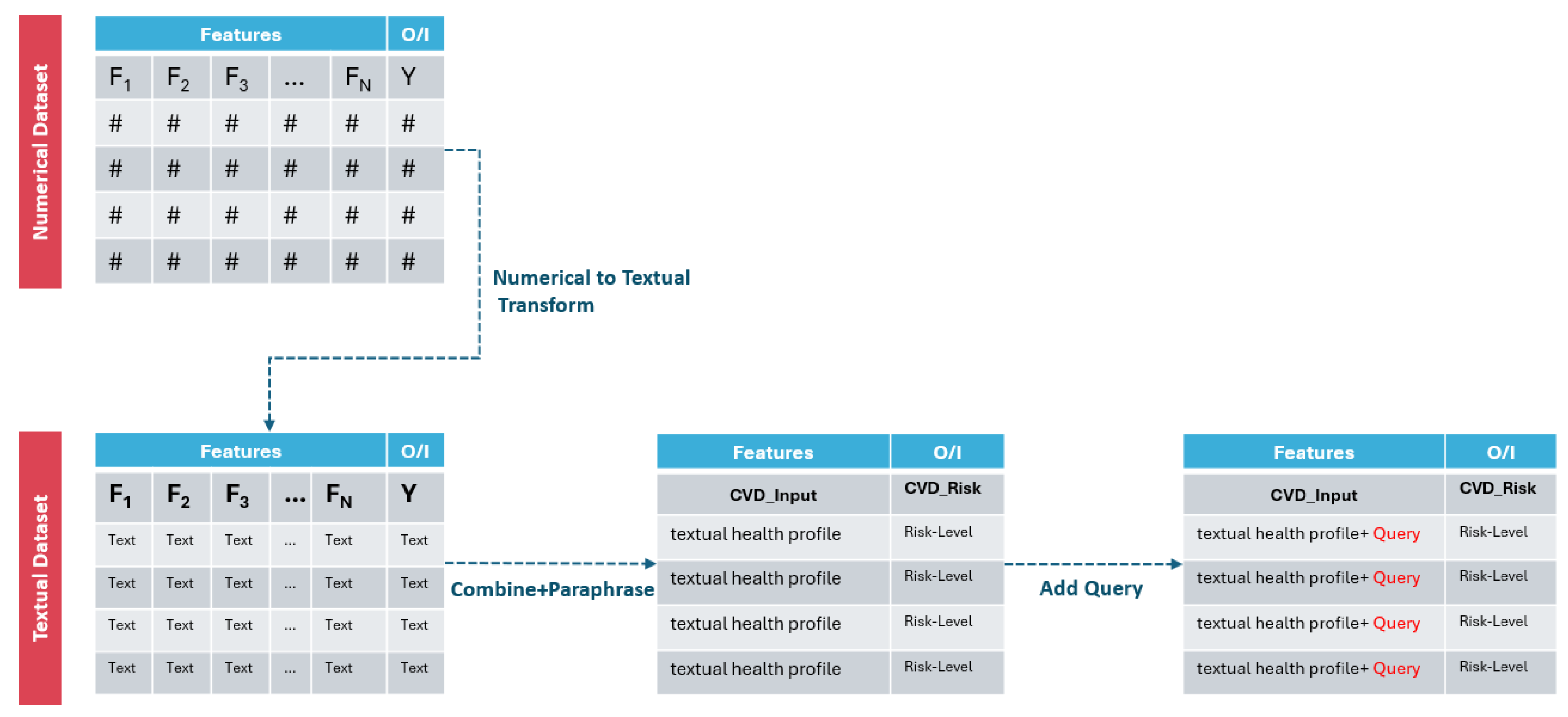
3.2.4. Data Textualization Descriptions
3.3. Model Development
3.3.1. Medical-Specific vs. General-Purpose LLMs for CVD Risk Prediction
Medical-Specific Models
- BioBert [27]: a pre-trained language model designed specifically for biomedical text, leveraging a large corpus of literature to excel in domain-specific tasks, with 110 million parameters.
- Meditron [28]: a model with 7 billion parameters; focuses on medical image captioning, generating descriptive text to support diagnostics.
- Med42 [29]: With 8 billion parameters, Med42 functions as a medical question-answering system, providing accurate responses based on an extensive biomedical knowledge base.
- MedAlpaca [30]: a medical chatbot trained on healthcare conversations and built on the LLaMA 2 base; features 7 billion parameters and emphasizes contextual understanding in dialogue.
