Multi-Fundus Diseases Classification Using Retinal Optical Coherence Tomography Images with Swin Transformer V2
Abstract
:1. Introduction
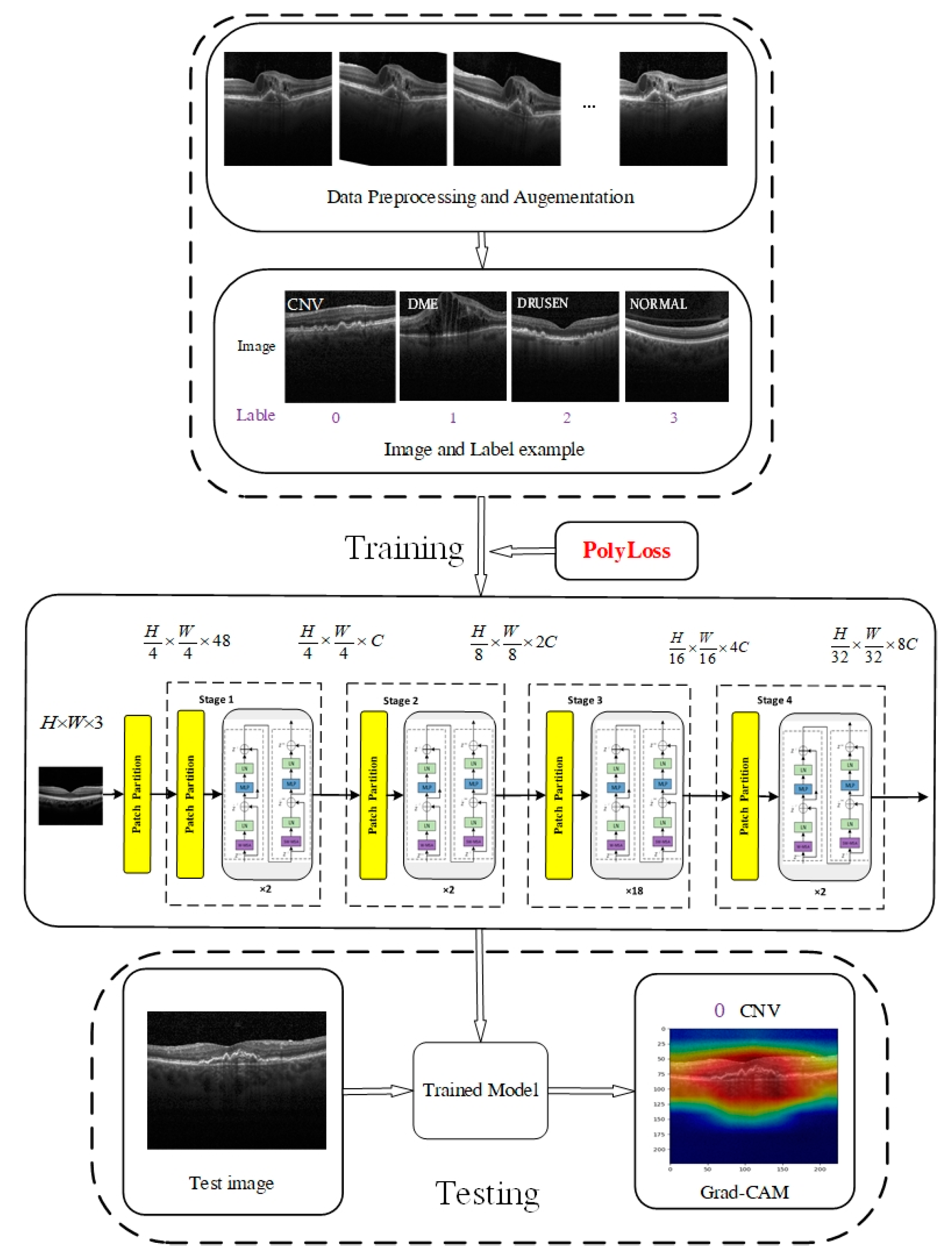
1.1. The Proposed Model
- The proposed method will first use the Swin Transformer V2 model to classify multiple diseases in retinal OCT images.
- Based on the Swin Transformer V2 model, its loss function is improved by introducing PolyLoss, which improves the model’s performance.
- Experimental validation was performed with two datasets, OCT2017 and OCT-C8, and using Grad-CAM visualization to help understand decision-making mechanisms in network models.
