Bio-Inspired Optimization of Transfer Learning Models for Diabetic Macular Edema Classification
Abstract
1. Introduction
- We have developed an innovative dataset by amalgamating the OCT 2017 and OCTC8 public datasets. This aids in comprehending the study’s generalizability.
- The parameters, such as learning rate, batch size, and dropout layer of the fully connected network, are further optimized using the Particle Swarm Optimization (PSO) approach, with comprehensive preprocessing.
- The methodology employs sophisticated transfer learning architectures, such as VGG16, VGG19, ResNet50, EfficientNetB7, EfficientNetV2S, InceptionV3, and InceptionResNetV2, for the examination of binary and multi-class OCT datasets.
- Explainable AI methodologies, particularly SHAP, offer clear insights into the decision-making processes of the model.
2. Literature Review
2.1. Diabetic Retinopathy
2.2. Diabetic Macular Edema
2.3. Glaucoma Detection
2.4. Pre-Processing Methods
2.5. Feature Extraction
2.6. Deep and Machine Learning Models
2.7. Optimization Methods
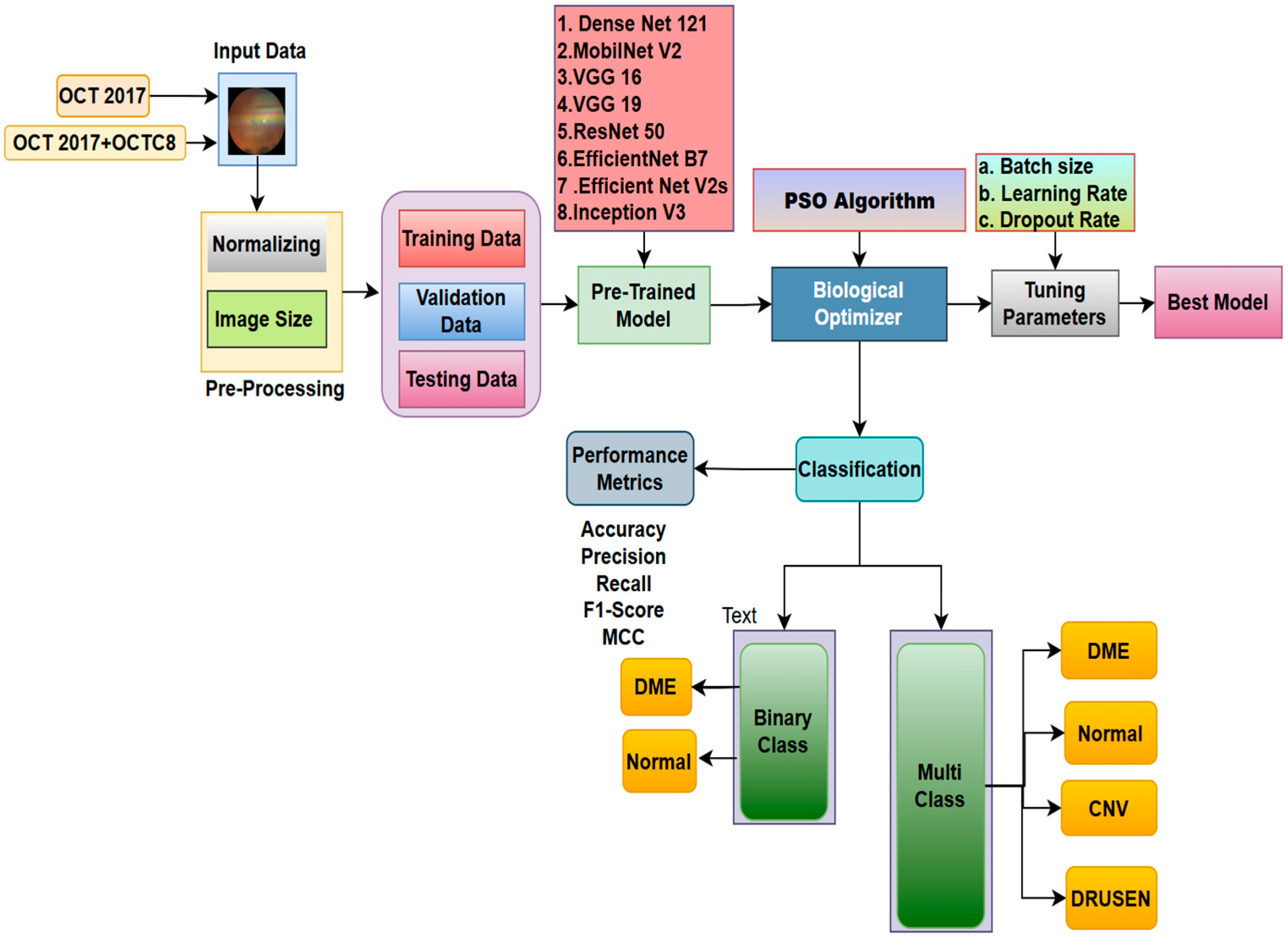
3. Methodology
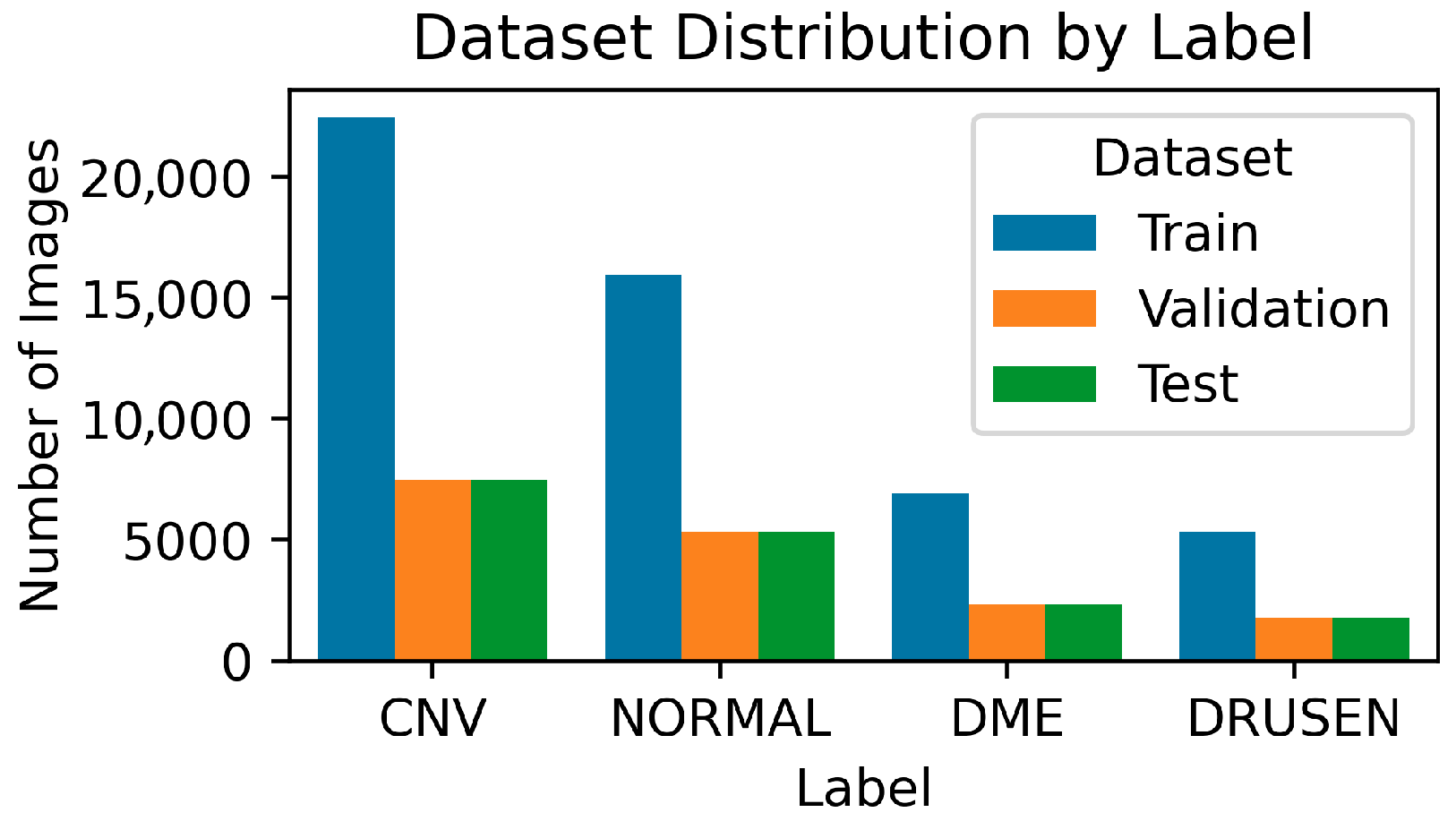
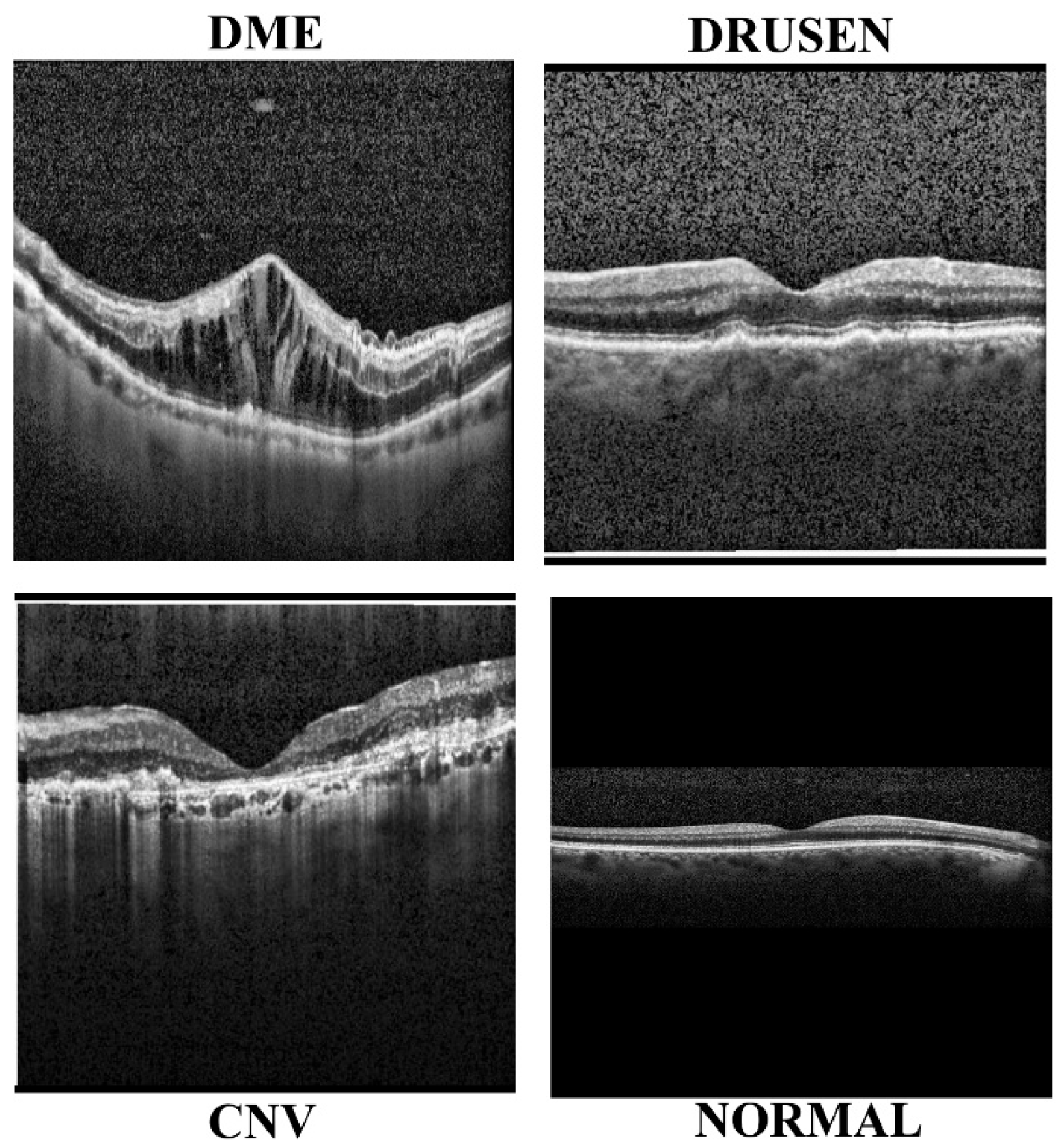
3.1. Data Collection
3.2. Model Architecture
3.2.1. DenseNet121
3.2.2. MobileNetV2
3.2.3. VGG16
3.2.4. VGG19
3.2.5. ResNet50
3.2.6. EfficientNetB7
3.2.7. EfficientNetV2S
3.2.8. InceptionV3
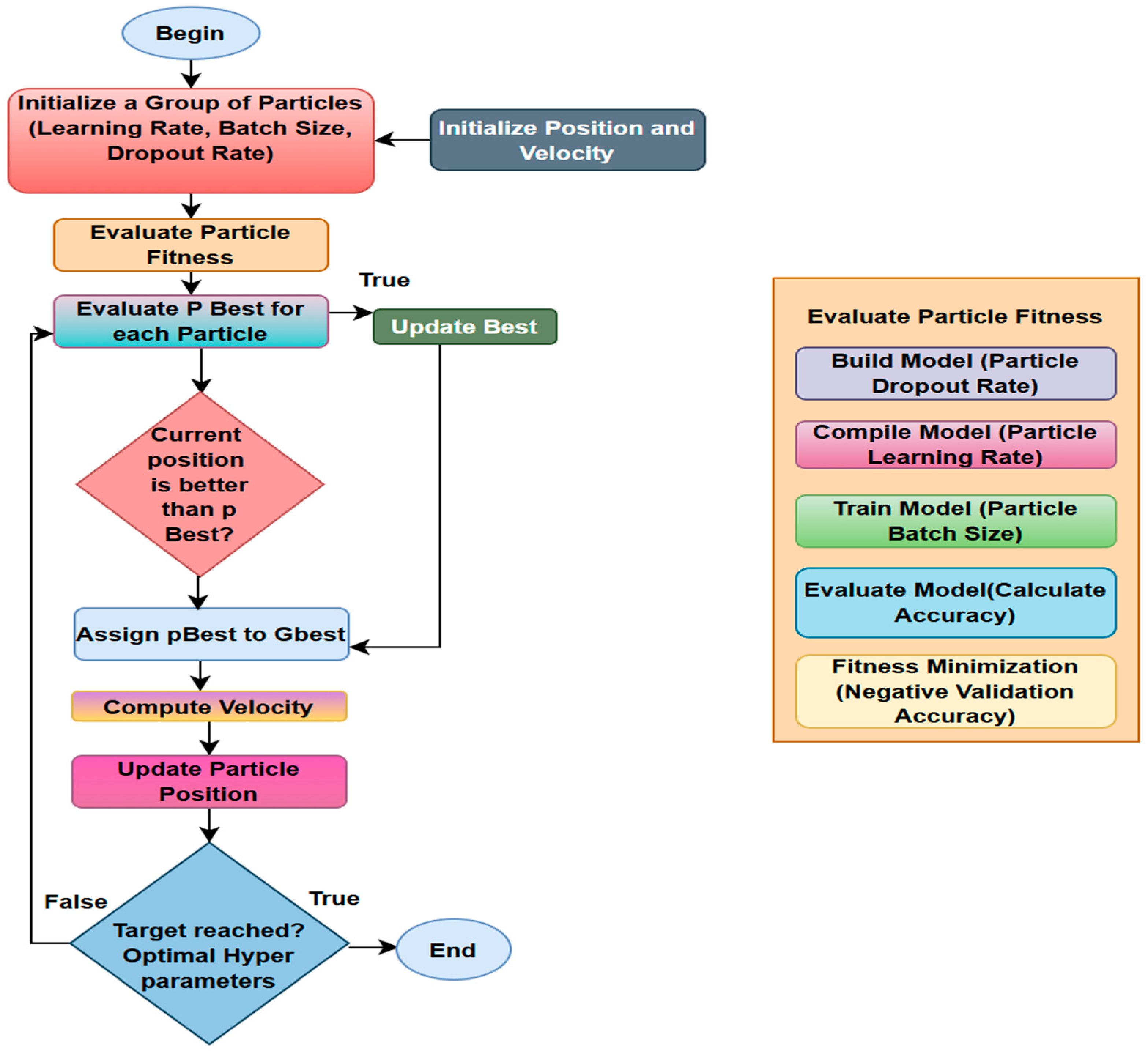
3.3. Biological Optimization: Particle Swarm
- Initialize: Generate a swarm of particles, each representing a random set of hyperparameter values. Set each particle’s best position (pBest) as well as the swarm’s overall best location (gBest). Evaluate by training the model and calculating the fitness function, which is the validation loss for each particle’s parameter set.