General-Purpose Models
- Gemma2 [31]: a versatile LLM with 2 billion parameters, capable of text generation, translation, and content creation.
- LLaMA2 [32]: a model with 7 billion parameters, designed for a wide range of natural language processing tasks.
- LLaMA3 [33]: the successor to LLaMA2, featuring 8 billion parameters and enhanced reasoning capabilities and improved performance across NLP benchmarks.
- Mistral [34]: a model with 7 billion parameters; focuses on improved reasoning and code generation, demonstrating robust general-purpose capabilities.
3.3.2. Fine-Tuning Approach
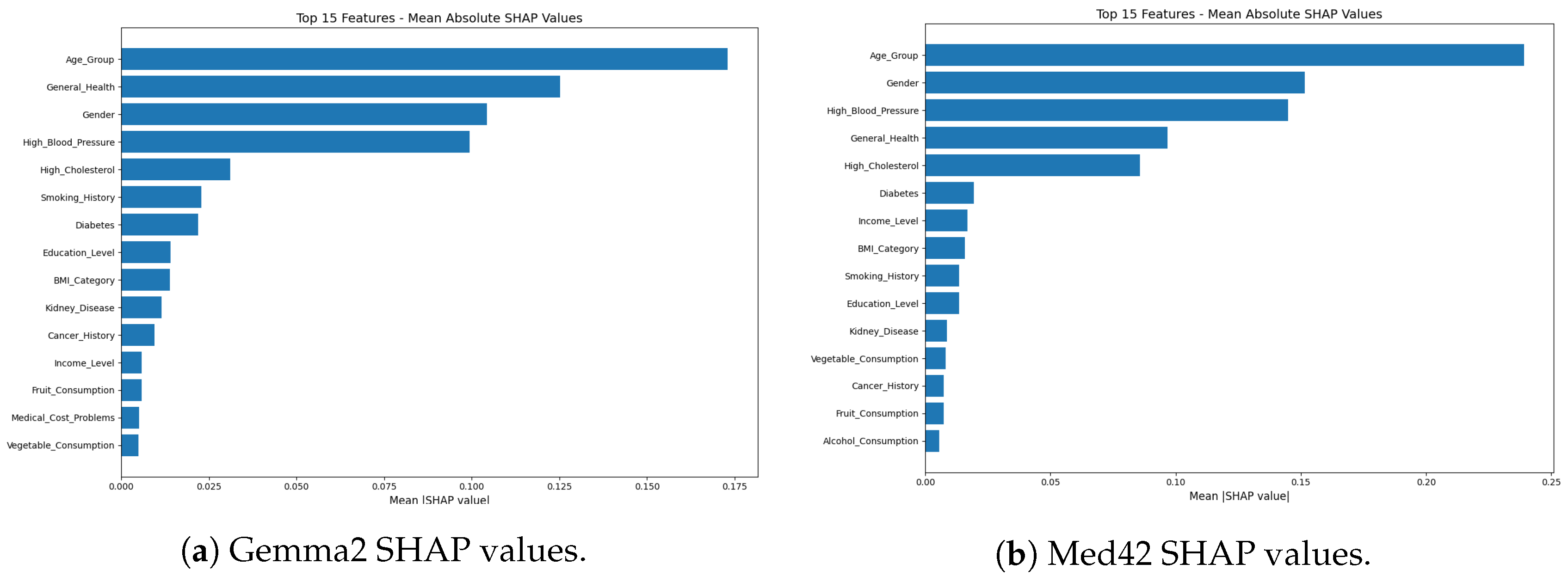
3.3.3. Feature Importance Analysis
- Feature Extraction: A subset of the test data was used, in which the textual health profiles (original inputs to the LLMs) were converted back into their structured, categorical feature representations (e.g., Age_Group, Gender, Smoking_History) using a rule-based feature extractor. This step enabled SHAP to operate on interpretable, human-understandable features.
- Prediction Function for SHAP: A wrapper function was created that accepts structured features as input, reconstructs the full textual health profile in the format expected by the fine-tuned LLM, and outputs the CVD risk probability—specifically, the likelihood of being classified as “High Risk”.
- SHAP Value Computation: Using the shap library, a SHAP explainer (e.g., Kernel-Explainer or another model-compatible variant) was initialized with a background dataset drawn from structured test instances. SHAP values were computed for each feature on a representative evaluation sample, with consideration for computational constraints (GPU/CPU).