1.2. Related Work
2. Materials and Methods
2.1. Architecture of Swin Transformer V2
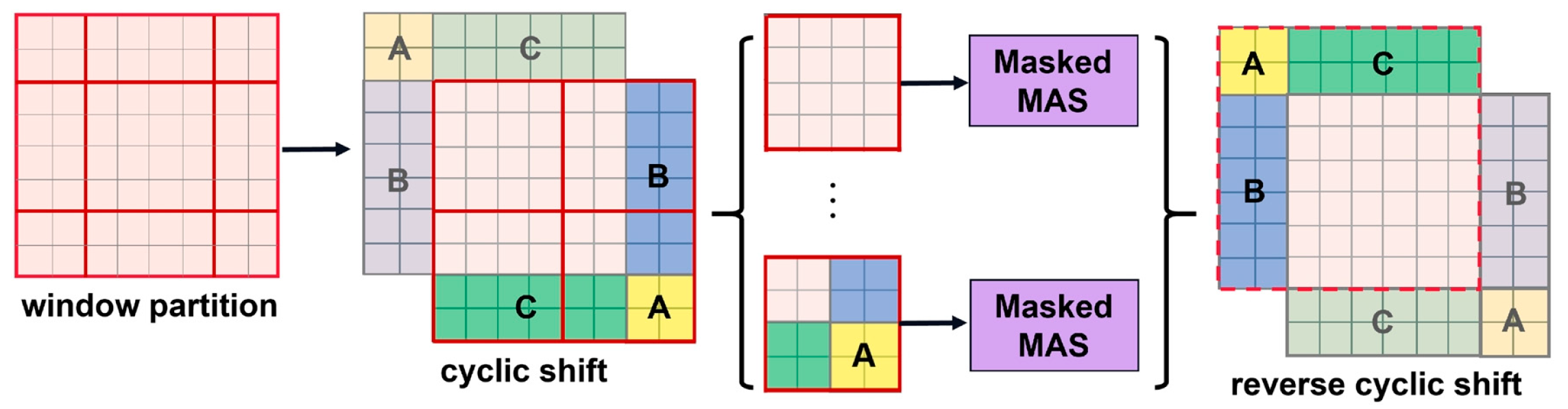
2.2. Shifted-Window-Based Self-Attention
2.3. PolyLoss
2.4. Datasets
2.5. Evaluation Metrics
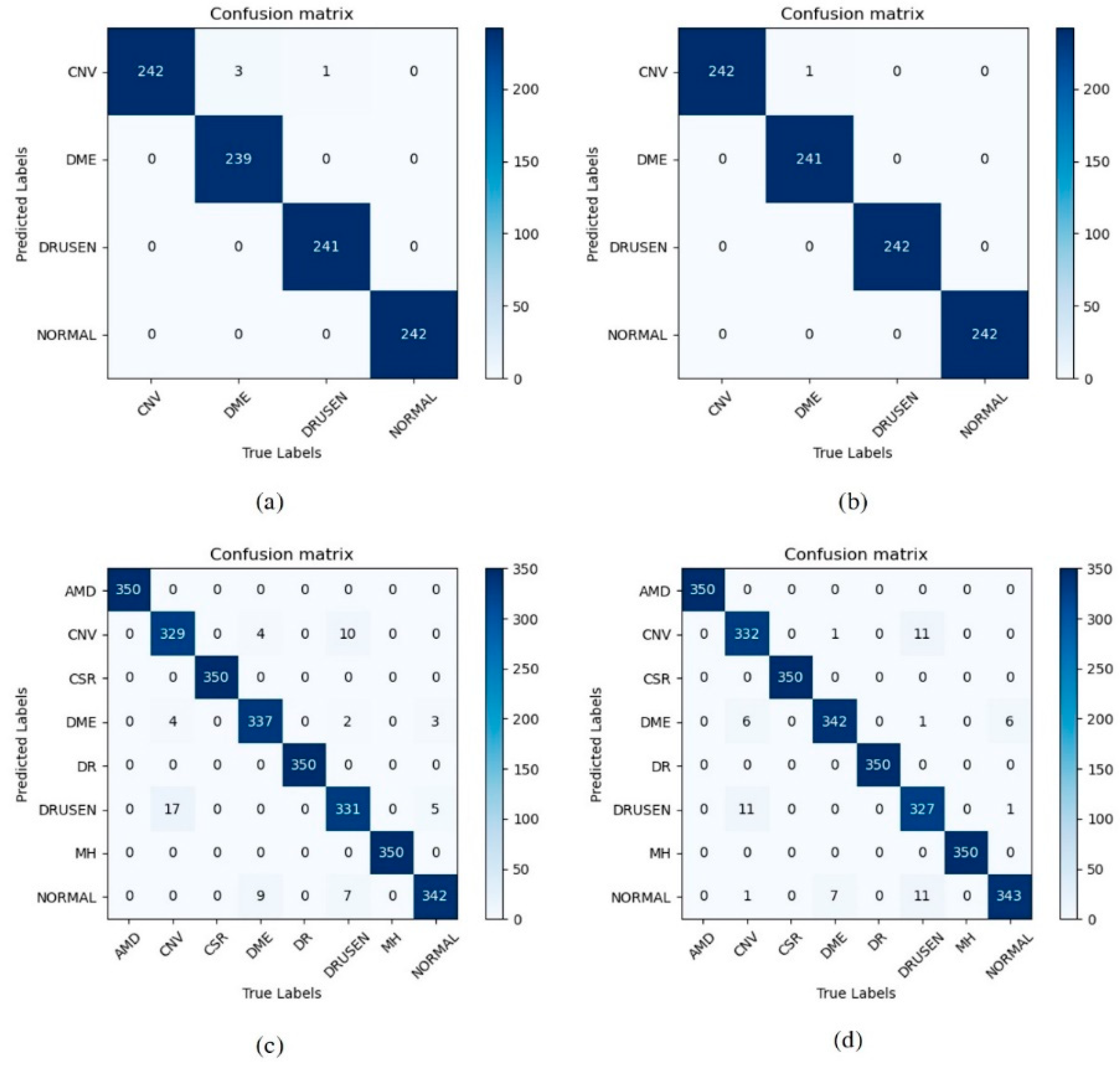
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prem Senthil, M.; Khadka, J.; Gilhotra, J.S.; Simon, S.; Pesudovs, K. Exploring the quality of life issues in people with retinal diseases: A qualitative study. J. Patient-Rep. Outcomes 2017, 1, 15. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Swanson, E.A.; Lin, C.P.; Schuman, J.S.; Stinson, W.G.; Chang, W.; Hee, M.R.; Flotte, T.; Gregory, K.; Puliafito, C.A. Optical coherence tomography. Science 1991, 254, 1178–1181. [Google Scholar] [CrossRef] [PubMed]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. Off. J. Comput. Med. Imaging Soc. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.Q.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Tan, M.X.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning (ICML), Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Annual Conference on Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Liu, Z.; Lin, Y.T.; Cao, Y.; Hu, H.; Wei, Y.X.; Zhang, Z.; Lin, S.; Guo, B.N. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proceedings of the 18th IEEE/CVF International Conference on Computer Vision (ICCV), Electr Network, Montreal, BC, Canada, 11–17 October 2021; pp. 9992–10002. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L.; et al. Swin Transformer V2: Scaling Up Capacity and Resolution. arXiv 2022, arXiv:2111.09883. [Google Scholar]
- Leng, Z.; Tan, M.; Liu, C.; Cubuk, E.D.; Shi, X.; Cheng, S.; Anguelov, D. PolyLoss: A Polynomial Expansion Perspective of Classification Loss Functions. arXiv 2022, arXiv:2204.12511. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
- Lee, C.S.; Baughman, D.M.; Lee, A.Y. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmology. Retina 2017, 1, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, L. On OCT Image Classification via Deep Learning. IEEE Photonics J. 2019, 11, 1–14. [Google Scholar] [CrossRef]
- Islam, K.T.; Wijewickrema, S.; Leary, S.O. Identifying Diabetic Retinopathy from OCT Images using Deep Transfer Learning with Artificial Neural Networks. In Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Cordoba, Spain, 5–7 June 2019; pp. 281–286. [Google Scholar]