- Update: Update pBest if the particle’s current position is superior. Update gBest if one of the particles’ pBest is the best overall.
- Move: Each particle adjusts its velocity and position in response to its pBest and the swarm’s gBest.
- Stop: Repeat until the maximum number of iterations is achieved or gBest no longer improves. The final gBest represents the optimal parameter set.
3.4. Experimental Setup
3.4.1. PSO Parameters
3.4.2. Hyperparameter Settings
4. Results
4.1. Binary Classification
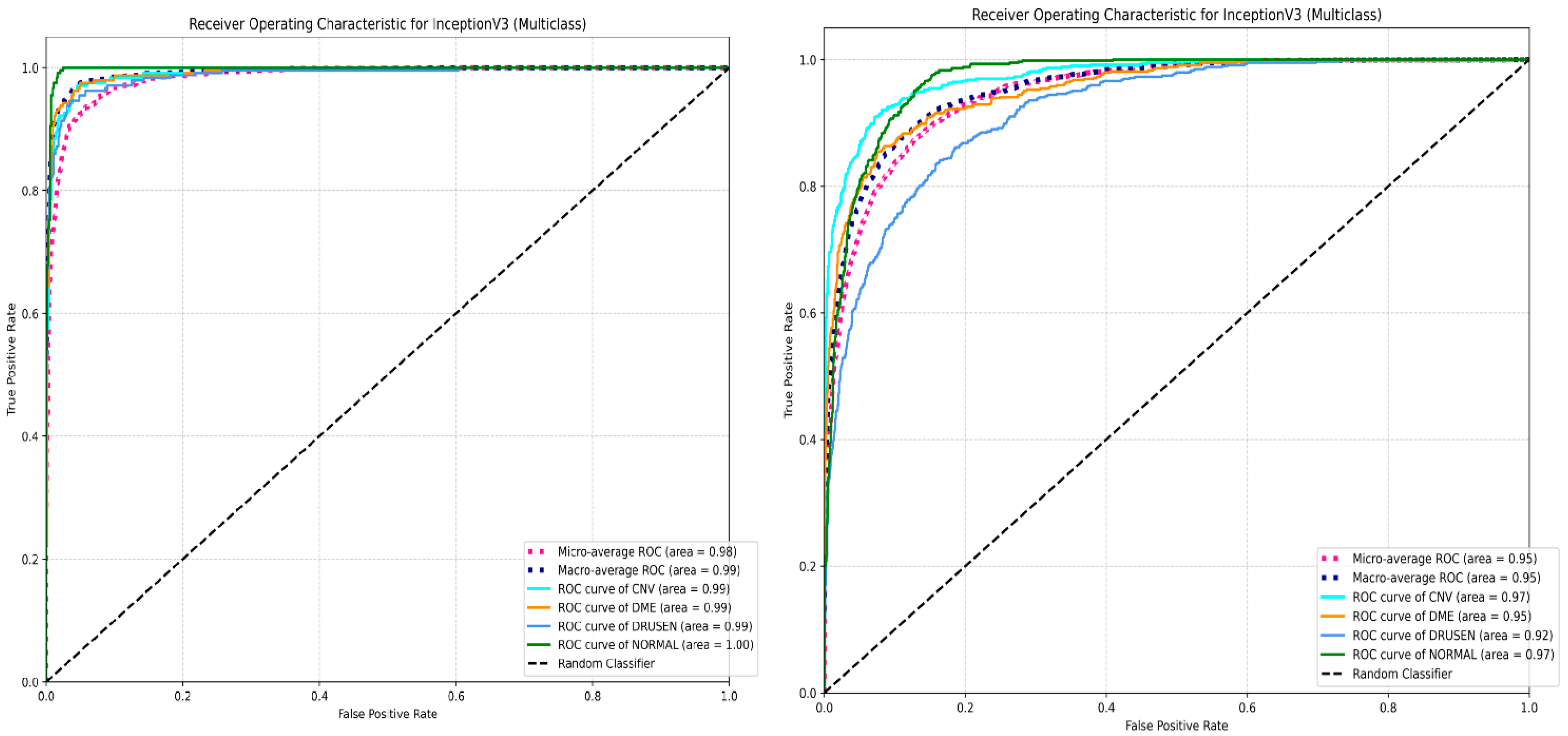
4.2. Multi-Class Classification
4.3. Wilcoxon Test
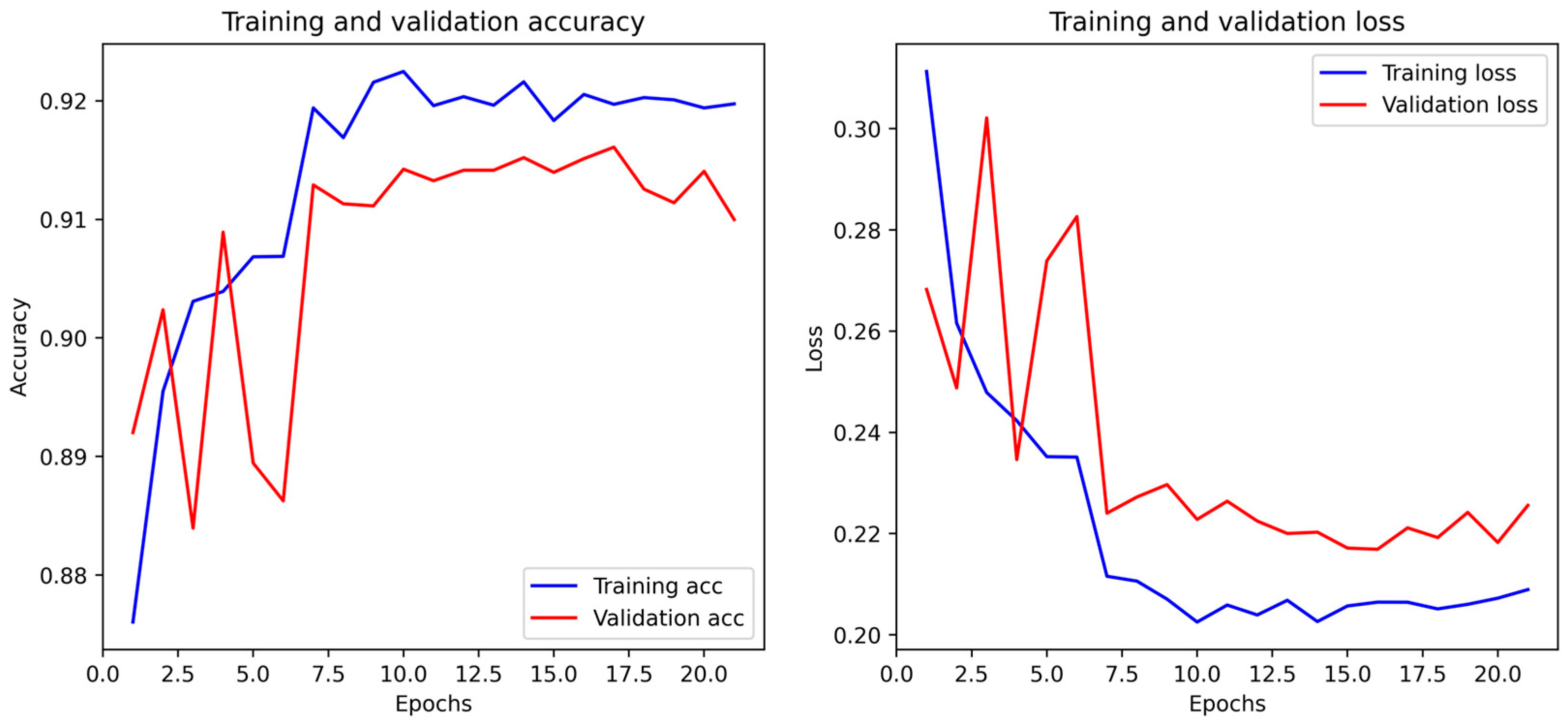
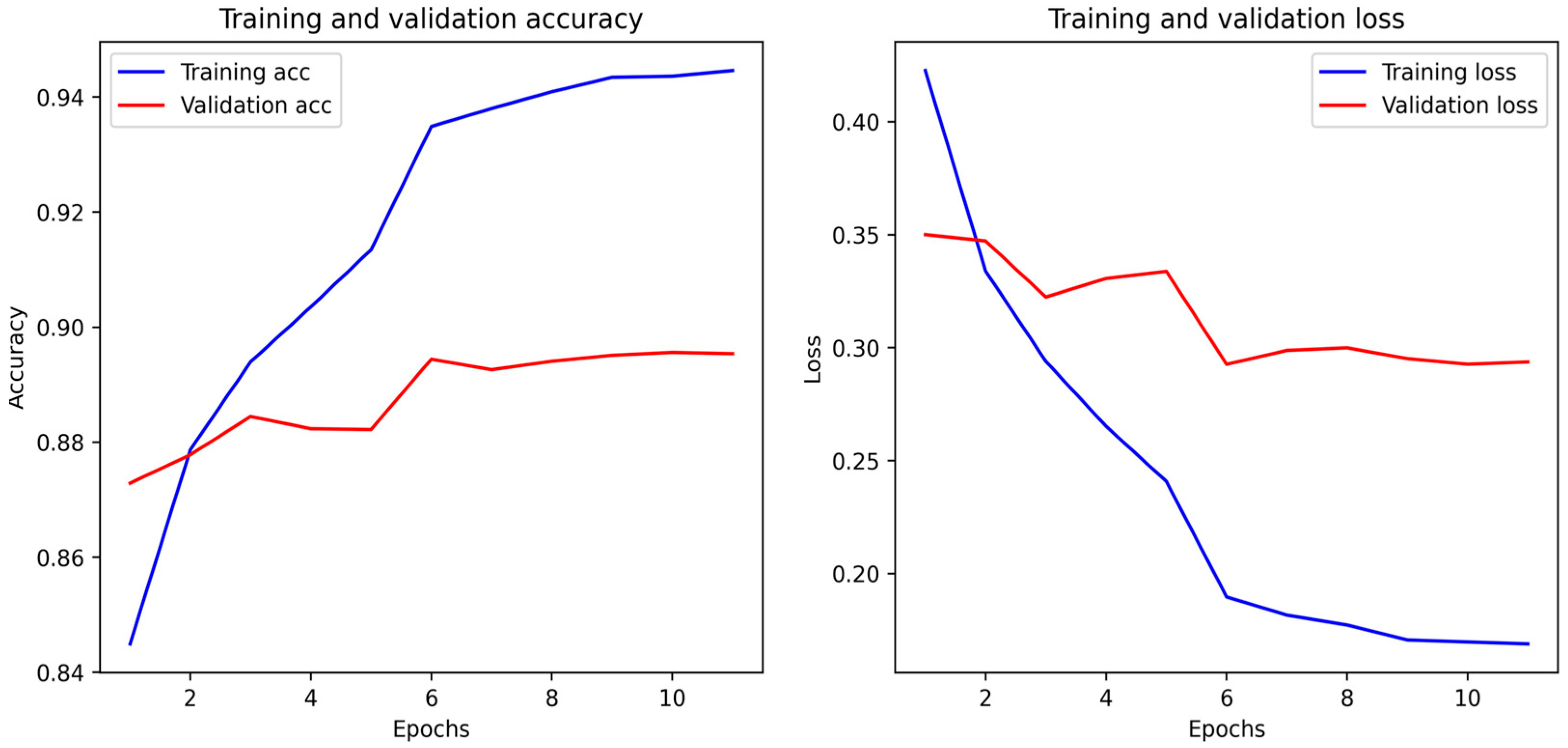
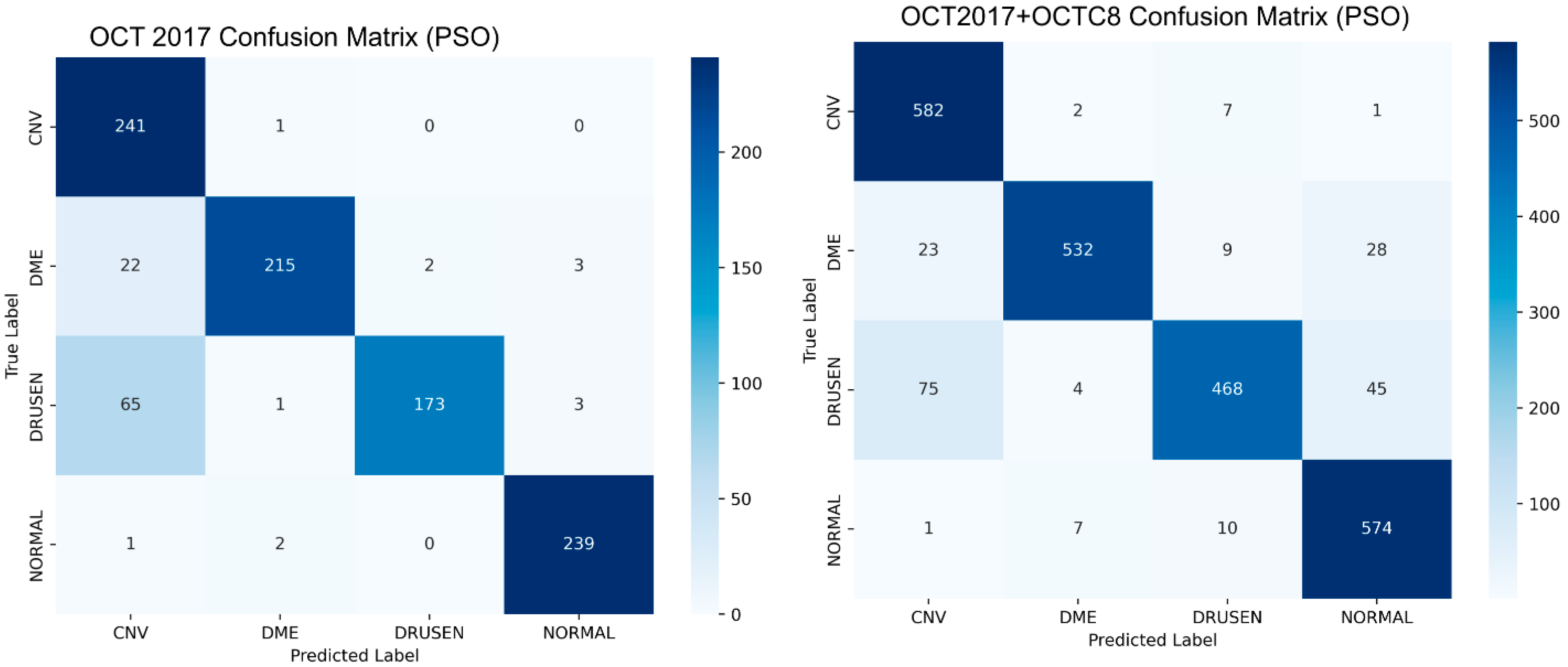
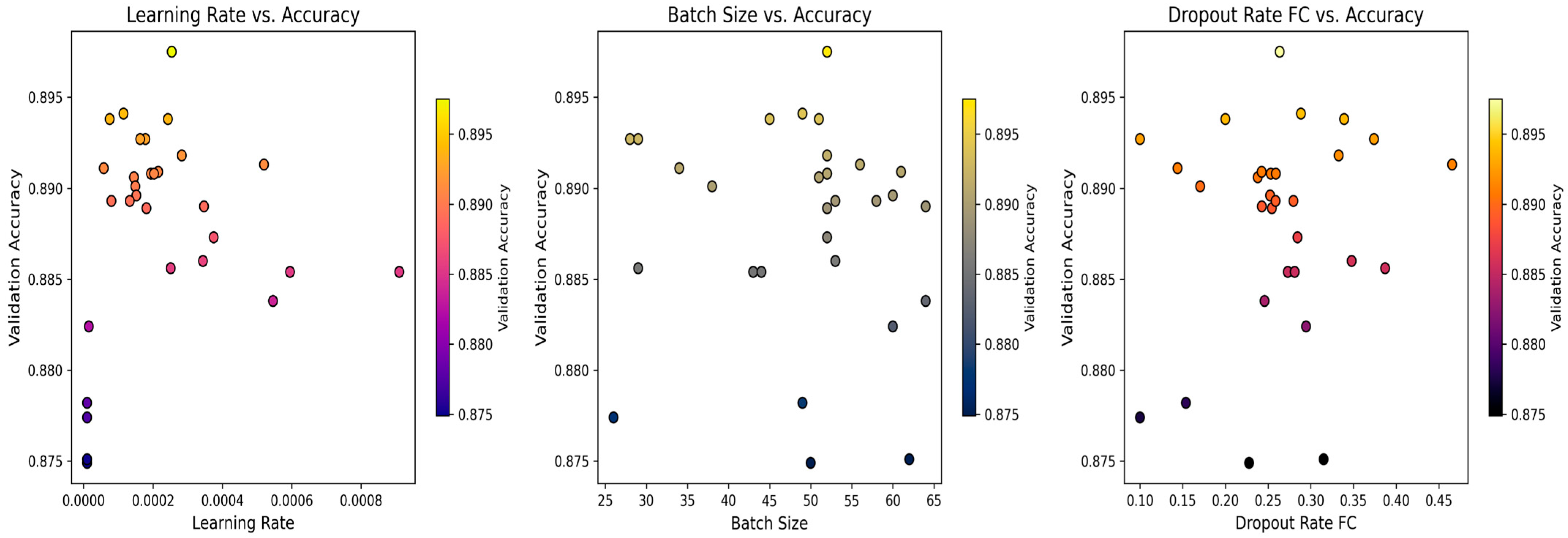
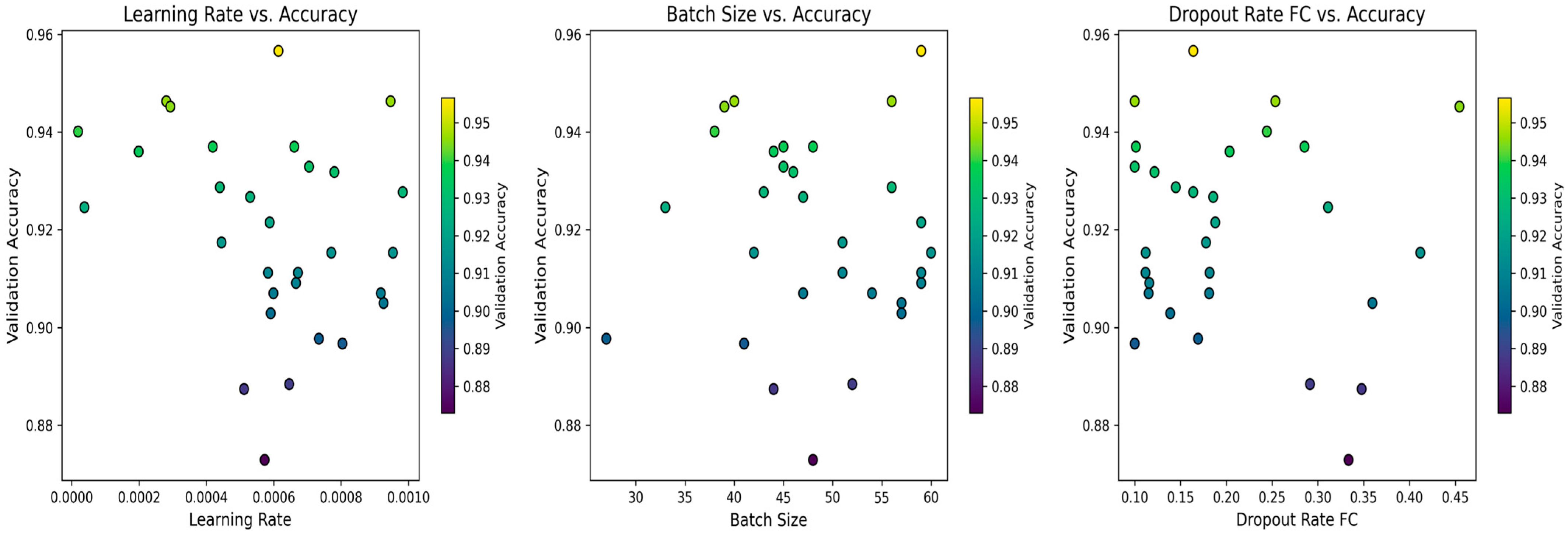
4.4. Biological Optimization: PSO
4.5. Score CAM
5. Discussion
5.1. Ablation Study: Impact of Dropout Layer
5.2. External Validation
6. Conclusions
Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sharma, N.; Lalwani, P. A multi model deep net with an explainable AI based framework for diabetic retinopathy segmentation and classification. Sci. Rep. 2025, 15, 8777. [Google Scholar] [CrossRef]
- Liu, J.A.; Che, H.; Zhao, A.; Li, N.; Huang, X.; Li, H.; Jiang, Z. SegDRoWS: Segmentation of diabetic retinopathy lesions by a whole-stage multi-scale feature fusion network. Biomed. Signal Process. Control 2025, 105, 107581. [Google Scholar] [CrossRef]
- Li, Z.-Q.; Fu, Z.-X.; Li, W.-J.; Fan, H.; Li, S.-N.; Wang, X.-M.; Zhou, P. Prediction of Diabetic Macular Edema Using Knowledge Graph. Diagnostics 2023, 13, 1858. [Google Scholar] [CrossRef] [PubMed]
- Moannaei, M.; Jadidian, F.; Doustmohammadi, T.; Kiapasha, A.M.; Bayani, R.; Rahmani, M.; Jahanbazy, M.R.; Sohrabivafa, F.; Asadi Anar, M.; Magsudy, A.; et al. Performance and limitation of machine learning algorithms for diabetic retinopathy screening and its application in health management: A meta-analysis. Biomed. Eng. OnLine 2025, 24, 34. [Google Scholar] [CrossRef]
- Li, Z.; Han, Y.; Yang, X. Multi-Fundus Diseases Classification Using Retinal Optical Coherence Tomography Images with Swin Transformer V2. J. Imaging 2023, 9, 203. [Google Scholar] [CrossRef]
- Yao, Z.; Yuan, Y.; Shi, Z.; Mao, W.; Zhu, G.; Zhang, G.; Wang, Z. FunSwin: A deep learning method to analysis diabetic retinopathy grade and macular edema risk based on fundus images. Front. Physiol. 2022, 13, 961386. (In English) [Google Scholar] [CrossRef]
- Phene, S.; Dunn, R.C.; Hammel, N.; Liu, Y.; Krause, J.; Kitade, N.; Schaekermann, M.; Sayres, R.; Wu, D.J.; Bora, A.; et al. Deep Learning and Glaucoma Specialists: The Relative Importance of Optic Disc Features to Predict Glaucoma Referral in Fundus Photographs. Ophthalmology 2019, 126, 1627–1639. [Google Scholar] [CrossRef]
- Wan, X.; Zhang, R.; Wang, Y.; Wei, W.; Song, B.; Zhang, L.; Hu, Y. Predicting diabetic retinopathy based on routine laboratory tests by machine learning algorithms. Eur. J. Med. Res. 2025, 30, 183. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Liu, K.; Yang, L.; Liu, R.; Xu, Y.; Xu, Y.; Zhang, Y.; Liang, D.; Sun, Y.; Hu, W. Predicting diabetic retinopathy based on biomarkers: Classification and regression tree models. Diabetes Res. Clin. Pract. 2025, 222, 112091. [Google Scholar] [CrossRef] [PubMed]
- Sunil, S.S.; Vindhya, A.S. Efficient diabetic retinopathy classification grading using GAN based EM and PCA learning framework. Multimed. Tools Appl. 2025, 84, 5311–5334. [Google Scholar] [CrossRef]
- Rajeshwar, S.; Thaplyal, S.; M., A.; G., S.S. Diabetic Retinopathy Detection Using DL-Based Feature Extraction and a Hybrid Attention-Based Stacking Ensemble. Adv. Public Health 2025, 2025, 8863096. [Google Scholar] [CrossRef]