- Global Feature Importance: Mean absolute SHAP values were calculated across the evaluation set to derive a global measure of each feature’s importance in influencing model predictions.
3.4. Application Development
- Data Acquisition and Processing: HTML content from the specified URLs is automatically downloaded and parsed, and relevant text is extracted.
- Chunking: The extracted text is segmented into coherent chunks of approximately 200–300 words to preserve context.
- Embedding: Each chunk is converted into a vector representation using SentenceTransformers [39], creating a semantic index of the knowledge base.
- Vector Storage: These embeddings are stored in a vector database to enable efficient similarity search during response generation.
| Given the following profile: ’{user_input}’, and considering this individual has a {risk_level} risk of CVD, first, identify the key risk factors mentioned. |
| Then, provide 3 UNIQUE and SPECIFIC ACTIONABLE recommendations to improve their cardiovascular health, drawing from the provided context. |
| Avoid repeating the same advice. |
| Ex: If overweight, suggest a weight loss goal. |
| If high cholesterol, recommend foods to avoid. |
| Present recommendations in a numbered list. |
| where {user_input} is replaced with the user’s health profile and {risk_level} with the predicted risk. |
- Risk Prediction: The input text is passed to the fine-tuned CVD risk prediction model, which classifies the user as “Low Risk” or “High Risk.”
- Query Generation: The structured query from the prompt template is constructed using the user’s information and risk level.
- Knowledge Retrieval: This query is then used to perform a similarity search in the vector database, retrieving the most relevant chunks of information from the knowledge base.
- Recommendation Synthesis: The retrieved information is synthesized using the LLM to generate personalized and actionable recommendations tailored to the user’s specific risk factors and profile. The LLM is prompted to avoid generic advice and instead provide concrete steps the user can take.