- Rong, Y.B.; Xiang, D.H.; Zhu, W.F.; Yu, K.; Shi, F.; Fan, Z.; Chen, X.J. Surrogate-Assisted Retinal OCT Image Classification Based on Convolutional Neural Networks. IEEE J. Biomed. Health Inform. 2019, 23, 253–263. [Google Scholar] [CrossRef]
- Fang, L.Y.; Wang, C.; Li, S.T.; Rabbani, H.; Chen, X.D.; Liu, Z.M. Attention to Lesion: Lesion-Aware Convolutional Neural Network for Retinal Optical Coherence Tomography Image Classification. IEEE Trans. Med. Imaging 2019, 38, 1959–1970. [Google Scholar] [CrossRef]
- Singh, A.; Rasheed, M.A.; Zelek, J.; Lakshminarayanan, V. Interpretation of deep learning using attributions: Application to ophthalmic diagnosis. In Proceedings of the Conference on Applications of Machine Learning, Electr Network, Online, 24 August–4 September 2020. [Google Scholar]
- Wang, C.; Jin, Y.; Chen, X.; Liu, Z. Automatic Classification of Volumetric Optical Coherence Tomography Images via Recurrent Neural Network. Sens. Imaging 2020, 21, 32. [Google Scholar] [CrossRef]
- Arefin, R.; Samad, M.D.; Akyelken, F.A.; Davanian, A.; Soc, I.C. Non-transfer Deep Learning of Optical Coherence Tomography for Post-hoc Explanation of Macular Disease Classification. In Proceedings of the 9th IEEE International Conference on Healthcare Informatics (IEEE ICHI), Electr Network, Victoria, BC, Canada, 9–12 August 2021; pp. 48–52. [Google Scholar]
- Latha, V.; Ashok, L.R.; Sreeni, K.G.; IEEE. Automated Macular Disease Detection using Retinal Optical Coherence Tomography images by Fusion of Deep Learning Networks. In Proceedings of the 27th National Conference on Communications (NCC), Electr Network, Kanpur, India, 27–30 July 2021; pp. 333–338. [Google Scholar]
- Liu, X.M.; Bai, Y.J.; Cao, J.; Yao, J.P.; Zhang, Y.; Wang, M. Joint disease classification and lesion segmentation via one-stage attention-based convolutional neural network in OCT images. Biomed. Signal Process. Control 2022, 71, 103087. [Google Scholar] [CrossRef]
- Esfahani, E.N.; Daneshmand, P.G.; Rabbani, H.; Plonka, G. Automatic Classification of Macular Diseases from OCT Images Using CNN Guided with Edge Convolutional Layer. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Glasgow, UK, 11–15 July 2022; pp. 3858–3861. [Google Scholar] [CrossRef]