- Lin, Y.; Dou, X.; Luo, X.; Wu, Z.; Liu, C.; Luo, T.; Wen, J.; Ling, B.W.-K.; Xu, Y.; Wang, W. Multi-view diabetic retinopathy grading via cross-view spatial alignment and adaptive vessel reinforcing. Pattern Recognit. 2025, 164, 111487. [Google Scholar] [CrossRef]
- Beuse, A.; Wenzel, D.A.; Spitzer, M.S.; Bartz-Schmidt, K.U.; Schultheiss, M.; Poli, S.; Grohmann, C. Automated Detection of Central Retinal Artery Occlusion Using OCT Imaging via Explainable Deep Learning. Ophthalmol. Sci. (Online) 2025, 5, 100630. [Google Scholar] [CrossRef] [PubMed]
- Tan-Torres, A.; Praveen, P.A.; Jeji, D.; Brant, A.; Yin, X.; Yang, L.; Singh, P.; Ali, T.; Traynis, I.; Jadeja, D.; et al. Validation of a Deep Learning Model for Diabetic Retinopathy on Patients with Young-Onset Diabetes. Ophthalmol. Ther. 2025, 14, 1147–1155. (In English) [Google Scholar] [CrossRef] [PubMed]
- Bilal, A.; Shafiq, M.; Obidallah, W.J.; Alduraywish, Y.A.; Tahir, A.; Long, H. Quantum chimp-enanced SqueezeNet for precise diabetic retinopathy classification. Sci. Rep. 2025, 15, 12890. [Google Scholar] [CrossRef] [PubMed]
- Zubair, M.; Umair, M.; Naqvi, R.A.; Hussain, D.; Owais, M.; Werghi, N. A comprehensive computer-aided system for an early-stage diagnosis and classification of diabetic macular edema. J. King Saud. Univ. Comput. Inf. Sci. 2023, 35, 101719. [Google Scholar] [CrossRef]
- Saini, D.J.B.; Sivakami, R.; Venkatesh, R.; Raghava, C.S.; Sandeep Dwarkanath, P.; Anwer, T.M.K.; Smirani, L.K.; Ahammad, S.H.; Pamula, U.; Amzad Hossain, M.; et al. Convolution neural network model for predicting various lesion-based diseases in diabetic macula edema in optical coherence tomography images. Biomed. Signal Process. Control 2023, 86, 105180. [Google Scholar] [CrossRef]
- Phan, C.; Hariprasad, S.M.; Mantopoulos, D. The Next-Generation Diagnostic and Therapeutic Algorithms for Diabetic Macular Edema Using Artificial Intelligence. Ophthalmic Surg. Lasers Imaging Retin. 2023, 54, 201–204. [Google Scholar] [CrossRef]
- Kumar, A.; Tewari, A.S. Classifying diabetic macular edema grades using extended power of deep learning. Multimed. Tools Appl. 2023, 83, 14151–14172. [Google Scholar] [CrossRef]
- Fu, Y.; Lu, X.; Zhang, G.; Lu, Q.; Wang, C.; Zhang, D. Automatic grading of Diabetic macular edema based on end-to-end network. Expert. Syst. Appl. 2023, 213, 118835. [Google Scholar] [CrossRef]
- Wang, T.-Y.; Chen, Y.-H.; Chen, J.-T.; Liu, J.-T.; Wu, P.-Y.; Chang, S.-Y.; Lee, Y.-W.; Su, K.-C.; Chen, C.-L. Diabetic Macular Edema Detection Using End-to-End Deep Fusion Model and Anatomical Landmark Visualization on an Edge Computing Device. Front. Med. 2022, 9, 851644. (In English) [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Tewari, A.S.; Singh, J.P. Classification of diabetic macular edema severity using deep learning technique. Res. Biomed. Eng. 2022, 38, 977–987. [Google Scholar] [CrossRef]
- Nasir, N.; Afreen, N.; Patel, R.; Kaur, S.; Sameer, M. A transfer learning approach for diabetic retinopathy and diabetic macular edema severity grading. Rev. d’Intelligence Artif. 2021, 35, 497–502. [Google Scholar] [CrossRef]
- Stino, H.; Birner, K.; Steiner, I.; Hinterhuber, L.; Gumpinger, M.; Schürer-Waldheim, S.; Bogunovic, H.; Schmidt-Erfurth, U.; Reiter, G.S.; Pollreisz, A. Correlation of point-wise retinal sensitivity with localized features of diabetic macular edema using deep learning. Can. J. Ophthalmol. 2025, 60, 297–305. (In English) [Google Scholar] [CrossRef] [PubMed]