3.5. Evaluation
3.5.1. Human Expert Assessment
3.5.2. Statistical Analysis
4. Results
4.1. Performance of Medical-Specific vs. General-Purpose LLMs in CVD Risk Assessment
4.2. Human Expert Assessment
4.3. Statistical Analysis
4.4. Feature Importance Analysis
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Demographic Variable Distributions
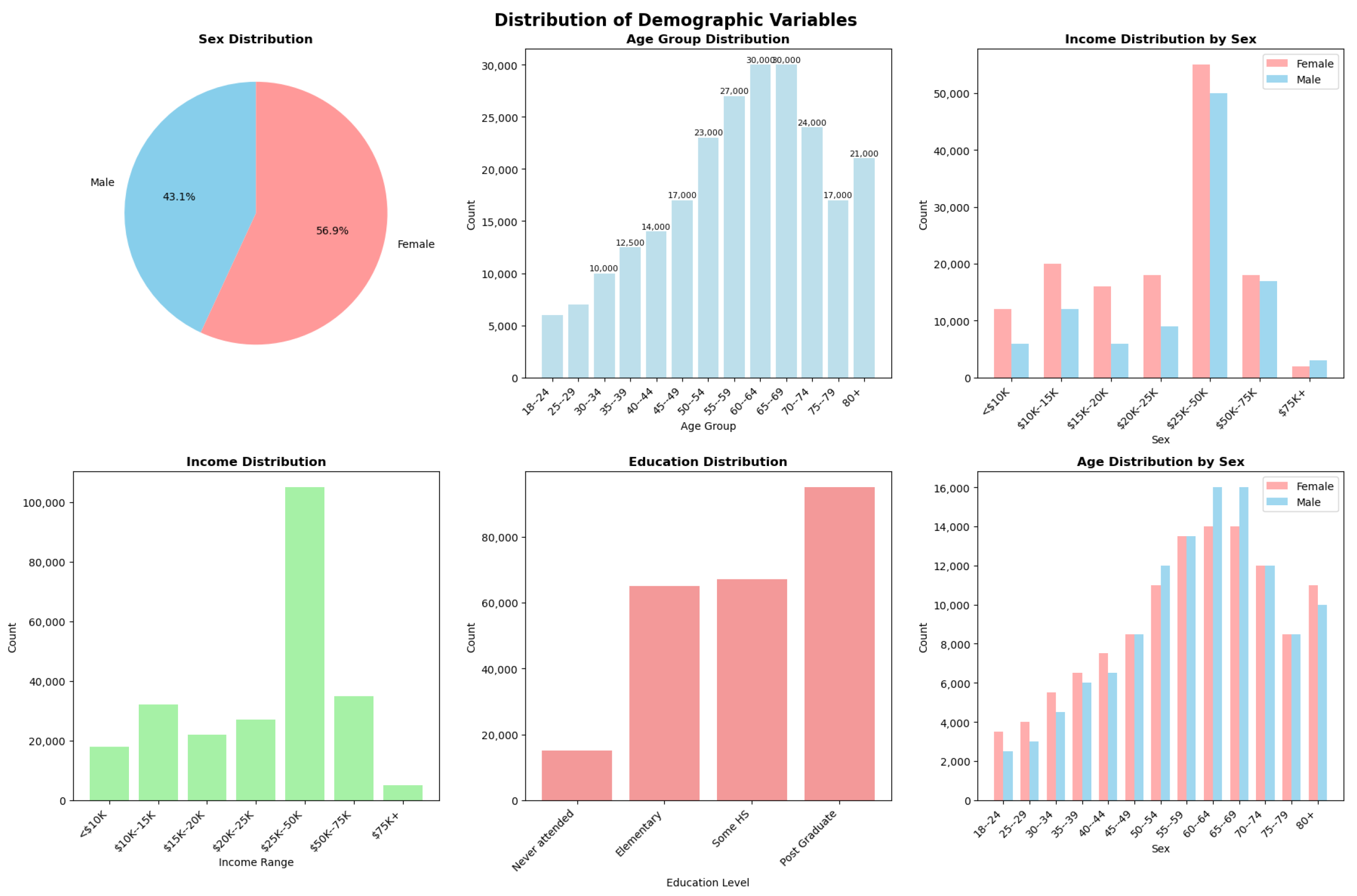

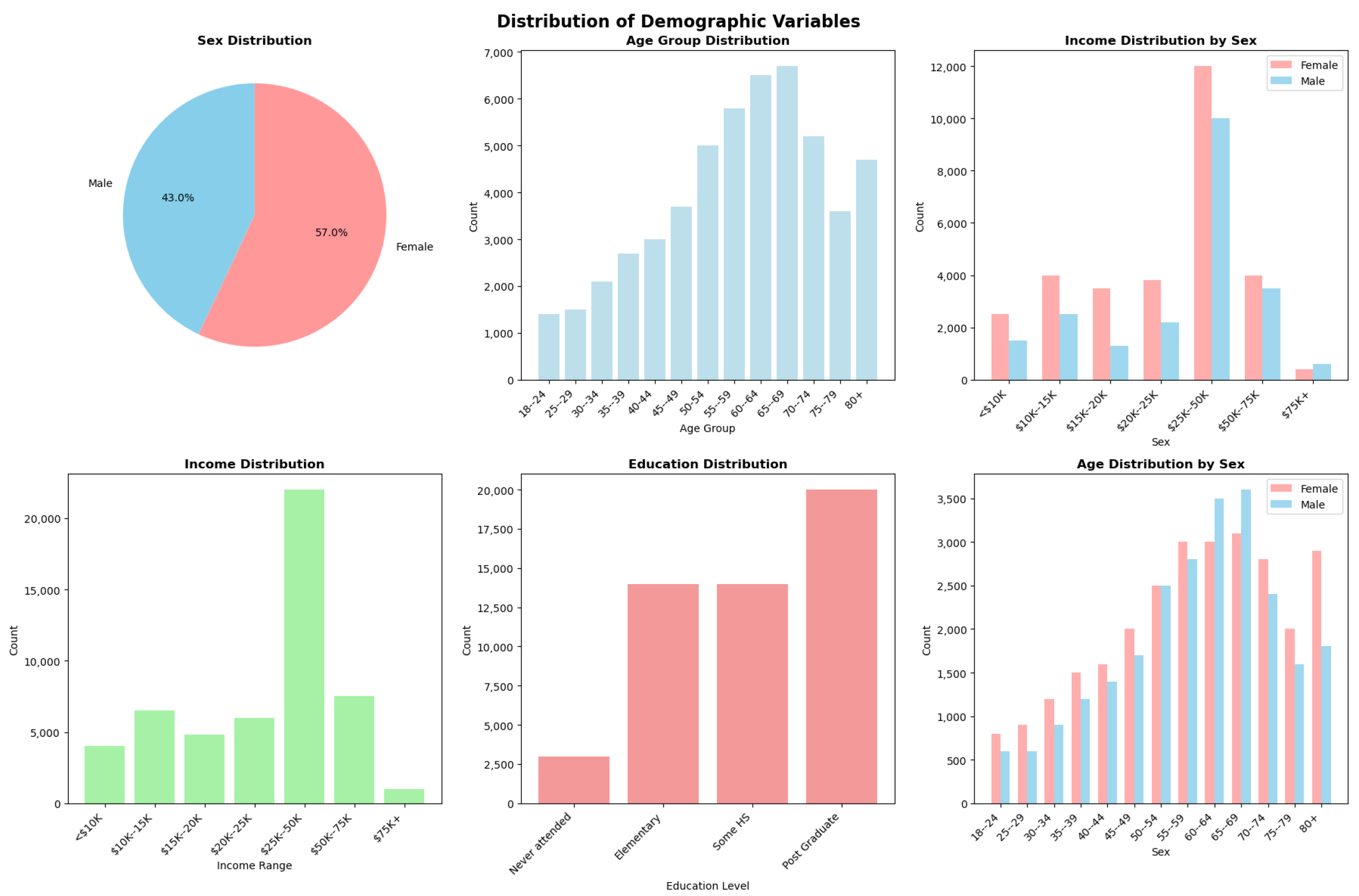
Appendix B. ChatCVD Interface Example

Appendix C. Confusion Matrix

References
- Rehman, S.; Rehman, E.; Ikram, M.; Jianglin, Z. Cardiovascular disease (CVD): Assessment, prediction and policy implications. BMC Public Health 2021, 21, 1299. [Google Scholar] [CrossRef]
- Boukhatem, C.; Youssef, H.Y.; Nassif, A.B. Heart disease prediction using machine learning. In Proceedings of the 2022 Advances in Science and Engineering Technology International Conferences (ASET), Dubai, United Arab Emirates, 21–24 February2022; pp. 1–6. [Google Scholar]
- Ananthajothi, K.; David, J.; Kavin, A. Cardiovascular Disease Prediction using Patient History and Real Time Monitoring. In Proceedings of the 2024 2nd International Conference on Intelligent Data Communication Technologies and Internet of Things (IDCIoT), Bengaluru, India, 4–6 January 2024; pp. 1226–1233. [Google Scholar]
- Zhao, D.; Liu, J.; Xie, W.; Qi, Y. Cardiovascular risk assessment: A global perspective. Nat. Rev. Cardiol. 2015, 12, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.D. Cardiovascular risk assessment: The foundation of preventive cardiology. Am. J. Prev. Cardiol. 2020, 1, 100008. [Google Scholar] [CrossRef]
- Li, Y.H.; Li, Y.L.; Wei, M.Y.; Li, G.Y. Innovation and challenges of artificial intelligence technology in personalized healthcare. Sci. Rep. 2024, 14, 18994. [Google Scholar] [CrossRef]
- Gunning, D.; Stefik, M.; Choi, J.; Miller, T.; Stumpf, S.; Yang, G.Z. XAI—Explainable artificial intelligence. Sci. Robot. 2019, 4, eaay7120. [Google Scholar] [CrossRef]