- He, J.Z.; Wang, J.X.; Han, Z.Y.; Ma, J.; Wang, C.J.; Qi, M. An interpretable transformer network for the retinal disease classification using optical coherence tomography. Sci. Rep. 2023, 13, 3637. [Google Scholar] [CrossRef]
- Ibrahim, M.R.; Fathalla, K.M.; Youssef, S.M. HyCAD-OCT: A Hybrid Computer-Aided Diagnosis of Retinopathy by Optical Coherence Tomography Integrating Machine Learning and Feature Maps Localization. Appl. Sci. 2020, 10, 4716. [Google Scholar] [CrossRef]
- Ai, Z.; Huang, X.; Feng, J.; Wang, H.; Tao, Y.; Zeng, F.X.; Lu, Y.P. FN-OCT: Disease Detection Algorithm for Retinal Optical Coherence Tomography Based on a Fusion Network. Front. Neuroinform. 2022, 16, 876927. [Google Scholar] [CrossRef] [PubMed]
- Arkin, E.; Yadikar, N.; Xu, X.B.; Aysa, A.; Ubul, K. A survey: Object detection methods from CNN to transformer. Multimed. Tools Appl. 2023, 82, 21353–21383. [Google Scholar] [CrossRef]
- Hendria, W.F.; Phan, Q.T.; Adzaka, F.; Jeong, C. Combining transformer and CNN for object detection in UAV imagery. ICT Express 2023, 9, 258–263. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.J.; Valentim, C.C.S.; Liang, H.Y.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.K.; Yan, F.B.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, M.; Shanmugavadivel, K.; Naren, O.S.; Premkumar, K.; Rankish, K. Classification of Retinal OCT Images Using Deep Learning. In Proceedings of the 2022 International Conference on Computer Communication and Informatics (ICCCI), Coimbatore, India, 25–27 January 2022; pp. 1–7. [Google Scholar]
- Subramanian, M.; Kumar, M.S.; Sathishkumar, V.E.; Prabhu, J.; Karthick, A.; Ganesh, S.S.; Meem, M.A. Diagnosis of Retinal Diseases Based on Bayesian Optimization Deep Learning Network Using Optical Coherence Tomography Images. Comput. Intell. Neurosci. 2022, 2022, 8014979. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.X.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training. In Proceedings of the International Conference on Machine Learning (ICML), Electr Network, Virtual Event, 18–24 July 2021; pp. 7102–7110. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z.; IEEE. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.L.