- Venkataiah, C.; Chennakesavulu, M.; Mallikarjuna Rao, Y.; Janardhana Rao, B.; Ramesh, G.; Sofia Priya Dharshini, J.; Jayamma, M. A novel eye disease segmentation and classification model using advanced deep learning network. Biomed. Signal Process. Control 2025, 105, 107565. [Google Scholar] [CrossRef]
- Ikram, A.; Imran, A. ResViT FusionNet Model: An explainable AI-driven approach for automated grading of diabetic retinopathy in retinal images. Comput. Biol. Med. 2025, 186, 109656. [Google Scholar] [CrossRef]
- Navaneethan, R.; Devarajan, H. Enhancing diabetic retinopathy detection through preprocessing and feature extraction with MGA-CSG algorithm. Expert. Syst. Appl. 2024, 249, 123418. [Google Scholar] [CrossRef]
- Muthusamy, D.; Palani, P. Deep learning model using classification for diabetic retinopathy detection: An overview. Artif. Intell. Rev. 2024, 57, 185. [Google Scholar] [CrossRef]
- Fu, Y.; Wei, Y.; Chen, S.; Chen, C.; Zhou, R.; Li, H.; Qiu, M.; Xie, J.; Huang, D. UC-stack: A deep learning computer automatic detection system for diabetic retinopathy classification. Phys. Med. Biol. 2024, 69, 045021. (In English) [Google Scholar] [CrossRef]
- Özbay, E. An active deep learning method for diabetic retinopathy detection in segmented fundus images using artificial bee colony algorithm. Artif. Intell. Rev. 2023, 56, 3291–3318. [Google Scholar] [CrossRef]
- Pugal Priya, R.; Saradadevi Sivarani, T.; Gnana Saravanan, A. Deep long and short term memory based Red Fox optimization algorithm for diabetic retinopathy detection and classification. Int. J. Numer. Method. Biomed. Eng. 2022, 38, e3560. (In English) [Google Scholar] [CrossRef]
- Berbar, M.A. Features extraction using encoded local binary pattern for detection and grading diabetic retinopathy. Health Inf. Sci. Syst. 2022, 10, 14. (In English) [Google Scholar] [CrossRef]
- Pan, Z.; Wu, X.; Li, Z. Central pixel selection strategy based on local gray-value distribution by using gradient information to enhance LBP for texture classification. Expert. Syst. Appl. 2019, 120, 319–334. [Google Scholar] [CrossRef]
- D S, R.; Saji, K.S. Hybrid deep learning framework for diabetic retinopathy classification with optimized attention AlexNet. Comput. Biol. Med. 2025, 190, 110054. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, R.; Gandomi, A.H. Adaptive isomap feature extractive gradient deep belief network classifier for diabetic retinopathy identification. Multimed. Tools Appl. 2025, 84, 6349–6370. [Google Scholar] [CrossRef]
- Bindu Priya, M.; Manoj Kumar, D. MHSAGGCN-BCOA: A novel deep learning based approach for diabetic retinopathy detection. Biomed. Signal Process. Control 2025, 105, 107569. [Google Scholar] [CrossRef]
- Venkaiahppalaswamy, B.; Prasad Reddy, P.; Batha, S. Hybrid deep learning approaches for the detection of diabetic retinopathy using optimized wavelet based model. Biomed. Signal Process. Control 2023, 79, 104146. [Google Scholar] [CrossRef]
- Sushith, M.; Lakkshmanan, A.; Saravanan, M.; Castro, S. Attention dual transformer with adaptive temporal convolutional for diabetic retinopathy detection. Sci. Rep. 2025, 15, 7694. [Google Scholar] [CrossRef]
- Kufel, J.; Bargieł-Łączek, K.; Kocot, S.; Koźlik, M.; Bartnikowska, W.; Janik, M.; Czogalik, Ł.; Dudek, P.; Magiera, M.; Lis, A. What is machine learning, artificial neural networks and deep learning?—Examples of practical applications in medicine. Diagnostics 2023, 13, 2582. [Google Scholar] [CrossRef]
- Vasireddi, H.K.; K, S.D.; G, N.V.R. Deep feed forward neural network-based screening system for diabetic retinopathy severity classification using the lion optimization algorithm. Graefes Arch. Clin. Exp. Ophthalmol. 2022, 260, 1245–1263. (In English) [Google Scholar] [CrossRef]