- Hussain, H.K.; Tariq, A.; Gill, A.Y.; Ahmad, A. Transforming Healthcare: The Rapid Rise of Artificial Intelligence Revolutionizing Healthcare Applications. BULLET J. Multidisiplin. Ilmu 2022, 1, 592216. [Google Scholar]
- Sushil, M.; Kennedy, V.E.; Mandair, D.; Miao, B.Y.; Zack, T.; Butte, A.J. CORAL: Expert-curated oncology reports to advance language model inference. NEJM AI 2024, 1, AIdbp2300110. [Google Scholar] [CrossRef]
- Hosseini, A.; Serag, A. Is synthetic data generation effective in maintaining clinical biomarkers? Investigating diffusion models across diverse imaging modalities. Front. Artif. Intell. 2025, 7, 1454441. [Google Scholar] [CrossRef]
- Hosseini, A.; Serag, A. Self-Supervised Learning Powered by Synthetic Data From Diffusion Models: Application to X-Ray Images. IEEE Access 2025, 13, 59074–59084. [Google Scholar] [CrossRef]
- Ben Rabah, C.; Petropoulos, I.N.; Malik, R.A.; Serag, A. Vision transformers for automated detection of diabetic peripheral neuropathy in corneal confocal microscopy images. Front. Imaging 2025, 4, 1542128. [Google Scholar] [CrossRef]
- Ben Rabah, C.; Sattar, A.; Ibrahim, A.; Serag, A. A Multimodal Deep Learning Model for the Classification of Breast Cancer Subtypes. Diagnostics 2025, 15, 995. [Google Scholar] [CrossRef] [PubMed]
- Helmy, H.; Rabah, C.B.; Ali, N.; Ibrahim, A.; Hoseiny, A.; Serag, A. Optimizing ICU Readmission Prediction: A Comparative Evaluation of AI Tools. In International Workshop on Applications of Medical AI; Springer: Berlin/Heidelberg, Germany, 2024; pp. 95–104. [Google Scholar]
- Ibrahim, A.; Hosseini, A.; Ibrahim, S.; Sattar, A.; Serag, A. D3: A Small Language Model for Drug-Drug Interaction prediction and comparison with Large Language Models. Mach. Learn. Appl. 2025, 20, 100658. [Google Scholar] [CrossRef]
- CDC. CDC—2015 BRFSS Survey Data and Documentation. Available online: https://www.cdc.gov/brfss/annual_data/annual_2015.html (accessed on 9 September 2024).