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Kamran, S.A.; Tavakkoli, A.; Zuckerbrod, S.L. Improving robustness using joint attention network for detecting retinal degeneration from optical coherence tomography images. In Proceedings of the IEEE International Conference on Image Processing (ICIP), Electr Network, Abu Dhabi, United Arab Emirates, 25–28 October 2020; pp. 2476–2480. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 30th IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Amit Kamran, S.; Saha, S.; Shihab Sabbir, A.; Tavakkoli, A. Optic-Net: A Novel Convolutional Neural Network for Diagnosis of Retinal Diseases from Optical Tomography Images. arXiv 2019, arXiv:1910.05672. [Google Scholar]
- Yoo, T.K.; Choi, J.Y.; Kim, H.K. Feasibility study to improve deep learning in OCT diagnosis of rare retinal diseases with few-shot classification. Med. Biol. Eng. Comput. 2021, 59, 401–415. [Google Scholar] [CrossRef]
- Sathishkumar, V.E.; Park, J.; Cho, Y. Seoul bike trip duration prediction using data mining techniques. IET Intell. Transp. Syst. 2020, 14, 1465–1474. [Google Scholar] [CrossRef]
- Nazir, T.; Nawaz, M.; Rashid, J.; Mahum, R.; Masood, M.; Mehmood, A.; Ali, F.; Kim, J.; Kwon, H.Y.; Hussain, A. Detection of Diabetic Eye Disease from Retinal Images Using a Deep Learning based CenterNet Model. Sensors 2021, 21, 5283. [Google Scholar] [CrossRef]


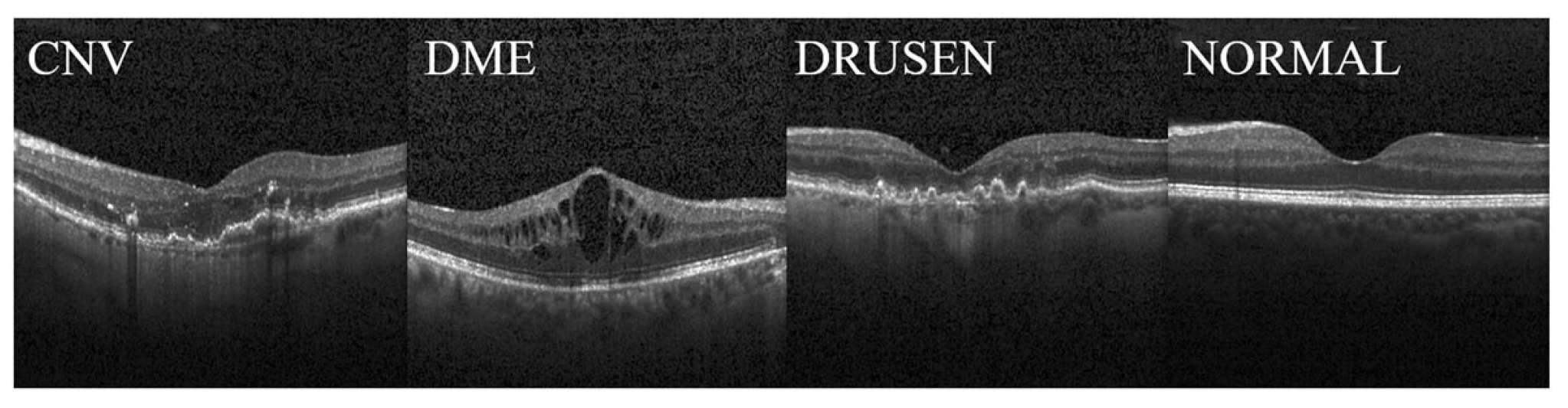

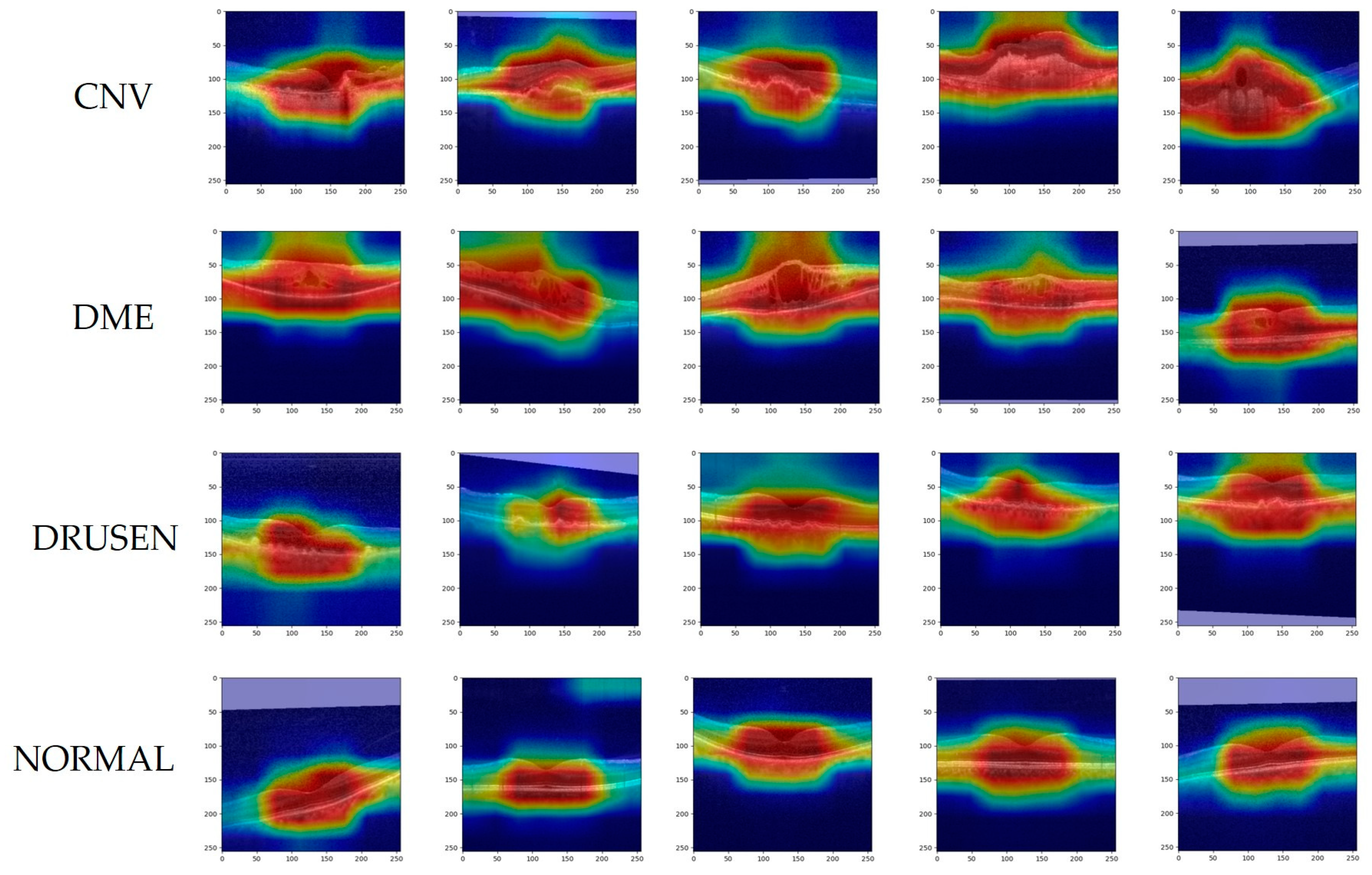
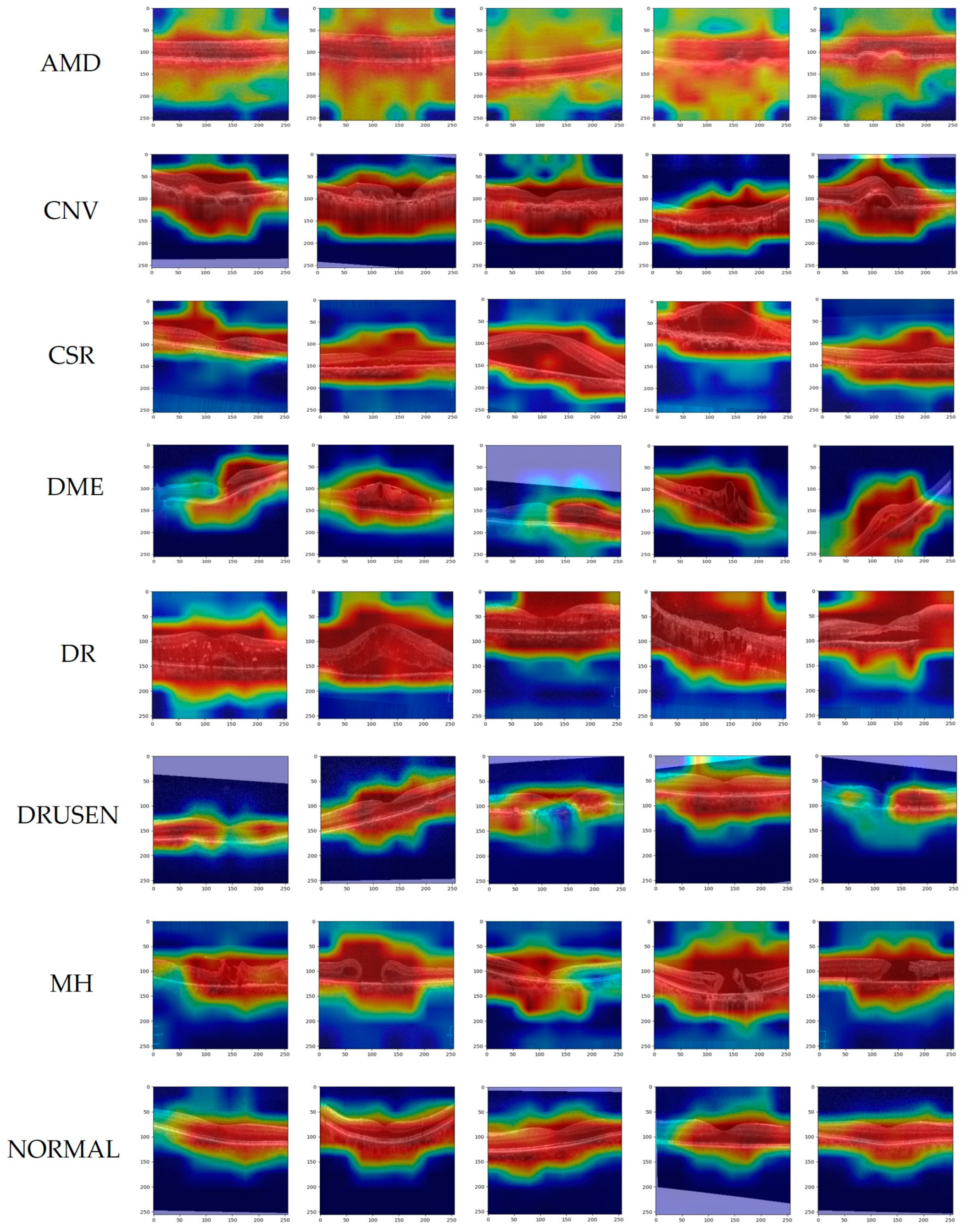
| Dataset | Class | Number | Train | Validation | Test |
|---|---|---|---|---|---|
| OCT2017 | CNV | 37,447 | 36,205 | 1000 | 242 |
| DME | 11,590 | 10,348 | 1000 | 242 | |
| DRUSEN | 8858 | 7616 | 1000 | 242 | |
| NORMAL | 26,557 | 25,315 | 1000 | 242 | |
| OCT-C8 | AMD | 3000 | 2300 | 350 | 350 |
| CNV | 3000 | 2300 | 350 | 350 | |
| CSR | 3000 | 2300 | 350 | 350 | |
| DME | 3000 | 2300 | 350 | 350 | |
| DR | 3000 | 2300 | 350 | 350 | |
| DRUSEN | 3000 | 2300 | 350 | 350 | |
| MH | 3000 | 2300 | 350 | 350 | |
| NORMAL | 3000 | 2300 | 350 | 350 |
| Dataset | Method | Class | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|---|---|
| OCT2017 | EfficientNetV2 | CNV | 0.975 | 0.913 | 0.996 | 0.968 | 0.953 |
| DME | 0.986 | 0.996 | 0.946 | 0.968 | 0.970 | ||
| DRUSEN | 0.977 | 1.0 | 0.909 | 0.999 | 0.952 | ||
| NORMAL | 0.988 | 0.953 | 1.0 | 0.983 | 0.976 | ||
| VIT | CNV | 0.950 | 0.839 | 0.992 | 0.937 | 0.909 | |