- Asif, S. DEO-Fusion: Differential evolution optimization for fusion of CNN models in eye disease detection. Biomed. Signal Process. Control 2025, 107, 107853. [Google Scholar] [CrossRef]
- Karthika, S.; Durgadevi, M. Improved ResNet_101 assisted attentional global transformer network for automated detection and classification of diabetic retinopathy disease. Biomed. Signal Process. Control 2024, 88, 105674. [Google Scholar] [CrossRef]
- Al-Kahtani, N.; Varela-Aldás, J.; Aljarbouh, A.; Ishak, M.K.; Mostafa, S.M. Discrete migratory bird optimizer with deep transfer learning aided multi-retinal disease detection on fundus imaging. Results Eng. 2025, 26, 104574. [Google Scholar] [CrossRef]
- Bhimavarapu, U. Diagnosis and multiclass classification of diabetic retinopathy using enhanced multi thresholding optimization algorithms and improved Naive Bayes classifier. Multimed. Tools Appl. 2024, 83, 81325–81359. [Google Scholar] [CrossRef]
- Reddy, S.R.G.; Varma, G.P.S.; Davuluri, R.L. Resnet-based modified red deer optimization with DLCNN classifier for plant disease identification and classification. Comput. Electr. Eng. 2023, 105, 108492. [Google Scholar] [CrossRef]
- Minija, S.J.; Rejula, M.A.; Ross, B.S. Automated detection of diabetic retinopathy using optimized convolutional neural network. Multimed. Tools Appl. 2023, 83, 21065–21080. [Google Scholar] [CrossRef]
- Kulyabin, M.; Zhdanov, A.; Nikiforova, A.; Stepichev, A.; Kuznetsova, A.; Ronkin, M.; Borisov, V.; Bogachev, A.; Korotkich, S.; Constable, P.A.; et al. Octdl: Optical coherence tomography dataset for image-based deep learning methods. Sci. Data 2024, 11, 365. [Google Scholar] [CrossRef] [PubMed]
- Naren, O.S. Retinal OCT Image Classification—C8. Kaggle 2021. [Google Scholar] [CrossRef]
- Bhimavarapu, U.; Battineni, G. Automatic Microaneurysms Detection for Early Diagnosis of Diabetic Retinopathy Using Improved Discrete Particle Swarm Optimization. J. Pers. Med. 2022, 12, 317. (In English) [Google Scholar] [CrossRef] [PubMed]
- Melin, P.; Sánchez, D.; Cordero-Martínez, R. Particle swarm optimization of convolutional neural networks for diabetic retinopathy classification. In Fuzzy Logic and Neural Networks for Hybrid Intelligent System Design; Springer: Berlin/Heidelberg, Germany, 2023; pp. 237–252. [Google Scholar]









| References | Data Set and No. of Subjects | Methods and Models Used | Evaluation Metrics | Crucial Findings | Research Challenges |
|---|---|---|---|---|---|
| [8] | T2DM-4259 External validation-N = 323 | eXtreme Gradient Boosting (XGBoost), support vector machine (SVM), gradient boosting decision tree (GBDT), neural network (NN), and logistic regression (LR). The Shapley Additive Explanation (SHAP) | AUC, sensitivity, specificity, F1-score, SHAP | 9:1 split ratio | Limited Models used can be tried with transformer models. |
| [38] | DRIVE JPEG 40 color fundus images, including 7 abnormal pathology cases. | ADTATC (Attention dual transformer with adaptive temporal convolutional) | Precision, Recall, Specificity, F1-ScoreAccuracy, kappa coefficient | Batch size −32, epoch −50, loss function-categorical entropy loss. Dropout rate −0.5. | Limited to supervised algorithms, Optimization algorithms can be used in the work. |
| [1] | DiaRetDB1, APTOS 2019 dataset, Kaggle EyePACS. | AGF and Contrast-Limited Adaptive Histogram Equalization (CLAHE)-Preprocessing modified U-Net Architecture-Segmentation DenseNet, and an OGRU enhanced by the SANGO algorithm -Classification | IoU, accuracy, precision, recall, F1-measure, and Matthew Correlation Coefficient (MCC), Negative Predictive Value (NPV), Intersection over Union (IoU), and Dice Similarity Coefficient (DSC). | K-Fold cross validation = 5 Self-adaptive Northern goshawk optimization for convergence rate | Optimized GRU Parameters tuned |