- Kee, D.; Wisnivesky, J.; Kale, M.S. Lung cancer screening uptake: Analysis of BRFSS 2018. J. Gen. Intern. Med. 2021, 36, 2897–2899. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yuan, Y.; Zhang, Z. Enhancing llm factual accuracy with rag to counter hallucinations: A case study on domain-specific queries in private knowledge-bases. arXiv 2024, arXiv:2403.10446. [Google Scholar]
- Xian, L.; Xu, J. Smart Guardian: An AI-Based Coronary Heart Disease Prediction System, Focusing on Your Cardiac Health! SSRN Electron. J. 2024. Available online: https://ssrn.com/abstract=4713171 (accessed on 10 January 2025).
- Akther, K.; Kohinoor, M.S.R.; Priya, B.S.; Rahaman, M.J.; Rahman, M.M.; Shafiullah, M. Multi-Faceted Approach to Cardiovascular Risk Assessment by Utilizing Predictive Machine Learning and Clinical Data in a Unified Web Platform. IEEE Access 2024, 12, 120454–120473. [Google Scholar] [CrossRef]
- Goel, R. Using text embedding models and vector databases as text classifiers with the example of medical data. arXiv 2024, arXiv:cs.IR/2402.16886. [Google Scholar]
- Gundabathula, S.K.; Kolar, S.R. PromptMind Team at MEDIQA-CORR 2024: Improving Clinical Text Correction with Error Categorization and LLM Ensembles. arXiv 2024, arXiv:cs.CL/2405.08373. [Google Scholar]
- Acharya, A.; Shrestha, S.; Chen, A.; Conte, J.; Avramovic, S.; Sikdar, S.; Anastasopoulos, A.; Das, S. Clinical risk prediction using language models: Benefits and considerations. J. Am. Med. Inform. Assoc. 2024, 31, 1856–1864. [Google Scholar] [CrossRef]
- McInerney, D.J.; Dickinson, W.; Flynn, L.; Young, A.; Young, G.; van de Meent, J.W.; Wallace, B.C. Towards Reducing Diagnostic Errors with Interpretable Risk Prediction. arXiv 2024, arXiv:2402.10109. [Google Scholar]
- Liu, S.M.; Chen, J.H.; Liu, Z. An empirical study of dynamic selection and random under-sampling for the class imbalance problem. Expert Syst. Appl. 2023, 221, 119703. [Google Scholar] [CrossRef]
- Hall, T.; Beecham, S.; Bowes, D.; Gray, D.; Counsell, S. A systematic literature review on fault prediction performance in software engineering. IEEE Trans. Softw. Eng. 2011, 38, 1276–1304. [Google Scholar] [CrossRef]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef]
- Yaseen Jabarulla, M.; Oeltze-Jafra, S.; Beerbaum, P.; Uden, T. MedDoc-Bot: A Chat Tool for Comparative Analysis of Large Language Models in the Context of the Pediatric Hypertension Guideline. arXiv 2024, arXiv:2405.03359. [Google Scholar]
- Christophe, C.; Kanithi, P.K.; Raha, T.; Khan, S.; Pimentel, M.A. Med42-v2: A suite of clinical llms. arXiv 2024, arXiv:2408.06142. [Google Scholar]
- Han, T.; Adams, L.C.; Papaioannou, J.M.; Grundmann, P.; Oberhauser, T.; Löser, A.; Truhn, D.; Bressem, K.K. MedAlpaca—An open-source collection of medical conversational AI models and training data. arXiv 2023, arXiv:2304.08247. [Google Scholar]
- Team, G.; Riviere, M.; Pathak, S.; Sessa, P.G.; Hardin, C.; Bhupatiraju, S.; Hussenot, L.; Mesnard, T.; Shahriari, B.; Ramé, A.; et al. Gemma 2: Improving open language models at a practical size. arXiv 2024, arXiv:2408.00118. [Google Scholar]
- Masalkhi, M.; Ong, J.; Waisberg, E.; Zaman, N.; Sarker, P.; Lee, A.G.; Tavakkoli, A. A side-by-side evaluation of Llama 2 by meta with ChatGPT and its application in ophthalmology. Eye 2024, 38, 1789–1792. [Google Scholar] [CrossRef]