| DME | 0.975 | 0.987 | 0.913 | 0.996 | 0.949 | ||
| DRUSEN | 0.951 | 0.990 | 0.814 | 0.997 | 0.893 | ||
| NORMAL | 0.982 | 0.934 | 1.0 | 0.977 | 0.966 | ||
| Swin Transformer | CNV | 0.995 | 0.980 | 1.0 | 0.993 | 0.990 | |
| DME | 0.999 | 1.0 | 0.996 | 1.0 | 0.998 | ||
| DRUSEN | 0.996 | 1.0 | 0.983 | 1.0 | 0.991 | ||
| NORMAL | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| Swin Transformer V2 | CNV | 0.996 | 0.984 | 1.0 | 0.994 | 0.992 | |
| DME | 0.997 | 1.0 | 0.988 | 1.0 | 0.994 | ||
| DRUSEN | 0.999 | 1.0 | 0.996 | 1.0 | 0.998 | ||
| NORMAL | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Dataset | Method | Class | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|---|---|
| OCT2017 | EfficientNetV2 | CNV | 0.971 | 0.896 | 1.0 | 0.961 | 0.945 |
| DME | 0.987 | 1.0 | 0.946 | 1.0 | 0.972 | ||
| DRUSEN | 0.976 | 1.0 | 0.905 | 1.0 | 0.950 | ||
| NORMAL | 0.992 | 0.968 | 1.0 | 0.980 | 0.984 | ||
| VIT | CNV | 0.952 | 0.845 | 0.992 | 0.939 | 0.913 | |
| DME | 0.978 | 0.987 | 0.926 | 0.996 | 0.956 | ||
| DRUSEN | 0.950 | 0.985 | 0.814 | 0.996 | 0.891 | ||
| NORMAL | 0.985 | 0.942 | 1.0 | 0.979 | 0.970 | ||
| Swin Transformer | CNV | 0.997 | 0.988 | 1.0 | 0.996 | 0.994 | |
| DME | 0.999 | 1.0 | 0.996 | 1.0 | 0.998 | ||
| DRUSEN | 0.998 | 1.0 | 0.992 | 1.0 | 0.996 | ||
| NORMAL | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| Ours | CNV | 0.999 | 0.996 | 1.0 | 0.996 | 0.994 | |
| DME | 0.999 | 1.0 | 1.0 | 1.0 | 0.998 | ||
| DRUSEN | 1.0 | 1.0 | 1.0 | 1.0 | 0.996 | ||
| NORMAL | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Dataset | Method | Class | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|---|---|
| OCT-C8 | VIT | AMD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| CNV | 0.965 | 0.873 | 0.846 | 0.982 | 0.859 | ||
| CSR | 0.993 | 0.958 | 0.986 | 0.994 | 0.972 | ||
| DME | 0.962 | 0.901 | 0.783 | 0.988 | 0.838 | ||
| DR | 0.989 | 0.954 | 0.954 | 0.993 | 0.954 | ||
| DRUSEN | 0.943 | 0.775 | 0.769 | 0.968 | 0.772 | ||
| MH | 0.991 | 0.977 | 0.951 | 0.997 | 0.964 | ||
| NORMAL | 0.959 | 0.787 | 0.920 | 0.964 | 0.848 | ||
| Swin Transformer | AMD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| CNV | 0.988 | 0.954 | 0.951 | 0.993 | 0.952 | ||
| CSR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DME | 0.990 | 0.968 | 0.957 | 0.996 | 0.962 | ||