| [10] | 2750 samples provided by the Kaggle platform 3 classes | Deep ConvNet | Specificity, Sensitivity, AUC ROC | 80% and 20% train-test split transfer learning, data augmentation, and class balancing. | optimizing the preprocessing techniques, optimizing the balancing techniques |
| [5] | The OCT2017 dataset contains 84,452 retinal OCT images with 4 classes | Swin Transformer V2 | Accuracy, Precision, Recall, F1-score | The loss function is improved by introducing PolyLoss | Data set limitations and requirements of experts in the field. |
| [6] | MESSIDOR dataset (1200 color numerical images) | FunSWIN | Accuracy, Precision, Recall, F1-score | Model convergence performance | Data Limitations |
| [13] | Eyepacs Aptos Messidor2 | RESNET, VGG, EFFICIENT NET | Accuracy, Precision, Recall, F1-score | Quadratic Weighted Kappa | Future work—Federated Learning (FL) |
| Model Name | Core Concept/Innovation | Key Architectural Features | Approx. Parameters (Base Model, Include Top = False) |
|---|---|---|---|
| DenseNet121 | Dense Connectivity, Feature Reuse | Each layer is connected to all subsequent layers within a dense block; transition layers for down-sampling. | ~7 M |
| MobileNetV2 | Lightweight, Mobile-First | Inverted residual blocks with linear bottlenecks; depth-wise separable convolutions. | ~2.2 M |
| VGG16 | Simplicity, Uniformity | Stacks of small 3 × 3 convolutional filters; max-pooling layers. Relatively deep (16 layers). | ~14.7 M |
| VGG19 | Deeper VGG Variant | Like VGG16, but with 19 layers (more 3 × 3 convolutional layers per block). | ~20.0 M |
| ResNet50 | Residual Learning, Skip Connections | Residual blocks that add input directly to the block output via “skip connections” to learn identity mappings. | ~23.5 M |
| EfficientNetB7 | Compound Scaling | Systematically scales network depth, width, and resolution using a compound coefficient; uses inverted residual blocks (MBConv) and Squeeze-and-Excitation blocks. | ~66 M |
| EfficientNetV2S | Faster Training, Improved Efficiency | Builds on Efficient Net with training-aware neural architecture search; uses Fused-MBConv layers in early stages for faster training; smaller expansion ratios and 3 × 3 kernels in MBConv. | ~21 M |
| InceptionV3 | Parallel Convolutions, Factorization | “Inception modules” performing parallel convolutions with different filter sizes (e.g., 1 × 1, 3 × 3, 5 × 5); factorization of convolutions (e.g., 5 × 5 into 1 × 5 and 5 × 1); auxiliary classifiers. | ~21.8 M |
| Parameters | Range |
|---|---|
| Swam Size | 5 |
| Maximum iteration | 5 |
| Lower Bounds | [0.00001, 16, 0.1] |
| Upper Bounds | [0.001, 64, 0.5] |
| Optimizer | ADAM |
| Parameters | Range |
|---|---|
| Image Size | (224, 224) |
| Batch Size | 32 |
| Callbacks | ES (10), RLROP (5) |
| Epochs | 60 |
| Optimizer | ADAM |
| Data Augmentation | Rescale, Flip, Rotate (20) |
| Train-test-split | 70:30 |
| Models | Accuracy | Precision | Recall | F1-Score | Misclassification Rate | Matthews Correlation Coefficient | Jaccard Index (IOU) | Kappa Coefficient |
|---|---|---|---|---|---|---|---|---|
| DESNET 121 | 0.960 | 0.960 | 0.960 | 0.960 | 0.072 | 0.704 | 0.545 | 0.667 |
| EfficentNetB7 | 0.500 | 0.250 | 0.500 | 0.330 | 0.500 | 0.00 | 0.500 | 0.000 |
| Efficient Net V2S | 0.770 | 0.830 | 0.770 | 0.760 | 0.229 | 0.600 | 0.682 | 0.541 |
| RESNET50 | 0.630 | 0.770 | 0.630 | 0.580 | 0.365 | 0.379 | 0.574 | 0.268 |
| VGG-16 | 0.940 | 0.950 | 0.940 | 0.940 | 0.055 | 0.894 | 0.899 | 0.888 |
| VGG-19 | 0.920 | 0.930 | 0.920 | 0.920 | 0.082 | 0.845 | 0.857 | 0.834 |