- Gupta, P.; Yau, L.Q.; Low, H.H.; Lee, I.; Lim, H.M.; Teoh, Y.X.; Koh, J.H.; Liew, D.W.; Bhardwaj, R.; Bhardwaj, R.; et al. WalledEval: A Comprehensive Safety Evaluation Toolkit for Large Language Models. arXiv 2024, arXiv:2408.03837. [Google Scholar]
- Jin, B.; Liu, G.; Han, C.; Jiang, M.; Ji, H.; Han, J. Large language models on graphs: A comprehensive survey. arXiv 2023, arXiv:2312.02783. [Google Scholar]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS 2017); Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 4768–4777. [Google Scholar]
- Streamlit, Inc. Available online: https://streamlit.io/ (accessed on 20 September 2024).
- National Heart Foundation of Australia. Heart Foundation. Available online: https://www.heartfoundation.org.au/for-professionals/guideline-for-managing-cvd (accessed on 20 September 2024).
- National Heart Foundation of Australia. CVD Risk Guideline. Available online: https://www.cvdcheck.org.au/using-the-calculator-to-assess-cvd-risk (accessed on 20 September 2024).
- Shi, L.; Kazda, M.; Sears, B.; Shropshire, N.; Puri, R. Ask-EDA: A Design Assistant Empowered by LLM, Hybrid RAG and Abbreviation De-hallucination. arXiv 2024, arXiv:2406.06575. [Google Scholar]
- Wang, J.; Shi, E.; Yu, S.; Wu, Z.; Ma, C.; Dai, H.; Yang, Q.; Kang, Y.; Wu, J.; Hu, H.; et al. Prompt Engineering for Healthcare: Methodologies and Applications. arXiv 2023, arXiv:2304.14670. [Google Scholar]
- Sufi, F.K. Addressing Data Scarcity in the Medical Domain: A GPT-Based Approach for Synthetic Data Generation and Feature Extraction. Information 2024, 15, 264. [Google Scholar] [CrossRef]


| Study | Focus Area | Methodology | Unique Contributions | Limitations |
|---|---|---|---|---|
| Xian et al. [19] | CVD Risk Assessment | Ensemble ML Models | Integrates GPT-3.5 for personalized health advice | Relies on traditional ML for risk prediction; GPT is used solely for advice generation, not for direct classification from BRFSS data. |
| Akther et al. [20] | CVD Risk Prediction | ML and DL | Developed a web-based application for personalized risk assessment | Uses traditional ML/DL models; personalized advice may not be as current or context-specific as that provided by RAG-based approaches. |
| Goel et al. [21] | Medical Data Classification | LLMs with Vector Databases | Focuses on LLMs for classification rather than risk assessment | General medical data classification; not specific to CVD risk from textualized BRFSS or personalized recommendations via RAG. |
| Gundabathula et al. [22] | Error Detection in Clinical Notes | Prompt-based Learning | Focuses on error correction, not risk assessment | Addresses error correction in clinical notes; does not focus on predictive CVD risk assessment or personalized recommendations. |
| Acharya et al. [23] | Clinical Risk Prediction | Fine-tuned LLMs | Investigates LLMs for clinical risk prediction but not specifically on BRFSS | Uses EHR data; does not compare medical vs. general LLMs on BRFSS for CVD or integrate RAG for advice. |
| McInerney et al. [24] | Risk Prediction for Reducing Errors | Neural Additive Models | Investigates LLMs for clinical risk prediction but not specifically on BRFSS | General risk estimation; may not be fine-tuned on BRFSS for binary CVD risk or compare LLM types with RAG for CVD advice. |