| DR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DRUSEN | 0.985 | 0.956 | 0.937 | 0.992 | 0.946 | ||
| MH | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| NORMAL | 0.987 | 0.945 | 0.977 | 0.992 | 0.961 | ||
| Swin Transformer V2 | AMD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| CNV | 0.988 | 0.959 | 0.940 | 0.994 | 0.949 | ||
| CSR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DME | 0.992 | 0.974 | 0.963 | 0.996 | 0.968 | ||
| DR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DRUSEN | 0.985 | 0.938 | 0.946 | 0.991 | 0.942 | ||
| MH | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| NORMAL | 0.991 | 0.955 | 0.977 | 0.993 | 0.966 |
| Dataset | Method | Class | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|---|---|
| OCT-C8 | VIT | AMD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| CNV | 0.967 | 0.893 | 0.834 | 0.986 | 0.862 | ||
| CSR | 0.994 | 0.961 | 0.991 | 0.994 | 0.976 | ||
| DME | 0.962 | 0.894 | 0.794 | 0.987 | 0.841 | ||
| DR | 0.989 | 0.957 | 0.957 | 0.994 | 0.957 | ||
| DRUSEN | 0.943 | 0.772 | 0.774 | 0.967 | 0.773 | ||
| MH | 0.992 | 0.985 | 0.954 | 0.998 | 0.969 | ||
| NORMAL | 0.958 | 0.781 | 0.917 | 0.963 | 0.844 | ||
| Swin Transformer | AMD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| CNV | 0.988 | 0.959 | 0.943 | 0.995 | 0.952 | ||
| CSR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DME | 0.991 | 0.974 | 0.957 | 0.996 | 0.965 | ||
| DR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DRUSEN | 0.988 | 0.954 | 0.940 | 0.993 | 0.947 | ||
| MH | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| NORMAL | 0.990 | 0.938 | 0.986 | 0.993 | 0.961 | ||
| Ours | AMD | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |
| CNV | 0.989 | 0.965 | 0.949 | 0.995 | 0.957 | ||
| CSR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DME | 0.992 | 0.963 | 0.977 | 0.995 | 0.970 | ||
| DR | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| DRUSEN | 0.988 | 0.965 | 0.934 | 0.995 | 0.949 | ||
| MH | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||
| NORMAL | 0.991 | 0.948 | 0.980 | 0.992 | 0.964 |
| Dataset | Method | Loss | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|---|---|
| OCT2017 | EfficientNetV2 | CrossEntropy | 0.982 | 0.966 | 0.963 | 0.980 | 0.963 |
| PolyLoss | 0.981 | 0.966 | 0.963 | 0.985 | 0.963 | ||