| MobileNetV2 | 0.980 | 0.980 | 0.980 | 0.980 | 0.024 | 0.951 | 0.952 | 0.950 |
| Inception V3 | 0.980 | 0.980 | 0.980 | 0.980 | 0.018 | 0.963 | 0.964 | 0.962 |
| Models | Accuracy | Precision | Recall | F1-Score | Misclassification Rate | Matthews Correlation Coefficient | Jaccard Index (IoU) | Kappa Coefficient |
|---|---|---|---|---|---|---|---|---|
| DESNET 121 | 0.90 | 0.91 | 0.90 | 0.90 | 0.01 | 0.87 | 0.81 | 0.86 |
| EfficentNetB7 | 0.25 | 0.06 | 0.25 | 0.10 | 0.75 | 0.0 | 0.06 | 0.86 |
| Efficient Net V2S | 0.53 | 0.71 | 0.53 | 0.43 | 0.41 | 0.41 | 0.30 | 0.86 |
| RESNET50 | 0.48 | 0.25 | 0.48 | 0.32 | 0.52 | 0.26 | 0.24 | 0.86 |
| VGG-16 | 0.88 | 0.90 | 0.88 | 0.87 | 0.12 | 0.84 | 0.77 | 0.86 |
| VGG-19 | 0.88 | 0.90 | 0.88 | 0.88 | 0.12 | 0.84 | 0.78 | 0.86 |
| MobileNetV2 | 0.91 | 0.92 | 0.91 | 0.91 | 0.08 | 0.88 | 0.84 | 0.86 |
| Inception V3 | 0.90 | 0.91 | 0.90 | 0.90 | 0.01 | 0.87 | 0.81 | 0.86 |
| Models | Accuracy | Precision | Recall | F1-Score | Misclassification Rate | Matthews Correlation Coefficient | Jaccard Index (IoU) | Kappa Coefficient |
|---|---|---|---|---|---|---|---|---|
| DESNET 121 | 0.809 | 0.831 | 0.809 | 0.804 | 0.190 | 0.755 | 0.676 | 0.7461 |
| EfficentNetB7 | 0.25 | 0.0625 | 0.2500 | 0.100 | 0.7500 | 0.00 | 0.0625 | 0.00 |
| Efficient Net V2S | 0.4882 | 0.60 | 0.4882 | 0.3677 | 0.5118 | 0.3280 | 0.2580 | 0.3176 |
| RESNET50 | 0.446 | 0.229 | 0.446 | 0.3008 | 0.553 | 0.230 | 0.216 | 0.2618 |
| VGG-16 | 0.7855 | 0.8140 | 0.7855 | 0.772 | 0.2145 | 0.7256 | 0.6401 | 0.7140 |
| VGG-19 | 0.7736 | 0.8040 | 0.7736 | 0.7654 | 0.2264 | 0.7109 | 0.6235 | 0.6982 |
| MobileNetV2 | 0.8970 | 0.9056 | 0.8970 | 0.8956 | 0.1030 | 0.8664 | 0.8126 | 0.8626 |
| Inception V3 | 0.909 | 0.9157 | 0.9096 | 0.9086 | 0.0904 | 0.8822 | 0.8335 | 0.8795 |
| Models | Accuracy | Precision | Recall | F1-Score | Misclassification Rate | Matthews Correlation Coefficient | Jaccard Index (IoU) | Kappa Coefficient |
|---|---|---|---|---|---|---|---|---|
| Inception V3 (OCT 2017) | 0.896 | 0.9196 | 0.8967 | 0.8967 | 0.1033 | 0.8727 | 0.8193 | 0.8622 |
| InceptionV3 (OCT 2017 + OCTC8) | 0.913 | 0.9160 | 0.9105 | 0.9080 | 0.089443 | 0.8810 | 0.8352 | 0.8807 |
| Models | Learning Rate | Batch-Size | Dropout Rate |
|---|---|---|---|
| Inception V3 | 0.000614 | 59 | 0.9566 |
| Mobile Net | 0.000254 | 52 | 0.2636 |
| Models | Accuracy | Precision | Recall | F1-Score | Misclassification Rate | Matthews Correlation Coefficient | Jaccard Index (IoU) | Kappa Coefficient |
|---|---|---|---|---|---|---|---|---|
| MobileNetV2 | 0.8003 | 0.8221 | 0.8003 | 0.7943 | 0.1997 | 0.7430 | 0.6628 | 0.7337 |
| Inception V3 | 0.7842 | 0.8018 | 0.7842 | 0.7787 | 0.2158 | 0.7199 | 0.6405 | 0.7123 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutawa, A.M.; Sabti, K.; Sundaram Thankaleela, B.S.; Raizada, S. Bio-Inspired Optimization of Transfer Learning Models for Diabetic Macular Edema Classification. AI 2025, 6, 269. https://doi.org/10.3390/ai6100269
Mutawa AM, Sabti K, Sundaram Thankaleela BS, Raizada S. Bio-Inspired Optimization of Transfer Learning Models for Diabetic Macular Edema Classification. AI. 2025; 6(10):269. https://doi.org/10.3390/ai6100269
Chicago/Turabian StyleMutawa, A. M., Khalid Sabti, Bibin Shalini Sundaram Thankaleela, and Seemant Raizada. 2025. "Bio-Inspired Optimization of Transfer Learning Models for Diabetic Macular Edema Classification" AI 6, no. 10: 269. https://doi.org/10.3390/ai6100269
APA StyleMutawa, A. M., Sabti, K., Sundaram Thankaleela, B. S., & Raizada, S. (2025). Bio-Inspired Optimization of Transfer Learning Models for Diabetic Macular Edema Classification. AI, 6(10), 269. https://doi.org/10.3390/ai6100269