| Feature | Description |
|---|---|
| Age | Five-year age categories |
| Sex | Gender of the respondent |
| Smoking History | History of smoking 100 cigarettes |
| Physical Activity Level | Level of physical activity |
| Fruit Consumption | Frequency of fruit consumption |
| Vegetable Consumption | Frequency of vegetable consumption |
| BMI | Body Mass Index category |
| General Health | Self-reported general health status |
| High Cholesterol | History of high cholesterol |
| Kidney Disease | History of kidney disease |
| Diabetes | History of diabetes |
| Blood Pressure | History of high blood pressure |
| Alcohol Consumption | Frequency of alcohol consumption |
| Cancer History | History of any type of cancer |
| Education | Highest level of education attained |
| Income | Annual income level |
| Medical Cost Issues | Did not meet a doctor due to financial issues |
| Dataset Phase | Class 0 (Low Risk) | Class 1 (High Risk) |
|---|---|---|
| Original Dataset | 308,947 | 34,663 |
| Pre-RUS Split | ||
| Training | 216,360 | 24,167 |
| Validation | 46,286 | 5255 |
| Test (Held-out) | 46,301 | 5241 |
| Post-RUS | ||
| Training | 24,167 | 24,167 |
| Validation | 5255 | 5255 |
| Model | LR | BS | EP | LR | LA |
|---|---|---|---|---|---|
| Med LLMs | 1 | 5 | 64 | 32 | |
| Gemma-2b | 1 | 5 | 64 | 32 | |
| Mistral | 8 | 5 | 64 | 32 | |
| Llama3 | 8 | 5 | 64 | 32 | |
| BioBERT | 8 | 3 | FT | FT | |
| Llama-2-7b | 8 | 5 | 2 | 16 |
| Category | Model | Accuracy | Precision | Recall | F1-Score | AUC |
|---|---|---|---|---|---|---|
| Medical | BioBERT | 0.732 | 0.672 | 0.908 | 0.772 | 0.82 |
| Meditron | 0.741 | 0.754 | 0.715 | 0.734 | 0.81 | |
| Med42 | 0.728 | 0.664 | 0.922 | 0.772 | 0.82 | |
| MedAlpaca | 0.710 | 0.685 | 0.779 | 0.729 | 0.77 | |
| General | Mistral | 0.763 | 0.792 | 0.713 | 0.750 | 0.84 |
| Gemma2 | 0.730 | 0.670 | 0.907 | 0.770 | 0.82 | |
| LlaMa2 | 0.763 | 0.793 | 0.711 | 0.750 | 0.84 | |
| LlaMa3 | 0.761 | 0.790 | 0.712 | 0.749 | 0.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lakhdhar, W.; Arabi, M.; Ibrahim, A.; Arabi, A.; Serag, A. ChatCVD: A Retrieval-Augmented Chatbot for Personalized Cardiovascular Risk Assessment with a Comparison of Medical-Specific and General-Purpose LLMs. AI 2025, 6, 163. https://doi.org/10.3390/ai6080163
Lakhdhar W, Arabi M, Ibrahim A, Arabi A, Serag A. ChatCVD: A Retrieval-Augmented Chatbot for Personalized Cardiovascular Risk Assessment with a Comparison of Medical-Specific and General-Purpose LLMs. AI. 2025; 6(8):163. https://doi.org/10.3390/ai6080163
Chicago/Turabian StyleLakhdhar, Wafa, Maryam Arabi, Ahmed Ibrahim, Abdulrahman Arabi, and Ahmed Serag. 2025. "ChatCVD: A Retrieval-Augmented Chatbot for Personalized Cardiovascular Risk Assessment with a Comparison of Medical-Specific and General-Purpose LLMs" AI 6, no. 8: 163. https://doi.org/10.3390/ai6080163
APA StyleLakhdhar, W., Arabi, M., Ibrahim, A., Arabi, A., & Serag, A. (2025). ChatCVD: A Retrieval-Augmented Chatbot for Personalized Cardiovascular Risk Assessment with a Comparison of Medical-Specific and General-Purpose LLMs. AI, 6(8), 163. https://doi.org/10.3390/ai6080163