| VIT | CrossEntropy | 0.965 | 0.938 | 0.930 | 0.977 | 0.917 | |
| PolyLoss | 0.966 | 0.940 | 0.933 | 0.978 | 0.933 | ||
| Swin Transformer | CrossEntropy | 0.998 | 0.995 | 0.995 | 0.998 | 0.995 | |
| Paper [30] | PolyLoss | 0.998 | 0.997 | 0.997 | 0.999 | 0.997 | |
| Swin Transformer V2 | CrossEntropy | 0.998 | 0.996 | 0.996 | 0.999 | 0.996 | |
| Ours | PolyLoss | 0.999 | 0.999 | 1.0 | 0.999 | 0.997 | |
| OCT-C8 | VIT | CrossEntropy | 0.975 | 0.903 | 0.901 | 0.986 | 0.901 |
| PolyLoss | 0.976 | 0.905 | 0.903 | 0.986 | 0.903 | ||
| Swin Transformer | CrossEntropy | 0.994 | 0.978 | 0.978 | 0.997 | 0.978 | |
| Paper [30] | PolyLoss | 0.994 | 0.978 | 0.978 | 0.997 | 0.978 | |
| Swin Transformer V2 | CrossEntropy | 0.995 | 0.978 | 0.978 | 0.997 | 0.978 | |
| Ours | PolyLoss | 0.995 | 0.980 | 0.980 | 0.997 | 0.980 |
| Dataset | Model | Accuracy | Sensitivity |
|---|---|---|---|
| OCT2017 | InceptionV3 [39] | 0.934 | 0.978 |
| MobileNet-v2 [40] | 0.985 | 0.994 | |
| ResNet50-v1 [9] | 0.993 | 0.993 | |
| Joint-Attention-Network ResNet-v1 [41] | 0.924 | ||
| Xception [42] | 0.997 | 0.997 | |
| OpticNet-71 [43] | 0.998 | 0.998 | |
| Swin Transformer V1 [30] | 0.998 | 0.998 | |
| Ours | 0.999 | 0.999 | |
| OCT-C8 | VIT | 0.975 | 0.986 |
| GAN [44] | 0.939 | ||
| Swin Transformer | 0.994 | 0.997 | |
| Deep CNN [45] | 0.938 | ||
| CenterNet [46] | 0.981 | ||
| Ours | 0.995 | 0.997 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Han, Y.; Yang, X. Multi-Fundus Diseases Classification Using Retinal Optical Coherence Tomography Images with Swin Transformer V2. J. Imaging 2023, 9, 203. https://doi.org/10.3390/jimaging9100203
Li Z, Han Y, Yang X. Multi-Fundus Diseases Classification Using Retinal Optical Coherence Tomography Images with Swin Transformer V2. Journal of Imaging. 2023; 9(10):203. https://doi.org/10.3390/jimaging9100203
Chicago/Turabian StyleLi, Zhenwei, Yanqi Han, and Xiaoli Yang. 2023. "Multi-Fundus Diseases Classification Using Retinal Optical Coherence Tomography Images with Swin Transformer V2" Journal of Imaging 9, no. 10: 203. https://doi.org/10.3390/jimaging9100203
APA StyleLi, Z., Han, Y., & Yang, X. (2023). Multi-Fundus Diseases Classification Using Retinal Optical Coherence Tomography Images with Swin Transformer V2. Journal of Imaging, 9(10), 203. https://doi.org/10.3390/jimaging9100203







